Abstract
Salmonella enterica subsp. enterica serovar 4,[5],12:i:− is a monophasic variant of S. enterica serovar Typhimurium (antigenic formula 4,[5],12:i:1,2). Worldwide, especially in several European countries and the United States, it has been reported among the 10 most frequently isolated serovars in pigs and humans. In the study reported here, 148 strains of the monophasic serovar isolated from pigs, pork, and humans in 2006 and 2007 in Germany were characterized by various phenotypic and genotypic methods. This characterization was done in order to investigate their clonality, the prevalence of identical subtypes in pigs, pork, and humans, and the genetic relatedness to other S. enterica serovar Typhimurium subtypes in respect to the pathogenic and resistance gene repertoire. Two major clonal lineages of the monophasic serovar were detected which can be differentiated by their phage types and pulsed-field gel electrophoresis (PFGE) profiles. Seventy percent of the strains tested belonged to definite phage type DT193, and those strains were mainly assigned to PFGE cluster B. Nineteen percent of the strains were typed to phage type DT120 and of these 86% belonged to PFGE cluster A. Sixty-five percent of the isolates of both lineages carried core multiresistance to ampicillin, streptomycin, tetracycline, and sulfamethoxazole encoded by the genes blaTEM1-like, strA-strB, tet(B), and sul2. No correlation to the source of isolation was observed in either lineage. Microarray analysis of 61 S. enterica serovar 4,[5],12:i:− and 20 S. enterica serovar Typhimurium isolates tested determining the presence or absence of 102 representative pathogenicity genes in Salmonella revealed no differences except minor variations in single strains within and between the serovars, e.g., by presence of the virulence plasmid in four strains. Overall the study indicates that in Germany S. enterica serovar 4,[5],12:i:− strains isolated from pig, pork, and human are highly related, showing their transmission along the food chain. Since the pathogenicity gene repertoire is highly similar to that of S. enterica serovar Typhimurium, it is essential that interventions are introduced at the farm level in order to limit human infection.
Salmonella enterica subsp. enterica serovar Typhimurium is a ubiquitous serovar that usually induces gastroenteritis in a broad range of unrelated host species. Following the White-Kauffmann-Le Minor scheme, the seroformula for S. enterica serovar Typhimurium is 4,[5],12:i:1,2 (14). Salmonella serotyping is based on antigenic variability of lipopolysaccharides (O antigen) and flagellar proteins (H1 and H2 antigens).
In the mid-1990s a monophasic S. enterica serovar with the seroformula 4,[5],12:i:− started to emerge in Europe (10). Initial characterization of isolates from pig samples in Spain in 1997 demonstrated that this serovar in comparison with S. enterica serovar Typhimurium (4,[5],12:i:1,2) lacked the fljB gene encoding the structural subunit of the phase two flagellar (H2) antigen (11). The predominant phage type was U302. Another DNA microarray-based typing study indicated that the monophasic serovar had a gene repertoire highly similar to that of S. enterica serovar Typhimurium, indicating a close genetic relatedness between the serovars (13). Similarly, multi-locus sequence typing showed that S. enterica serovar 4,[5],12:i:− and S. enterica serovar Typhimurium represent a highly clonal group (23).
Within the last years S. enterica serovar 4,[5],12:i:− has increasingly been implicated in human disease worldwide (1, 10, 24, 25). Recently, larger outbreaks caused by this serovar have been reported from Luxembourg and the United States (5, 19). A European Union (EU) baseline survey on the prevalence of Salmonella in slaughter-age pigs in 2006 to 2007 revealed that the monophasic serovar was isolated from pigs in 9 of 25 participating member states (12). At the EU level, S. enterica serovar 4,[5],12:i:− was the fourth most prevalent serovar in slaughter-age pigs. In Germany it was the second most prevalent serovar after S. enterica serovar Typhimurium (12). Between 1999 and 2008 the proportion of S. enterica serovar 4,[5],12:i:− isolates among all S. enterica isolates received by the German National Reference Laboratory for Salmonella increased from 0.1% to 8.3% (305 isolates in 2008), with the most remarkable increase between 2006 and 2007. Most of these strains (48% on average between 2006 and 2008) were isolated from pigs, followed by cattle (13%), poultry (5%), and other isolates sporadically found in the environment, wildlife, and reptiles. Remarkably, the annual proportion of the monophasic serovar among all S. enterica serovar 4,[5],12:i:− and S. enterica serovar Typhimurium isolates increased from 0.3% to 32.7% in the same decade. Interestingly, the number of S. enterica serovar 4,[5],12:i:− strains isolated from humans and sent on voluntary basis to the National Reference Centre for Salmonella and other Enterics increased from 0.1% in 1999 to 14.0% (456 isolates) in 2008. Likewise, the proportion of the monophasic serovar among all S. enterica serovar 4,[5],12:i:− and S. enterica serovar Typhimurium isolates increased from 0.3% to 42.8% in the same time because of declining numbers of S. enterica serovar Typhimurium isolates.
In the present study a collection of S. enterica serovar 4,[5],12:i:− strains isolated from pigs, pork, and humans in Germany during the years 2006 and 2007 was examined using phenotypic and molecular methods. The aim of the analyses was to gain a better understanding of the clonality of the serovar and of the ability of its subtypes to be transmitted to humans via pigs and pork. Additionally, the genetic relatedness as well as the pathogenicity and antimicrobial resistance gene repertoire of S. enterica serovar 4,[5],12:i:− was compared with selected S. enterica serovar Typhimurium strains representing corresponding phage types in order to estimate the potential health risk for humans.
MATERIALS AND METHODS
Selection of strains.
