Abstract
Biologically active, passive treatment systems are commonly employed for removing high concentrations of dissolved Mn(II) from coal mine drainage (CMD). Studies of microbial communities contributing to Mn attenuation through the oxidation of Mn(II) to sparingly soluble Mn(III/IV) oxide minerals, however, have been sparse to date. This study reveals a diverse community of Mn(II)-oxidizing fungi and bacteria existing in several CMD treatment systems.
Acidic, metal-laden mine drainage is a significant problem for many regions in the United States and throughout the world. In Appalachia, centuries of coal mining has left thousands of abandoned mines that are discharging waters containing elevated levels of metals—particularly Mn, with concentrations as high as 150 mg liter−1 (see reference 9 and references therein and reference 23). In the eastern United States, one of the most common methods to remediate coal mine drainage (CMD) is the use of biologically active limestone treatment beds. In essence, dissolved metals, such as Mn(II), are immobilized in the treatment bed via precipitation of sparingly soluble oxide minerals (23, 24) that effectively remove other metal contaminants (e.g., Ni, Co, and Zn) through coprecipitation and surface adsorption reactions (26, 31, 47).
The importance of microbial activity in the remediation of Mn-contaminated waters has frequently been observed (6, 20, 21, 24, 25). Several strains of Mn(II)-oxidizing bacteria have even been used for treating manganiferous mine waters (49). Recently Mariner et al. (32) identified Mn(II)-oxidizing fungi, in addition to bacteria, successfully growing in a Mn-attenuating bioreactor for treatment of mine waters. We also observed that the addition of fungicides inhibited Mn(II) oxidation in laboratory-based CMD treatment simulations (W. D. Burgos, H. Tan, C. M. Santelli, and C. M. Hansel, presented at the National Meeting of the American Society of Mining and Reclamation, Pittsburgh, PA, 5 to 11 June 2010), suggesting a role for fungal activity in Mn remediation. The identities, growth characteristics, and oxidation mechanisms of the microbial community contributing to CMD remediation, however, remain largely unresolved. The objective of this study was to define the Mn(II)-oxidizing microbial community existing in passive treatment systems designed to remove dissolved Mn(II) from CMD. Because the mechanisms of microbial Mn(II) oxidation are not fully elucidated and are not genetically tractable (13, 18), we initiated an extensive culture survey to identify microorganisms that catalyze Mn(II) oxidation and precipitate Mn(III/IV) oxide minerals. These results provide the foundation for future explorations identifying the key players in CMD remediation and factors impacting their activity.
In October 2007, we sampled four Mn attenuation beds—Saxman Run (SRC1) and DeSale phases I, II, and III (DS1 to DS3)—in central Pennsylvania that are currently treating exceptionally high Mn concentrations (up to 119 mg liter−1; 2.2 mM) generated from abandoned coal mines. Each system has a slightly different design (for a further description, see reference 21) and treatment load; however, all systems have wetlands and limestone-filled beds (to raise pH) in series. At the time of sampling, waters flowing into SRC1 had a near-neutral pH (6.6) and dissolved Mn was 28 mg liter−1 (500 μM). SRC1 was highly effective in removing dissolved Mn where the effluent had <0.05 mg liter−1 Mn (data courtesy of the Pennsylvania Department of Environmental Protection). Systems DS1 to DS3 treat waters with a pH range of 5.7 to 6.3, containing 46 to 119 mg liter−1 (0.85 to 2.2 mM) Mn, and each attenuates about 50% of the total Mn load.
Culture enrichments were initiated using Mn oxide-coated limestone and debris from the treatment systems. Serial dilutions to 1/104 for each sample were plated on 7 types of agar-solidified media with 20 mM HEPES (pH 7) and 200 μM MnCl2: AY (34); K, M, and Leptothrix (46); J and J plus acetate (22); and medium 3 (11). Mn(II)-oxidizing microorganisms were transferred to fresh media until cultures were deemed axenic. Mn(III/IV) oxides were confirmed using the LBB colorimetric assay (28) and electron microscopy (not shown).
Fungal isolates were identified using a combination of phylogenetic analysis and morphological characterization (Table 1), and bacteria were identified through phylogenetic analysis. For morphological characterizations, fungi were grown on AY, potato dextrose agar, and malt extract agar for up to 12 weeks to induce sporulation. Isolates producing conidiogenous structures were examined using light microscopy to confirm species-level identifications. For phylogenetic analysis, genomic DNA of each culture was isolated, amplified by PCR, and sequenced. Bacterial 16S rRNA was amplified with the primers 8F/1492R using protocols described previously (38). The 18S rRNA gene, 28S rRNA gene, and ITS1-5.8S rRNA-ITS2 region (referred to as ITS) were amplified from fungal isolates using primer pairs NS1/NS302 and NS3/NS5 (42), LR0R/LR5-F (45), and ITS1F/ITS4 (35), respectively. Sequences were imported into the BLAST nucleotide search program (2), and ARB software (30) was used to align sequences (isolates and related organisms) and construct phylogenetic trees for representative species (99 to 100% sequence similarity). 18S rRNA analysis was used for related species comparisons, whereas ITS served as a bar code for species-level resolution when a sufficient database was present (reference 40 and references therein).
TABLE 1.
Mn(II)-oxidizing fungi and bacteria isolated from CMD passive treatment systems
| Species identification | Representative isolate | Morphological description for species-level identification of fungus | No. of isolates from site |
Mn2+ tolerance (mM) | Growth rateb at Mn2+ concn (mM) of: |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| DS1 | DS2 | DS3 | SRC1 | 0 | 0.5 | 1 | ||||
| Fungi | ||||||||||
| Plectosphaerella cucumerina | DS2psM2a2 | Conidiophores (tapered, 1-septate) after 2 weeks growth | 9 | 13 | —a | 27 | >10 | 4.6 | 4.7 | 4.6 |
| Microdochium bolleyi | SRC1dJ1a | Pink with chlamydospores (chain) and conidia (straight to slight crescent shape, ∼8-μm length) | — | — | — | 10 | 1 | 7.9 | 8.4 | 7.7 |
| Stilbella aciculosa | DS2rAY2a | Hyphal conidia (∼4 by 12-15 μm, cylindrical, sometimes septate); synemmata with conidia (nonseptate, tapered, ellipsoid, ∼6-μm length) | 2 | 11 | — | — | >10 | 4.3 | 4.3 | 4.1 |
| Pyrenochaeta sp. | DS3sAY3a | Pycnidia (dark-brown/black, setae near the osteole) filled with conidia (∼2.4 μm by 4.8 μm, near globose, slipper-shaped, two-gutulate) | — | — | 2 | — | >10 | 3.3 | 3.4 | 3.4 |
| Stagonospora sp. | SRC1lsM3a | No spores or reproductive structures produced after 5 months | — | — | — | 1 | >10 | 1.9 | 2.1 | 2.5 |
| Alternaria alternata | SRC1lrK2f | Chains (long, often branching) of ornamented conidia; conidiophores (long, 1- to 3-septate, straight to curved) | — | — | — | 2 | >10 | 8.3 | 7.8 | 7.8 |
| Acremonium strictum | DS1bioAY4a | Slimy and orange mycelial mat with gelatinous masses filled with conidia (∼4-5-μm length) | 4 | 1 | — | — | >10 | 1.2 | 1.4 | 1.9 |
| Pithomyces chartarum | DS1bioJ1b | Conidia (dark brown/black, muriform, verruculose, ∼22-24-μm length) | 1 | — | — | — | 5 | 6.9 | 7.4 | 7.7 |
| Phoma sp. | DS1wsM30b | Chlamydospores (slightly pigmented, nearly globose, ∼4-15-μm width, longitudinal septa absent) | 1 | — | — | — | >10 | 7.4 | 7.2 | 7.2 |
| Bacteria | ||||||||||
| Agrobacterium sp. | SRC1K2fb | — | — | — | 1 | >10 | 0.03 | 0.04 | 0.03 | |
| Bacillus sp. | DS3sK3a | — | 3 | 2 | — | >10 | 0.04 | 0.04 | 0.04 | |
| Flavobacterium sp. | DS2psK4b | — | 1 | — | — | >10 | 0.05 | 0.05 | 0.05 | |
| Pseudomonas sp. | DS3sK1h | — | — | 2 | — | >10 | 0.05 | 0.03 | 0.03 | |
—, no isolates obtained.