Fifty-two S. enterica serovar 4,[5],12:i:− strains were isolated from porcine lymph nodes during an EU monitoring study in 2006 and 2007 on the prevalence of Salmonella in slaughter-age pigs (3). Lymph nodes were selected because they are a marker of asymptomatic intestinal carriage, and cross-contamination is strongly reduced compared to samples from carcass swabs. In addition, 30 monophasic strains isolated from pork in the same time frame were selected and sent to the National Reference Laboratory for Salmonella (NRL-BFR), Berlin, Germany, on a routine basis. Moreover, 66 clinical strains from epidemiologically unrelated human gastroenteritis cases, also isolated in 2006 and 2007 and provided by the National Reference Centre for Salmonella and other Enterics (NRZ-RKI), Wernigerode, Germany, were included in the study (Table 1). Strains were selected to represent various geographical origins in Germany as well as to cover different seasons. A subset of 61 S. enterica serovar 4,[5],12:i:− strains representing a maximum of different combinations of phage types, resistance, multilocus variable-number tandem repeat (VNTR) assay (MLVA) results, and pulsed-field gel electrophoresis (PFGE) profiles were further selected for microarray analysis (Table 2 ). For genotypic comparison of the monophasic S. enterica serovar 4,[5],12:i:−, an additional 20 S. enterica serovar Typhimurium strains (10 from pig and 10 from human) were selected to provide coverage for the corresponding phage types (6 DT193, 11 DT120, 1 RDNC [for reaction did not conform to definite or provisional types], 1 DT104, and 1 DT029 phage type strains) or tetraresistance pattern of ampicillin, sulfamethoxazole, streptomycin, and tetracycline (AMP-SMX-STR-TET, respectively) as found among the S. enterica serovar 4,[5],12:i:− strains. S. enterica serovar Typhimurium strains from pig were also obtained from the EU monitoring study in 2006 and 2007. S. enterica serovar Typhimurium strains from human were isolated from gastroenteritis cases and sent to NRZ-RKI.
TABLE 1.
Source of isolates used in this study
| Isolate source (n)a | No. of isolates by date |
|
|---|---|---|
| 2006 | 2007 | |
| Primary production (52) | 9 | 43 |
| Pork (30) | 9 | 21 |
| Human (66) | 32 | 34 |
Isolates (total of 148) were obtained from all sources between October 2006 and September 2007 in agreement with the EU monitoring study (12). n, number of isolates.
TABLE 2.
S. enterica strains selected for DNA microarray and phenotypic analysis
| Strain no. | German federal state | Origin | Serotype | Phage typed | Resistanceb | PFGE cluster | MLVAc | Size of plasmid(s) (kb) |
|---|---|---|---|---|---|---|---|---|
| 07-00040 | Schleswig-Holstein | Pork | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-7-6-0-2 | None |
| 07-04070 | North Rhine-Westphalia | Pork | 4,5,12:i:− | RDNC | AMP, SMX, STR, TET | B | 2-4-4-0-2 | None |
| 07-00711 | Thuringia | Pork | 4,5,12:i:−a | DT59 | AMP, SPE, STR, SXT, TET, TMP | A | 2-8-5-0-2 | 121, 6, 3 |
| 07-01548 | North Rhine-Westphalia | Pork | 4,5,12:i:− | DT193 | AMP, CHL, FFN, NAL, SMX, STR, TET | C | 2-5-4-0-1 | 106 |
| 07-02081 | Bavaria | Pork | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-6-19-0-2 | None |
| 07-02603 | Lower Saxony | Pork | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-7-4-0-2 | None |
| 07-03608 | Lower Saxony | Pork | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-6-20-0-2 | 21 |
| 07-00009 | North Rhine-Westphalia | Pork | 4,12:i:− | DT120 | AMP, SMX, STR, TET | A | 2-5-7-0-2 | None |
| 07-01272 | Saxony-Anhalt | Pork | 4,12:i:− | DT120 | AMP, SMX, STR, TET | A | 2-5-8-0-2 | 3, 2 |
| 07-01585 | Schleswig-Holstein | Pork | 4,12:i:− | DT193 | AMP, CHL, FFN, NAL, SMX, STR, TET | C | 2-5-4-0-3 | None |
| 07-02902 | Baden-Württemberg | Pork | 4,12:i:− | RDNC | AMP, TET | B | 2-5-5-0-3 | 111 |
| 07-03017 | Lower Saxony | Pork | 4,12:i:−a | NT | AMP, CHL, FFN, SMX, STR, TET | D | 2-7-5-0-2 | 2 |
| 07-03371 | North Rhine-Westphalia | Pork | 4,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-4-4-0-2 | None |
| 06-04525 | North Rhine-Westphalia | Pork | 4,5,12:i:−a | DT120 | AMP, SMX, STR, TET | A | 2-7-5-0-2 | None |
| 06-04991 | Brandenburg | Pork | 4,5,12:i:−a | DT120 | AMP, KAN, NEO, SMX, SPE, STR, SXT, TET, TMP, | A | 2-7-5-0-2 | 131, 3 |
| 06-04115 | Saxony | Pork | 4,12:i:− | NT | SMX, SPE, STR, SXT, TET, TMP | A | 2-2-7-0-2 | 43, <2 |
| 06-04419 | Lower Saxony | Pig | 4,12:i:−a | NT | TET | A | 2-5-5-0-3 | 2.2, 2 |
| 06-04446 | North Rhine-Westphalia | Pig | 4,5,12:i:− | RDNC | Susceptible | E | 1-3-20-2-3 | 94 |
| 06-05055 | Brandenburg | Pig | 4,12:i:− | DT193 | AMP,KAN, NEO, SMX, SPE, STR, TET | A | 2-5-19-0-2 | 100, 73, 5, <2 |
| 06-05089 | Lower Saxony | Pig | 4,12:i:− | DT193 | AMP, CHL, FFN, SMX, SPE, STR, SXT, TET, TMP | B | 2-7-19-0-2 | 54 |
| 07-00244 | Lower Saxony | Pig | 4,5,12:i:−a | RDNC | AMP, SMX, STR, TET | D | 2-7-2-0-2 | None |