Growth rates for fungi (mm/day) were determined by measuring the increase in diameter with time. Growth rates for bacteria (OD600/h) were determined by measuring the optical density at 600 nm (OD600) with 200 μM ascorbic acid to dissolve Mn(III/IV) oxides. Rates were determined during exponential growth phase.
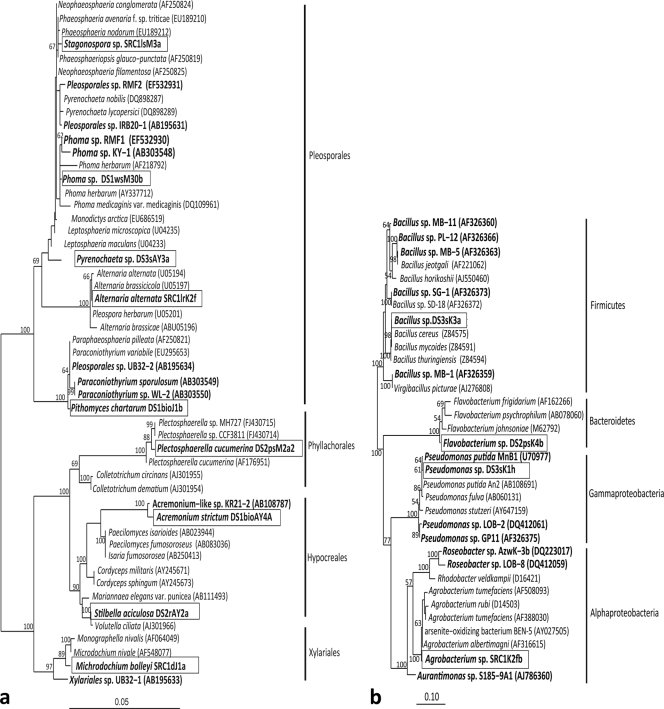
Results of the culture survey show that a diversity of Mn(II)-oxidizing fungi and bacteria exist in Mn attenuation systems that actively treat CMD. Mn(II)-oxidizing fungi, isolated from each of the Mn removal beds, represent ∼90% of the nearly 100 axenic cultures obtained from the enrichments (Table 1). Phylogenetic analysis of these fungi reveals 9 different species (Fig. 1 a and Table 1; see also Fig. S1 to S4 in the supplemental material) belonging to two classes (Sordariomycetes and Dothideomycetes) of the phylum Ascomycota. The two most widely recovered Mn(II)-oxidizing fungi are isolate DS2psM2a2, identified as Plectosphaerella cucumerina (Table 1; see also Fig. S1a), and isolate DS2rAY2a, identified as Stilbella aciculosa (Table 1; see also Fig. S1b). Although both of these Sordariomycetes are common, well studied (4, 29; for examples, see references 36 and 50) soil-inhabiting microorganisms (10), neither has previously been demonstrated to oxidize Mn(II), nor is either phylogenetically related to known Mn(II)-oxidizing species (Fig. 1a). Two additional Sordariomycetes were obtained, Acremonium strictum (isolate DS1bioAY4a) (Table 1; see also Fig. S1b) and Microdochium bolleyi (isolate SRC1dJ1a) (Table 1; see also Fig. S2a). Interestingly, both of these fungal isolates are closely related (based on ITS and 18S rRNA sequence analysis) to other known Mn(II)-oxidizing species, suggesting that some Mn(II)-oxidizing species have a cosmopolitan distribution. Microdochium bolleyi and Acremonium strictum sequences are very similar (Fig. 1a) to those of Mn(II)-oxidizing Xylariales sp. UB32-1 (98% similarity, 18S rRNA) and Acremonium sp. KR21-2 (100% similarity, ITS), respectively, isolated from a stream bed in Japan (33, 34).
FIG. 1.
Maximum-likelihood (ML) trees showing the phylogenetic relationships of Mn(II)-oxidizing fungi (a) or bacteria (b) isolated from Mn treatment systems using 18S rRNA and 16S rRNA gene analysis, respectively. Isolates from this study are shown in bold and boxed, and previously identified Mn(II)-oxidizing isolates are in bold and unboxed. Both ML trees were constructed with the PhyML software package (19) in ARB using the generalized time reversible (GTR) nucleotide substitution model with 1,000 bootstrap replicates. Bootstrap values for nodes with greater than 50% support are displayed as percentages. GenBank accession numbers are in parentheses. Scale bar, 0.05 (a) or 0.10 (b) substitutions per nucleotide site.
All other Mn(II)-oxidizing fungal isolates are classified as Pleosporales (Dothideomycetes class), although neither phylogenetic analysis nor morphological characterization could confirm the identification of several of these beyond the genus level. Two of these isolates do not have sequences with similarity to those of known Mn(II)-oxidizing species: DS3sAY3a, a Pyrenochaeta sp., and SRC1lrK2f, identified as Alternaria alternata (Table 1 and Fig. 1a; see also Fig. S4a and S4b in the supplemental material). Conversely, most of these species are related to other known Mn(II)-oxidizing strains. For example, isolate SRC1lsM3a, a Stagonospora sp. (Table 1; see also Fig. S2b), is related to two different Mn-oxidizing strains (Fig. 1a, 99% similarity): Pleosporales sp. IRB20-1 (33), isolated from a Japanese stream bed, and Pleosporales sp. RMF2, isolated from a prototype bioreactor treating Mn-contaminated mine waste in Wales, United Kingdom (32). Likewise, isolate DS1wsM30b, a Phoma sp. (Table 1; see also Fig. S3a), has high sequence similarity (≥98%, 18S) with Phoma sp. KMF1 from the Mn bioreactor and Phoma sp. KY-1, isolated from stream sediment (42). Pithomyces chartarum isolate DS1bioJ1b (Table 1; see also Fig. S3b) is 99% similar to Mn(II)-oxidizing Paraconiothyrium sp. WL-2, isolated from an artificial wetland in Japan (42), and Pleosporales sp. UB32-2, isolated from a Japanese stream bed (33); however, ITS analysis shows only 88% similarity to Paraconiothyrium sp. WL-2 (the ITS sequence for UB32-2 is not publically available).
Although relatively few Mn(II)-oxidizing bacteria were recovered from the treatment systems, these isolates present similar phylogenetic diversity: 9 bacterial isolates represent 4 taxa (Fig. 1b and Table 1) belonging to three different phyla: Firmicutes, Bacteroidetes, and Proteobacteria (Alpha- and Gammaproteobacteria). Bacillus spp., represented by isolate DS3sK3a, are the most commonly recovered bacteria (Table 1). It is not surprising to recover Bacillus species from the treatment systems, because they are the most commonly isolated Mn(II)-oxidizing bacteria (12, 16) for marine systems as well. Isolate DS3sK1h, a Pseudomonas sp., is similarly related to other previously identified Mn(II)-oxidizing bacteria, such as P. putida strain MnB1 and Pseudomonas sp. LOB-2, isolated from the deep ocean (≤97% sequence similarity; Fig. 1b) (46).
Two previously unreported Mn(II)-oxidizing bacterial species were also recovered: Agrobacterium sp. (isolate SRC1K2fb), an alphaproteobacterium, and Flavobacterium sp. (isolate DS2psK4b), a member of the Bacteroidetes (Table 1 and Fig. 1b). The isolation of a Flavobacterium sp. further expands the taxonomic representation of Mn oxidizers—to our knowledge, members of the Bacteroidetes have never been demonstrated to oxidize Mn(II). Alphaproteobacteria, on the other hand, account for a large portion of cultured Mn(II)-oxidizing bacterial representatives (3, 15, 22, 44, 46) and other metal-oxidizing bacteria (39).