| 07-00404 | Lower Saxony | Pig | 4,12:i:− | DT120 | AMP, SMX, STR, TET | A | 2-5-5-0-2 | None |
| 07-00528 | Lower Saxony | Pig | 4,12:i:−a | DT120 | AMP, SMX, STR, TET | A | 2-7-5-0-2 | 5, 3 |
| 07-00679 | Saxony-Anhalt | Pig | 4,5,12:i:− | DT120 | AMP, SMX, STR, SXT, TET, TMP | A | 2-5-20-0-2 | None |
| 07-00768 | Lower Saxony | Pig | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-4-20-0-2 | None |
| 07-00769 | Lower Saxony | Pig | 4,5,12:i:− | DT193 | TET | B | 2-5-4-0-2 | 94 |
| 07-01173 | Lower Saxony | Pig | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-6-4-0-1 | None |
| 07-01526 | Lower Saxony | Pig | 4,5,12:i:−a | DT120 | Susceptible | A | 2-6-4-0-2 | None |
| 07-01536 | North Rhine-Westphalia | Pig | 4,12:i:− | DT193 | CHL, FFN, NAL, TET | B | 2-5-5-0-2 | None |
| 07-01577 | North Rhine-Westphalia | Pig | 4,12:i:−a | DT120 | TET | A | 2-6-5-0-2 | None |
| 07-01798 | Lower Saxony | Pig | 4,5,12:i:− | DT120 | KAN, NEO | B | 2-6-4-0-2 | 3 |
| 07-02006 | Lower Saxony | Pig | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-6-4-0-2 | None |
| 07-02199 | Lower Saxony | Pig | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-4-5-0-2 | 2.2, 2 |
| 07-02432 | Lower Saxony | Pig | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-6-5-0-2 | None |
| 07-02684 | Lower Saxony | Pig | 4,12:i:− | DT193 | AMP, SMX, STR, SXT, TET, TMP | B | 2-6-20-0-1 | 5, <2 |
| 07-02736 | Lower Saxony | Pig | 4,5,12:i:− | DT193 | AMP, SMX,STR, TET | B | 2-6-17-0-2 | None |
| 07-02781 | Rheinland-Pfalz | Pig | 4,5,12:i:− | DT193 | Susceptible | D | 18-6-3-2-2 | 94 |
| 07-02789 | Thuringia | Pig | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-5-4-0-2 | None |
| 07-02809 | North Rhine-Westphalia | Pig | 4,5,12:i:− | DT120 | AMP, SMX, STR, TET | A | 2-5-4-0-2 | 2.4, 2.2 |
| 07-03136 | Lower Saxony | Pig | 4,5,12:i:−a | DT120 | KAN, NEO, SMX, SPE, STR, SXT, TET, TMP | D | 2-4-6-0-2 | 111, 94 |
| 07-03327 | Lower Saxony | Pig | 4,5,12:i:− | DT193 | AMP, SMX, SPE, STR, SXT, TET, TMP | B | 2-6-7-0-2 | 102 |
| 07-03466 | Lower Saxony | Pig | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-7-3-0-2 | None |
| 08-03968 | Hamburg | Human | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-6-4-0-2 | None |
| 08-03969 | Hamburg | Human | 4,5,12:i:− | DT7 | TET | A | 2-5-5-0-2 | 5 |
| 08-03972 | Hamburg | Human | 4,5,12:i:−a | DT120 | AMP, CHL, FFN, SMX, TET | D | 2-7-3-4-3 | 94, 5 |
| 08-03974 | Hamburg | Human | 4,5,12:i:− | DT120 | AMP, SMX, STR, TET | A | 2-7-19-0-2 | None |
| 08-03979 | Berlin | Human | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-4-5-0-2 | None |
| 08-03985 | Lower Saxony | Human | 4,12:i:− | U302 | AMP, CIP, NAL, SMX, STR, TET | C | 2-5-4-0-3 | 65 |
| 08-03987 | Baden-Württemberg | Human | 4,5,12:i:−a | DT120 | AMP, SMX, STR, TET | A | 2-7-5-0-2 | None |
| 08-03988 | North Rhine-Westphalia | Human | 4,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-4-4-0-2 | 54 |
| 08-03990 | Saxony | Human | 4,5,12:i:− | DT193 | TET | B | 2-6-4-0-2 | None |
| 08-03996 | Lower Saxony | Human | 4,12:i:− | DT193 | AMP SMX STR TET | B | 2-4-6-0-2 | None |
| 08-03997 | Lower Saxony | Human | 4,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-6-20-0-1 | None |
| 08-03999 | Saxony-Anhalt | Human | 4,5,12:i:− | DT120 | AMP, SMX, STR, TET | A | 2-5-4-0-2 | None |
| 08-04002 | Schleswig-Holstein | Human | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-4-4-0-2 | None |
| 08-04009 | Mecklenburg Western Pomerania | Human | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-6-5-0-2 | None |
| 08-04018 | Saxony-Anhalt | Human | 4,5,12:i:− | DT193 | AMP, SMX, STR, TET | B | 2-5-4-0-2 | None |
| 08-04019 | Mecklenburg Western Pomerania | Human | 4,5,12:i:− | NT | AMP, SMX, STR | B | 2-6-4-0-2 | None |
| 08-04024 | Thuringia | Human | 4,12:i:− | DT193 | Susceptible | A | 2-6-7-0-2 | 2, <2 |
| 08-04030 | Saxony-Anhalt | Human | 4,5,12:i:−a | DT193 | AMP, SMX, STR, TET | B | 2-5-5-0-2 | None |
| 08-04031 | Rhineland-Palatinate | Human | 4,5,12:i:− | NT | AMP, TET | A | 2-6-20-0-2 | 6, 3 |
| 06-04998 | North Rhine-Westphalia | Pig | 4,5,12:i:1,2 | DT104 | AMP, SMX, STR, TET | D | 5-2-8-0-2 | 3 |
| 07-00577 | Lower Saxony | Pig | 4,12:i:1,2 | DT120 | AMP, CHL, GEN, SMX, STR, TET | A | 2-5-4-0-2 | 132,30 |
| 07-00635 | Lower Saxony | Pig | 4,12:i:1,2 | DT120 | AMP, CHL, KAN, NEO, SMX, STR, TET | A | 2-6-5-0-2 | 3, 9, 30 |
| 07-01010 | Lower Saxony | Pig | 4,5,12:i:1,2 | DT120 | AMP, KAN, NEO, SMX, SPE, STR, SXT, TET, TMP | A | 2-6-19-0-2 | 170, 114, 4 |
| 07-01529 | Lower Saxony | Pig | 4,5,12:i:1,2 | DT120 | AMP, SMX, STR | A | 2-5-4-0-2 | 132, 6, 3 |
| 07-02186 | Lower Saxony | Pig | 4,12:i:1,2 | DT029 | AMP, SMX, STR, SXT, TET, TMP | A | 2-6-0-0-2 | 70 |
| 07-02788 | Thuringia | Pig | 4,5,12:i:1,2 | DT120 | AMP, SMX, SPE, STR, SXT, TMP | A | 2-8-5-0-2 | 120 |
| 07-03137 | Lower Saxony | Pig | 4,5,12:i:1,2 | DT193 | KAN, NEO, SMX, SPE, STR, SXT, TET, TMP, | A | 2-6-4-0-2 | 140, 4 |