Microbial communities living in CMD treatment systems are exposed to widely fluctuating environmental conditions and metal concentrations; therefore, the ability to tolerate and grow in various Mn concentrations was tested for all isolates. Fungi and bacteria were grown in media supplemented with Mn2+ at the following concentrations: 0, 0.5, 1, 5, and 10 mM MnCl2. Our results show that all isolates, with the exception of Microdochium bolleyi and Pithomyces chartarum, grew and oxidized Mn(II) at Mn concentrations greater than those observed in the treatment systems (Table 1). Metal tolerance of bacterial strains was somewhat unexpected since it is generally believed that fungi are more tolerant than bacteria to high concentrations of heavy metals, often leading to a prevalence of fungi in metal-contaminated soils (7, 8, 27, 37). It is possible that these bacterial strains have developed metal tolerance in the treatment systems; the development of Mn tolerance has been observed previously (1) in the Mn(II)-oxidizing bacterium Leptothrix discophora.
Mn(II) exposure experiments also show that growth rates (Table 1) remain constant with various metal concentrations for many of the isolates (e.g., Microdochium bolleyi and Bacillus sp.), even at 10 mM Mn concentrations (data not shown). Some species, however, grow fastest at lower Mn(II) concentrations (e.g., Stilbella aciculosa), whereas others had increased growth rates at high Mn concentrations that were even more pronounced at 10 mM Mn (e.g., Stagonospora sp., 3.5 mm/day). Decreased (5) or similar (34, 41) growth rates at higher dissolved Mn(II) concentrations have previously been observed for many nonoxidizing and Mn-oxidizing ascomycota and bacteria (14, 22), suggesting a possible toxic effect of high metal concentrations. Faster growth at very high Mn(II) concentrations, however, is somewhat surprising, although small concentrations have stimulated growth of some Mn-oxidizing bacteria relative to metal-free conditions (14, 15). Since fungi and all known Mn(II)-oxidizing bacteria are heterotrophs and therefore do not gain energy from the oxidation reaction, these organisms may be benefiting from either the presence of Mn oxide minerals (e.g., scavenging of nutrients, immobilization of metals, or UV protection) or potentially an increased uptake of dissolved Mn(II) for cellular functions or scavenging of reactive oxygen species (see reference 43 and references therein).
Here we introduce new bacterial and fungal players in the oxidation of Mn(II). We also reveal a diversity and predominance of fungi within culturable Mn(II)-oxidizing communities in CMD passive treatment systems. It is not entirely unexpected to obtain fungi in such environments, since fungi often possess multiple mechanisms to tolerate environmental stresses (e.g., nutrient fluctuations, desiccation, or high metal loading). Consequently, it is becoming increasingly evident that fungi represent great (and often overlooked) potential for the remediation of a wide range of pollutants, including metals (for examples, see references 17 and 48). The results in this and previous studies (32; Burgos et al., presented at the National Meeting of the American Society of Mining and Reclamation, 2010) suggest fungi also contribute to the remediation of Mn-contaminated mine drainage, warranting continued investigations of the cultivated Mn(II)-oxidizing fungi. Future investigations of these organisms revealing the mechanisms of Mn(II) oxidation and the factors influencing optimal growth and activity will greatly aid the engineering of efficient systems for CMD bioremediation and likely beyond.
Nucleotide sequence accession numbers.
Sequences were submitted to the GenBank database with the following accession numbers: 16S rRNA genes, HM216202 to HM216205; 18S rRNA genes, HM216184 to HM216192; 28S rRNA genes, HM216193 to HM216201; and ITS region, HM216206 to HM216214.
Supplementary Material
Acknowledgments
We thank Brent Means (OSM), Slippery Rock Watershed Coalition, Stream Restoration Inc., and the Pennsylvania Department of Environmental Protection for assistance and access to sample sites. We also thank Anne Pringle (Harvard) for project advice and resources.