| 07-03250 | North Rhine-Westphalia | Pig | 4,12:i:1,2 | DT120 | AMP, SMX, STR, SXT, TET, TMP | A | 2-5-20-0-2 | 120 |
| 07-03714 | Lower Saxony | Pig | 4,12:i:1,2 | RDNC | AMP, SMX, STR, TET | D | 2-6-5-0-2 | 25, 3 |
| 09-01035 | Schleswig-Holstein | Human | 4,5,12:i:1,2 | DT193 | AMP, SMX, STR, TET | D | 5-2-5-0-2 | 91, 2 |
| 09-01036 | Mecklenburg Western Pomerania | Human | 4,5,12:i:1,2 | DT120 | AMP SMX STR TET | A | 2-5-20-0-2 | 3, 4 |
| 09-01037 | Mecklenburg Western Pomerania | Human | 4,5,12:i:1,2 | DT120 | AMP, SMX, STR, TET | A | 2-5-20-0-2 | 6, 4, 3 |
| 09-01039 | Saxony-Anhalt | Human | 4,5,12:i:1,2 | DT193 | AMP, SMX, STR, TET | D | 5-2-17-0-2 | 91 |
| 09-01041 | Saxony-Anhalt | Human | 4,5,12:i:1,2 | DT120 | AMP, SMX, STR, TET | A | 2-8-20-0-2 | 4 |
| 09-01043 | Mecklenburg Western Pomerania | Human | 4,5,12:i:1,2 | DT193 | AMP, SMX, STR, TET | D | 4-2-3-0-2 | 91 |
| 09-01044 | Mecklenburg Western Pomerania | Human | 4,5,12:i:1,2 | DT193 | AMP, SMX, STR, TET | D | 4-2-2-0-2 | 91 |
| 09-01045 | Mecklenburg Western Pomerania | Human | 4,12:i:1,2 | DT193 | AMP, SMX, STR, TET | A | 2-7-20-0-2 | 108 |
| 09-01046 | Saxony | Human | 4,5,12:i:1,2 | DT120 | AMP, SMX, STR, TET | A | 2-5-20-0-2 | None |
| 09-01047 | Saxony-Anhalt | Human | 4,12:i:1,2 | DT120 | AMP, SMX, STR | A | 2-5-20-0-2 | None |
PCR positive for fljB_1,2.
Abbreviations are given in Materials and Methods.
Order of VNTR loci: STTR9-STTR5-STTR6-STTR10-STTR3. According to Larsson et al. (16) STTR3 allele number 2 corresponds to number 211, number 1 corresponds to 111, and number 3 corresponds to 311.
NT, nontypeable.
Serotyping.
All strains were previously serotyped according to the White-Kauffmann-Le Minor scheme (14) by slide agglutination with O and H antigen-specific sera (Sifin Diagnostics, Berlin, Germany).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility of strains was tested against 17 antimicrobials or antimicrobial combinations by determining the MIC using the CLSI broth microdilution method (6) in combination with the semiautomatic Sensititre system (TREK Diagnostic Systems, Cleveland, OH). Breakpoints were applied as previously described (22). Antimicrobials tested were amoxicillin-clavulanic acid (AMC), AMP, chloramphenicol (CHL), ciprofloxacin (CIP), colistin (COL), florfenicol (FLO), gentamicin (GEN), kanamycin (KAN), neomycin (NEO), nalidixic acid (NAL), spectinomycin (SPE), STR, SMX, trimethoprim-sulfamethoxazole (SXT), TET, trimethoprim (TMP), and ceftiofur (XNL).
Phage typing.
Phage typing was performed according to the phage typing scheme developed by Callow (4) and extended by Anderson et al. (2). Phage patterns not included in the scheme are designated RDNC.
DNA purification.
Strains were grown aerobically in Luria-Bertani broth (Merck, Darmstadt, Germany) with shaking at 38°C for 16 to 18 h. DNA isolation was carried out using a DNeasy Blood and Tissue Kit (Qiagen GmbH, Germany) according to the manufacturer's protocol with the addition of 25 μl of proteinase K instead of 20 μl and extended lysis for 3.5 h. The quality and quantity of DNA were measured spectrophotometrically, and a minimum of 4 μg of high-quality DNA was used for labeling.
PCR.
All phenotypically monophasic S. enterica serovar 4,[5],12:i:− strains were tested by multiplex PCR with specific oligonucleotides for the presence of the antigenic genes rfbJ (O:4 antigen), fliC (H:i antigen), and fljB (H:1,2 antigen) according to Lim et al. (17). Another PCR using oligonucleotides Fsa2 and rFsa2 resulting in a product of 1,478 bp was also used in order to detect almost the entire DNA sequence of the fljB gene (7).
Plasmid analysis.
Plasmid DNA was extracted by an alkaline denaturation method described previously (15a), with minor modifications. Following extraction, plasmids were electrophoretically separated in horizontal 0.9% agarose gels at 100 V for 3.5 h in Tris-borate-EDTA buffer. For the determination of plasmid sizes the Escherichia coli reference plasmids R27 (112 MDa), R1 (62 MDa), RP4 (36 MDa), and ColE1 (4.2 MDa) and a supercoiled DNA ladder (Invitrogen Life Technologies, Karlsruhe, Germany) were included. The plasmids were stained with an aqueous solution of ethidium bromide (10 μg/ml; Sigma, Deisenhofen, Germany) and imaged under UV illumination.
Multilocus variable-number tandem-repeat analysis.
MLVA using an ABI 310 Genetic Analyzer and VNTR allele number assignment was performed as described by Lindstedt et al. (18).
Pulsed-field gel electrophoresis.
PFGE was carried out after digestion of genomic DNA with the restriction enzyme XbaI according to the PulseNet protocol (21). Gel images were analyzed in BioNumerics, version 5.1 (Applied Maths, Sint-Martens-Latem, Belgium), and compared by cluster analysis using the Dice coefficient and unweighted pair group method with arithmetic mean (UPGMA) with a position tolerance of 1.5% and optimization of 1.0%.