This project was funded by the Office of Surface Mining (cooperative agreement S07AP12478) and the National Science Foundation (grants EAR-0846715 [to C.M.H.] and CHE-0431328 [to W.D.B.]).
Footnotes
Published ahead of print on 21 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, L. F., and W. C. Ghiorse. 1985. Influence of manganese on growth of a sheathless strain of Leptothrix discophora. Appl. Environ. Microbiol. 49:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, C. R., G. J. Dick, M.-L. Chu, J.-C. Cho, R. E. Davis, S. L. Brauer, and B. M. Tebo. 2009. Aurantimonas manganoxydans, sp. nov. and Aurantimonas litoralis, sp. nov.: Mn(II)-oxidizing representatives of a globally distributed clade of alpha-Proteobacteria from the order Rhizobiales. Geomicrobiol. J. 26:189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkins, S., I. Clark, D. Sosnowska, P. Hirsch, and B. Kerry. 2003. Detection and quantification of Plectosphaerella cucumerina, a potential biological control agent of potato cyst nematodes, by using conventional PCR, real-time PCR, selective media, and baiting. Appl. Environ. Microbiol. 69:4788-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babich, H., and G. Stotzky. 1981. Manganese toxicity to fungi: influence of pH. Bull. Environ. Contam. Toxicol. 27:474-480. [DOI] [PubMed] [Google Scholar]
- 6.Bamforth, S., D. Manning, I. Singleton, P. Younger, and K. Johnson. 2006. Manganese removal from mine waters investigating the occurrence and importance of manganese carbonates. Appl. Geochem. 21:1274-1287. [Google Scholar]
- 7.Chander, K., J. Dyckmans, H. Hoeper, R. Joergensen, and M. Raubuch. 2001. Long-term effects on soil microbial properties of heavy metals from industrial exhaust deposition. J. Plant Nutr. Soil Sci. 164:657-663. [Google Scholar]
- 8.Chander, K., J. Dyckmans, R. Joergensen, B. Meyer, and M. Raubuch. 2001. Different sources of heavy metals and their long-term effects on soil microbial properties. Biol. Fertil. Soils 34:241-247. [Google Scholar]
- 9.Cravotta, C., III. 2008. Dissolved metals and associated constituents in abandoned coal-mine discharges, Pennsylvania, USA. 1. Constituent concentrations and correlations. Appl. Geochem. 23:166-2002. [Google Scholar]
- 10.de Hoog, G., J. Guarro, J. Gene, and M. Figueras. 2000. Atlas of clinical fungi, 2nd ed. Centraalbureau voor Schimmelcultures, Utrecht, Netherlands.
- 11.de la Torre, M. A., and G. Gomez-Alarcon. 1994. Manganese and iron oxidation by fungi isolated from building stone. Microb. Ecol. 27:177-188. [DOI] [PubMed] [Google Scholar]
- 12.Dick, G. J., Y. E. Lee, and B. M. Tebo. 2006. Manganese(II)-oxidizing Bacillus spores in Guaymas Basin hydrothermal sediments and plumes. Appl. Environ. Microbiol. 72:3184-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dick, G. J., S. Podell, H. A. Johnson, Y. Rivera-Espinoza, R. Bernier-Latmani, J. K. McCarthy, J. W. Torpey, B. G. Clement, T. Gaasterland, and B. M. Tebo. 2008. Genomic insights into Mn(II) oxidation by the marine alphaproteobacterium Aurantimonas sp. strain SI85-9A1. Appl. Environ. Microbiol. 74:2646-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes, S., K. Krishnan, V. Khedekar, and P. Loka Bharathi. 2005. Manganese oxidation by bacterial isolates from the Indian Ridge System. BioMetals 18:483-492. [DOI] [PubMed] [Google Scholar]
- 15.Francis, C., E. Co, and B. Tebo. 2001. Enzymatic manganese(II) oxidation by a marine alpha-proteobacterium. Appl. Environ. Microbiol. 67:4024-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis, C. A., and B. Tebo. 2002. Enzymatic manganese(II) oxidation by metabolically dormant spores of diverse Bacillus species. Appl. Environ. Microbiol. 68:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadd, G. M. 2007. Geomycology: biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol. Res. 111:3-49. [DOI] [PubMed] [Google Scholar]
- 18.Geszvain, K., and B. M. Tebo. 2010. Identification of a two-component regulatory pathway essential for Mn(II) oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 76:1224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:694-704. [DOI] [PubMed] [Google Scholar]
- 20.Haack, E. A., and L. Warren. 2003. Biofilm hydrous manganese oxyhydroxides and metal dynamics in acid rock drainage. Environ. Sci. Technol. 37:4138-4147. [DOI] [PubMed] [Google Scholar]
- 21.Hallberg, K., and D. Johnson. 2005. Biological manganese removal from acid mine drainage in constructed wetlands and prototype bioreactors. Sci. Total Environ. 338:115-124. [DOI] [PubMed] [Google Scholar]
- 22.Hansel, C. M., and C. A. Francis. 2006. Coupled photochemical and enzymatic Mn(II) oxidation pathways of a planktonic Roseobacter-like bacterium. Appl. Environ. Microbiol. 72:3543-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herlihy, A., P. Kaufmann, M. Mitch, and D. Brown. 1990. Regional estimates of acid mine drainage impact on streams in the Mid-Atlantic and Southeastern United States. Water Air Soil Pollut. 50:91-107. [Google Scholar]
- 24.Johnson, K., A. Baker, and D. Manning. 2005. Passive treatment of Mn-rich mine water: using fluorescence to observe microbiological activity. Geomicrobiol. J. 22:141-149. [Google Scholar]
- 25.Johnson, K. L., and P. L. Younger. 2005. Rapid manganese removal from mine waters using an aerated packed-bed bioreactor. J. Environ. Qual. 34:987-993. [DOI] [PubMed] [Google Scholar]
- 26.Kay, J., M. Conklin, C. Fuller, and P. O'Day. 2001. Processes of nickel and cobalt uptake by a manganese oxide forming sediment in Pinal Creek, globe mining district, Arizona. Environ. Sci. Technol. 35:4719-4725. [DOI] [PubMed] [Google Scholar]
- 27.Kelly, J. J., M. Haggblom, and R. L. TateIii. 1999. Changes in soil microbial communities over time resulting from one time application of zinc: a laboratory microcosm study. Soil Biol. Biochem. 31:1455-1465. [Google Scholar]
- 28.Krumbein, W. E., and H. J. Altman. 1973. A new method for detection and enumeration of manganese-oxidizing and -reducing microorganisms. Helgol. Wiss. Meeresunters. 25:4985-4999. [Google Scholar]
- 29.Larkin, R. P., and D. R. Fravel. 1998. Efficacy of various fungal and bacterial biocontrol organisms for control of fusarium wilt of tomato. Plant Dis. 82:1022-1028. [DOI] [PubMed] [Google Scholar]
- 30.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, A. Yadhukumar, T. Lai, S. Steppi, G. Jobb, and W. FoÈrster. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manceau, A., N. Tamura, M. A. Marcus, A. MacDowell, R. S. Celestre, R. E. Sublett, G. Sposito, and H. A. Padmore. 2002. Deciphering Ni sequestration in soil ferromanganese modules by combining X-ray fluorescence, absorption, and diffraction at micrometer scales of resolution. Am. Mineral. 87:1494-1499. [Google Scholar]
- 32.Mariner, R., D. Johnson, and K. Hallberg. 2008. Characterisation of an attenuation system for the remediation of Mn (II) contaminated waters. Hydrometallurgy 94:100-104. [Google Scholar]
- 33.Miyata, N., K. Maruo, Y. Tani, H. Tsuno, H. Seyama, M. Soma, and K. Iwahori. 2006. Production of biogenic manganese oxides by anamorphic ascomycete fungi isolated from streambed pebbles. Geomicrobiol. J. 23:63-73. [Google Scholar]