DNA microarray analysis.
The DNA microarray used in this study was as previously described (15). A set of 275 gene-specific 57- to 60-mer oligonucleotide probes derived from Salmonella sequences deposited in the GenBank at NCBI (http://www.ncbi.nlm.nih.gov/) were designed using the program Array Designer, version 4.1 (Premier Biosoft, Palo Alto, CA). The probes were assigned to seven different marker groups depending on the functionality of the corresponding gene sequence (number of probes): pathogenicity (80), resistance (49), serotyping (33), fimbriae (22), DNA mobility (57), metabolism (21), and prophages (13). In addition, three 57- to 61-mer oligonucleotides derived from the Arabidopsis thaliana genes RCA (M86720), RCP1 (NM_12175), and PRKASE (X58149) were designed as negative-control probes. Virulence determinants for each strain analyzed were categorized according to their locations on the Salmonella genome: Salmonella pathogenicity islands (SPIs), prophages, plasmid, islets, and fimbrial clusters. Microarray signals, which were assigned as “uncertain” by microarray analysis, were reanalyzed by PCR using primers as previously described (15). Following PCR testing, an individual decision was made for the presence or absence of this target. Analysis of the DNA microarray data was performed as previously described (15). A comparison between strains was done in BioNumerics, version 5.1, after importing data, gene present or absent, as the character type.
Statistical methods.
To assess the discriminatory power of phage type, PFGE, and MLVA, Simpson's index of diversity (ID) and the 95% confidence intervals (CI) were calculated using the Comparing Partitions website (http://darwin.phyloviz.net/ComparingPartitions/index.php?link=Tool ).
RESULTS
Phenotypic characteristics of S. enterica serovar 4,[5],12:i:−.
Seventy percent (104 strains) of the 148 selected monophasic Salmonella strains expressed the O:5 antigen (S. enterica serovar 4,5,12:i:−) while for 30% the O:5 antigen was serologically undetectable (S. enterica serovar 4,12:i:−). The O:5 antigen-negative strains were found at 35% prevalence in isolates from primary production (lymph nodes) and pork and with a 24% prevalence in isolates of human origin. Phage typing revealed that 70% (104) of the strains were assigned as DT193, and 19% (28) were assigned as DT120. A smaller proportion was tested as RDNC (4%) or was not typeable ([NT] 5%). Only 2% of the strains belonged to other phage types (Table 3). The Simpson's index of diversity for phage typing was 47.8 (95% CI, 39.2 to 56.5). No statistically significant difference (P < 0.05) was found between phage types and the source of strains.
TABLE 3.
Phage type distribution in 148 S. enterica serovar 4,[5],12:i:− strains
| Phage typea | % Distribution of different phage type by isolate source (n)b |
|||
|---|---|---|---|---|
| Primary production (52) | Pork (30) | Human (66) | Total (148) | |
| DT193 | 75 | 57 | 71 | 70 |
| DT120 | 19 | 23 | 17 | 19 |
| RDNC | 4 | 10 | 1 | 4 |
| Other (DT59, DT7, U302, NT) | 2 | 10 | 11 | 7 |
RDNC, reaction did not conform; NT, nontypeable.
n, number of isolates.
Twenty-seven different antimicrobial resistance profiles were identified, with 90% (133) of the strains resistant to more than one antimicrobial and 81% (120) resistant to four or more antimicrobials. The predominant resistance pattern observed was the combination of ampicillin, sulfamethoxazole, streptomycin, and tetracycline (AMP-SMX-STR-TET). This pattern of resistance was observed in 65% (96) of the strains and approximately equally distributed among isolates from primary production, meat, and human origin (Table 4). Sixteen percent of the strains were resistant to other antimicrobials in addition to the tetraresistance group.
TABLE 4.
Resistance profiles in 148 S. enterica serovar 4,[5],12:i:− strains
| Resistancea | % Distribution of different resistance profiles by isolate source (n)b |
|||
|---|---|---|---|---|
| Primary production (52) | Pork (30) | Human (66) | Total (148) | |
| AMP, SMX, STR, TET | 58 | 63 | 71 | 65 |
| TET | 8 | 10 | 6 | 7 |
| AMP, SMX, STR | 4 | 0 | 9 | 5 |
| AMP, SMX, STR, SXT, TET, TMP | 6 | 0 | 0 | 2 |
| AMP, TET | 2 | 3 | 1.5 | 2 |
| Susceptible | 6 | 0 | 1.5 | 3 |
| Other | 16c | 24d | 11e | 16f |
See Materials and Methods for abbreviations.
n, number of isolates.
Containing eight different resistance profiles.
Containing six different resistance profiles.
Containing seven different resistance profiles.
Containing 21 different resistance profiles
Typing of S. enterica serovar 4,[5],12:i:− by pulsed-field gel electrophoresis (PFGE).
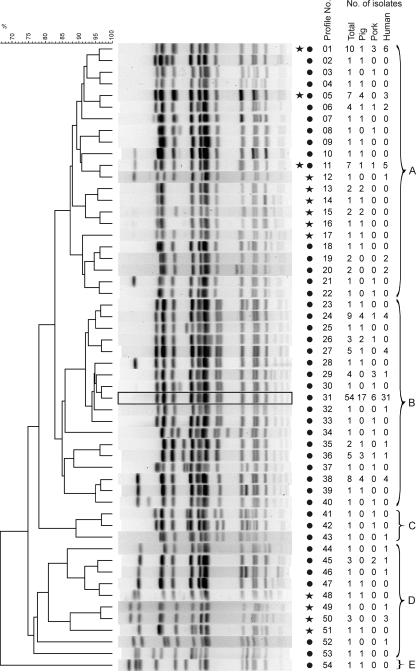
Among the 148 S. enterica serovar 4,[5],12:i:− strains, 44 different XbaI profiles (ID, 85.4 [95% CI, 80.1 to 90.8]) were identified (Fig. 1). The 20 S. enterica serovar Typhimurium strains selected for genotypic comparison revealed 13 different XbaI profiles (Fig. 1). Three profiles (X01, X05, and X11) were found in both serovars. The profiles were used to generate a UPGMA tree using the Dice similarity coefficient. The tree distinguished the strains into five PFGE clusters (A, B, C, D, and E) (Fig. 1). Sixty-eight percent of the monophasic strains (100 out of 148) belonged to cluster B, dominated by phage type DT193 strains (96 out of 100). The most prevalent PFGE profile was X31, which corresponds to STYMXB.0131 (designation according to PulseNet Europe). It was observed in 36% (54 strains) of all monophasic strains analyzed. No statistically significant difference (P < 0.05) was found in cluster B for the source of strains.