- 34.Miyata, N., Y. Tani, K. Iwahori, and M. Soma. 2004. Enzymatic formation of manganese oxides by an Acremonium-like hyphomycete fungus, strain KR 21-2. FEMS Microbiol. Ecol. 47:101-109. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien, H. E., J. L. Parrent, J. A. Jackson, J. M. Moncalvo, and R. Vilgalys. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol. 71:5544-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palm, M., W. Gams, and H. Nirenberg. 1995. Plectosporium, a new genus for Fusarium tabacinum, the anamorph of Plectosphaerella cucumerina. Mycologia 87:397-406. [Google Scholar]
- 37.Rajapaksha, R. M. C. P., M. A. Tobor-Kaplon, and E. Baath. 2004. Metal toxicity affects fungal and bacterial activities in soil differently. Appl. Environ. Microbiol. 70:2966-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santelli, C. M., B. N. Orcutt, E. Banning, W. Bach, C. L. Moyer, M. L. Sogin, H. Staudigel, and K. J. Edwards. 2008. Abundance and diversity of microbial life in ocean crust. Nature 453:653-657. [DOI] [PubMed] [Google Scholar]
- 39.Santini, J., L. Sly, A. Wen, D. Comrie, P. De Wulf-Durand, and J. Macy. 2002. New arsenite-oxidizing bacteria isolated from Australian gold mining environments—phylogenetic relationships. Geomicrobiol. J. 19:67-76. [Google Scholar]
- 40.Seifert, K. A. 2009. Progress towards DNA barcoding of fungi. Mol. Ecol. Res. 9:83-89. [DOI] [PubMed] [Google Scholar]
- 41.Shao, Z., and F. Sun. 2007. Intracellular sequestration of manganese and phosphorus in a metal-resistant fungus Cladosporium cladosporioides from deep-sea sediment. Extremophiles 11:435-443. [DOI] [PubMed] [Google Scholar]
- 42.Takano, K., Y. Itoh, T. Ogino, K. Kurosawa, and K. Sasaki. 2006. Phylogenetic analysis of manganese-oxidizing fungi isolated from manganese-rich aquatic environments in Hokkaido, Japan. Limnology 7:219-223. [Google Scholar]
- 43.Tebo, B., W. Ghiorse, L. van Waasbergen, P. Siering, and R. Caspi. 1997. Bacterially mediated mineral formation; insights into manganese (II) oxidation from molecular genetic and biochemical studies, p. 225-266. In J. F. Banfield and K. H. Nealson (ed.), Geomicrobiology: interactions between microbes and minerals. Mineralogical Society of America, Washington, DC.
- 44.Tebo, B., H. Johnson, J. McCarthy, and A. Templeton. 2005. Geomicrobiology of manganese (II) oxidation. Trends Microbiol. 13:421-428. [DOI] [PubMed] [Google Scholar]
- 45.Tedersoo, L., T. Jairus, B. Horton, K. Abarenkov, T. Suvi, I. Saar, and U. Kıljalg. 2008. Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol. 180:479. [DOI] [PubMed] [Google Scholar]
- 46.Templeton, A. S., H. Staudigel, and B. M. Tebo. 2005. Diverse Mn(II)-oxidizing bacteria isolated from submarine basalts at Loihi Seamount. Geomicrobiol. J. 22:127-139. [Google Scholar]
- 47.Toner, B., A. Manceau, S. M. Webb, and G. Sposito. 2006. Zinc sorption to biogenic hexagonal-birnessite particles within a hydrated bacterial biofilm. Geochim. Cosmochim. Acta 70:27-43. [Google Scholar]
- 48.Tortella, G., M. Diez, and N. Duran. 2005. Fungal diversity and use in decomposition of environmental pollutants. Crit. Rev. Microbiol. 31:197-212. [DOI] [PubMed] [Google Scholar]
- 49.Vail, W., and R. Riley. 2000. The pyrolusite process: a bioremediation process for the abatement of acid mine drainage. Green Lands 30:40-46. [Google Scholar]
- 50.Zhang, W., M. Sulz, and K. L. Bailey. 2001. Growth and spore production of Plectosporium tabacinum. Can. J. Bot. 79:1297-1306. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.