FIG. 1.
UPGMA dendrogram of PFGE profiles identified in 148 S. enterica serovar 4,[5],12:i:− and 20 S. enterica serovar Typhimurium strains after digestion with XbaI. At the right of the PFGE profiles, a star indicates the presence of the profile in S. enterica serovar Typhimurium, and a filled circle indicates its presence in S. enterica serovar 4,[5],12:i:−. Profiles were designated X01 to X54. The numbers of isolates belonging to each source (total, pig, pork, and human) are also shown. Designated clusters A to E are indicated by curly brackets. A rectangle highlights the most prominent PFGE profile, X31, also found in an outbreak in Luxembourg (STYMXB.0131; designation according to PulseNet Europe).
Typing of S. enterica serovar 4,[5],12:i:− by MLVA.
Table 5 summarizes the MLVA typing for the 148 S. enterica serovar 4,[5],12:i:− strains. Thirty-eight different MLVA profiles (ID, 91.8 [95% CI, 98.6 to 94.1]) were identified, with most variation noted in loci STTR5 (8 different alleles) and STTR6 (10 different alleles). For locus STTR9, the allele number 2 was observed in all but two strains, and locus STTR10pl located on the Salmonella virulence plasmid was absent in all but three strains. The most prominent combination of alleles was 2-6-4-0-2 (order of loci, STTR9-STTR5-STTR6-STTR10pl-STTR3) which was found in 20% of the S. enterica serovar 4,[5],12:i:− strains tested. Statistically significant differences (P < 0.05) were calculated for MLVA profile 2-6-4-0-2 between isolates from pork and human as well as for profile 2-7-5-0-2 between isolates from pork and both primary production and human.
TABLE 5.
Distribution of different MLVA profiles in S. enterica serovar 4,[5],12:i:− strains
| MLVA allelesa | % Distribution of different MLVA profiles by isolate source (n)b |
|||
|---|---|---|---|---|
| Primary production (52) | Pork (30) | Human (66) | Total (148) | |
| 2-6-4-0-2 | 17 | 10 | 26 | 20 |
| 2-4-4-0-2 | 13 | 10 | 14 | 13 |
| 2-5-4-0-2 | 13 | 7 | 12 | 11 |
| 2-7-5-0-2 | 6 | 23 | 3 | 8 |
| 2-6-5-0-2 | 12 | 3 | 6 | 7 |
| 2-6-20-0-2 | 4 | 10 | 2 | 4 |
| 2-4-5-0-2 | 4 | 0 | 4 | 3 |
| 2-5-5-0-2 | 4 | 0 | 3 | 3 |
| 2-6-7-0-2 | 2 | 0 | 4 | 3 |
| 2-7-19-0-2 | 2 | 3 | 3 | 3 |
| Other | 23c | 34d | 23e | 25f |
Order of loci: STTR9-STTR5-STTR6-STTR10pl-STTR3. In all strains STTR3 allele number 2 corresponds to number 211 according to Larsson et al. (16).
n, number of isolates.
Containing 12 different MLVA profiles.
Containing 10 different MLVA profiles.
Containing 13 different MLVA profiles.
Containing 28 different MLVA profiles.
Characterization of clonal lineages in S. enterica serovar 4,[5],12:i:−.
The most prominent combination of phenotypic characteristics was that 74 of the 103 O:5 antigen-positive strains belonged to phage type DT193. Among these strains 77% (57 strains) harbored the most prevalent tetraresistance pattern AMP-SMX-STR-TET, and 84% of these (48 strains) also corresponded to the most frequently found PFGE cluster B. These characteristics were united in 32% of the 148 strains selected. Further discrimination of the 48 strains by MLVA revealed 15 different MLVA profiles, with 50% of the strains assigned to the combined MLVA profile 2-X-4-0-2 (X represents five various STTR5 alleles). These isolates were obtained from primary production, pork, and human samples.
For the second-most-prevalent phage type DT120 (28 strains), 57% (16 strains) of the DT120 strains also demonstrated AMP-SMX-STR-TET tetraresistance. Interestingly, eight tetraresistant strains and another eight multiresistant phage type DT120 strains were positive by PCR for the second-phase flagellum gene fljB_1,2 although phenotypically the H1,2 antigen was repeatedly not detected. In contrast to the phage type DT193 strains, nearly all phage type DT120 strains belonged to PFGE cluster A, with further discrimination into 10 different MLVA profiles and a maximum of three strains belonging to one pattern. Similarly to phage type DT193, isolates originated again from pigs, pork, and humans.
Determination of pathogenicity gene repertoire in S. enterica serovar 4,[5],12:i:−.
For 57 of the 61 strains tested (Table 2), an identical virulence gene profile was observed with all probes positive for Salmonella pathogenicity islands SPI1 to SPI5 and probes negative for SPI7. Additionally, genes for Gifsy-1 (gipA and gogB) and Gifsy-2 (gtgA, sodC1, and sseI) prophage were present while sspH1 (encoding a Salmonella-secreted protein) located in Gifsy-3 and sodCIII (encoding putative Cu/Zn superoxide dismutase) located in Fels-1 were absent. Also other genes (hldD_DT104, irsA, and sopE1) harbored by prophages were absent (data not shown).
Four strains shared a different pathogenicity gene profile. Three genes associated with the Salmonella virulence plasmid pSLT (spvC, spvR, and rck) gave positive signals but the fimbrial gene pefA usually harbored by the plasmid was absent. One of the three isolates (isolate 08-03972; phage type DT120) harbored hldD typically on a prophage in S. enterica serovar Typhimurium phage type DT104 as well as sopE1 (prophage-encoded effector protein). The same strain lacked gipA (encoding the Peyer's patch-specific virulence factor GipA), which was also absent in another of the three isolates (isolate 07-02781; phage type DT193). The fourth, nontypeable phage type isolate (06-04115) lacked the pipA and pipD genes present on SPI5.
Microarray analysis of serotype marker genes in S. enterica serovar 4,[5],12:i:−.
The three genes fljA, fljB_1,x (where x represents various antigenic markers), and hin consecutively ordered on the Salmonella chromosome encode the structural gene (fljB) and are important for phase variation (fljA and hin) of the second-phase flagellum antigen. Five combinations for these markers were found within the test set of S. enterica serovar 4,[5],12:i:− isolates. Of 61 strains, 42 were negative when tested with the three probes for fljA, fljB_1,x, and hin. Three isolates harbored only fljA while three were positive only for hin. Of the remaining 13 serologically monophasic strains, four possessed genes for fljA and fljB_1,x but were negative for hin while nine isolates were positive for all three genes. All 13 strains were positive for a specific fljB_1,2 DNA fragment. Additional PCRs to amplify the complete fljB revealed seven positive results, of which six gave the expected DNA fragment size of 1,478 bp, while for one strain (07-00711) a fragment larger than 2 kb was amplified. The majority of strains showing variation within this region belonged to phage type DT120 or other rare phage types (e.g., DT7 or DT59).
Determination of the antimicrobial resistance determinant repertoire.
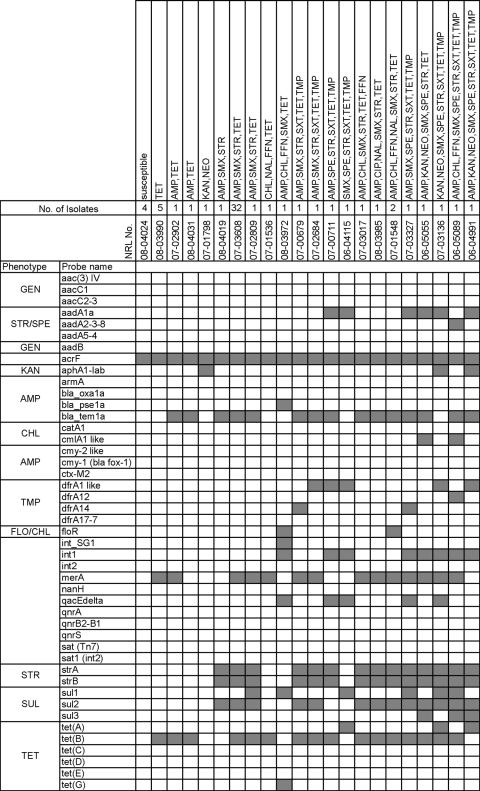
Twenty-two different genotypic resistance combinations were found within the 61 test strains of S. enterica serovar 4,[5],12:i:− analyzed (Fig. 2). Nine additional genotypic combinations were identified in the 20 S. enterica serovar Typhimurium strains that were selected for genetic relatedness studies (see below). Generally, the antimicrobial phenotype corresponded with the antimicrobial genotype with two exceptions. Strains 07-03017 (phage type NT) and 07-01536 (phage type DT193) were negative for floR although phenotypically conferring resistance to florfenicol and chloramphenicol. The main phenotypic tetraresistance pattern AMP-SMX-STR-TET was encoded by genes blaTEM1-like (encoding ß-lactamase), sul2 (encoding dihydropteroate synthase), strA-strB (encoding aminoglycoside phosphotransferase), and tet(B) (encoding an efflux pump), respectively.
FIG. 2.
Phenotypic and corresponding genotypic resistance profiles identified for the 61 S. enterica serovar 4,[5],12:i:− strains tested. At the top of the figure the NRL strain numbers, numbers of isolates with the same genetic resistance profile, and the corresponding phenotypic resistance patterns are indicated. On the left-hand side, the probes for genes related to antimicrobial resistance phenotypes (see Materials and Methods for abbreviations) or to other resistance elements, e.g., integron-associated integrases (int1, int2, and int_SG1) are listed in alphabetical order. The graph shows the hybridization result of each strain. A gray box indicates the presence of the target sequence in the strain; a white box indicates its absence.
One S. enterica serovar 4,5,12:i:− phage type DT120 strain (08-03972) isolated from human harbored the combination of int1, qacEΔ, sul, tet(G) and floR typical for Salmonella genomic island 1 (SGI-1). The strain was genotypically identified as S. enterica serovar Typhimurium (probes for hin, fljA, and fljB_1,2 positive).
Genetic relatedness of S. enterica serovar 4,[5],12:i:− to S. enterica serovar Typhimurium.
The genetic relatedness of S. enterica serovar 4,[5],12:i:− and S. enterica serovar Typhimurium strains was determined by MLVA, PFGE, and DNA microarray. For the comparative studies 20 S. enterica serovar Typhimurium strains isolated from humans and pigs were selected. Of the 20 isolates selected, 30% were assigned to phage type DT193, 55% to DT120, 5% to RDNC, and 10% to other phage types. The isolates belonged to eight different phenotypic resistance profiles including the tetraresistance AMP-SMX-STR-TET profile in 50% of the strains. In MLVA 14 different allele combinations were found in the 20 S. enterica serovar Typhimurium strains, with the most prominent combination 2-5-20-0-2 in 25% of the strains belonging to phage type DT120. This combination was found only in one (phage type DT120) of the 148 S. enterica serovar 4,[5],12:i:− strains tested. In both serovars VNTR loci STTR3 and STTR9 were mainly assigned to allele number 2. Only one of the phage type DT120 strains of both serovars was positive for the VNTR locus STTR10pl typically located on the pSLT plasmid. Seventy percent (14/20) of the S. enterica serovar Typhimurium strains were grouped to PFGE cluster A. The majority of these strains (79%) belonged to phage type DT120. The remaining 30% of the S. enterica serovar Typhimurium isolates were assigned to cluster D and belonged mainly to DT193.
Twenty-seven of the 61 S. enterica serovar 4,[5],12:i:− strains analyzed harbored at least one plasmid ranging between 110 kb and <2 kb (Table 2). Of these, only three strains were also positive for the spvC, spvR, and rck genes typically located on the Salmonella virulence plasmid pSLT. In comparison, only two S. enterica serovar Typhimurium strains harbored no plasmid while most strains revealed one (nine strains), two (five strains), or four plasmids (four strains). Although plasmids of 91-kb size (virulence plasmid size of S. enterica serovar Typhimurium strain LT2, 94 kb) that were positive for spvC, spvR, and rck were found in four phage type DT193 strains, the plasmid-associated pef gene and MLVA VNTR locus STTR10pl were not identified, indicating a possible variant of the virulence plasmid.
The pathogenicity gene repertoire analyzed by DNA microarray in 14 out of the 20 S. enterica serovar Typhimurium strains was identical to the pattern of virulence determinants found in the S. enterica serovar 4,[5],12:i:− strains. Additionally four other strains shared the virulence genes spvC, spvR, and rck harbored on the plasmid Furthermore, one of the four strains was negative for the gipA gene. One strain (06-04998) was positive for spvC but negative for rck and gipA. Another strain (07-02186; phage type DT193) was positive for sopE1 (phage-encoded effector protein) and lacked gipA and gogB. All S. enterica serovar Typhimurium and S. enterica serovar 4,[5],12:i:− strains shared the same set of fimbrial genes.
The resistance gene repertoire often differed between both serovars despite the identical phenotypic profiles including tetraresistance (AMP-SMX-STR-TET). In S. enterica serovar Typhimurium phage type DT193 strains (five out of six), tetracycline resistance was encoded by tet(A) instead of tet(B) as detected in S. enterica serovar 4,[5],12:i:− phage type DT193 strains. Sulfamethoxazole resistance was sometimes additionally encoded by the gene sul1 in addition to sul2. Ampicillin resistance was uniquely encoded by blaTEM1-like, and streptomycin was mainly encoded by strA-strB but sometimes in combination with aadA1.
DISCUSSION
S. enterica serovar 4,[5],12:i:− is a worldwide emerging monophasic serovar (24). In Europe, especially, pork and its products contaminated with this serovar were identified as sources for human Salmonella infections (8, 10, 19). Several studies indicated that the serovar is a monophasic variant of S. enterica serovar Typhimurium lacking a genomic region that harbors the structural and regulating genes fljA, fljB, and hin encoding the second-phase flagellum (1, 8, 11, 13, 23).
The studies reported here support the observation that S. enterica serovar 4,[5],12:i:− is an emerging hazard for humans and that this hazard is directly linked to the consumption of contaminated pork. A substantial number of strains isolated from pig, pork, and human were extensively characterized to understand the clonality, resistance patterns, and pathogenicity gene repertoire, and their genetic relatedness to the classical biphasic S. enterica serovar Typhimurium was studied. This is the first study which comprehensively compares S. enterica serovar 4,[5],12:i:− isolates obtained from the food chain and from clinical cases of gastroenteritis in human.
A main lineage of S. enterica serovar 4,[5],12:i:− was identified in the isolates which primarily belonged to phage type DT193 and exhibited at least the tetraresistance pattern AMP-SMX-STR-TET encoded by blaTEM1-like, sul2, strA-strB, and tet(B), respectively. The second independently evolved lineage was phage type DT120. It was striking that 57% of the phenotypically monophasic phage type DT120 strains were positive by PCR for fljB_1,2, fljA, and hin. Furthermore, the DT193 and DT120 strains investigated revealed a number of other different genetic properties, e.g., different clustering by PFGE and MLVA. This indicates that in Germany monophasic phage type DT120 strains have formed an additional clonal lineage different from that of phage type DT193 strains. However, further studies are required to elucidate if the monophasic phage type DT120 lineage occurs also in other European countries or worldwide. Independent of their phage type, approximately 30% of the strains investigated did not express the O:5 antigen although the antigen-encoding gene oafA was present in their genomes. Preliminary experiments showed that a small 7-bp deletion or an interruption by an insertion (IS) element within the oafA open reading frame was responsible for the abolition of the O:5 antigen expression in these strains (E. Hauser et al., unpublished data). The lack of the O:5 antigen did not correlate with the source of the isolates.
An outbreak of human gastroenteritis associated with consumption of pork products in Luxembourg was found to be caused by S. enterica serovar 4,[5],12:i:− phage type DT193 with the tetraresistance pattern and the PFGE profile STYMXB.0131 (18). This was also the most prominent PFGE profile for the phage type DT193 strains in this study. It may be that an expansion of this clonal lineage has begun within Europe. However, no reliable data confirming this hypothesis have been published from other European countries; this is likely to be because of the lack of subtyping data for this monophasic serovar. Identical traits were found in isolates from pigs, pork, and humans. Consequently, the serovar is able to transmit via the food chain to humans. Isolates from feeding stuff were not received at the NRL-BFR before 2007. The role feeding stuff may play in dissemination remains to be elucidated.
Previous studies from Spain revealed that monophasic phage type U302 strains were isolated mainly from pigs (8). In these studies U302 strains exhibited resistance to chloramphenicol (CHL) in addition to the tetraresistance within the phage type DT193 strains. An Italian study of S. enterica serovar 4,[5],12:i:− strains from humans indicated that the tetraresistance of S. enterica serovar 4,[5],12:i:− was associated with phage type U302 and nontypeable strains (9). Therefore, the DT193 strains from Germany probably originated from a different clonal lineage than the Spanish U302 strains. The difference in phage types reported in Spain and Italy from those reported in Germany indicates a change within the last years in the subtype of S. enterica serovar 4,[5],12:i:− spreading throughout Europe because currently phage type DT193 is much more frequently observed than the phage type U302 initially observed in Spain. A recent study comparing Spanish and U.S. isolates identified also at least two clonal lines in S. enterica serovar 4,[5],12:i:− (23). They differ in a deletion surrounding fljAB. However, the German isolates investigated here represent yet another clonal lineage compared to the U.S. isolates because those were mainly pan-susceptible and represented by other PFGE profiles, as described in this study.
In 42 S. enterica serovar 4,[5],12:i:− strains tested by microarray, a chromosomal deletion including hin, fljB, and fljA was responsible for the monophasic phenotype. This deletion was previously described in association with the Spanish monophasic phage type U302 strain, where 16 genes were implicated (13). Furthermore, several variants of partial deletions within this region were detected, including partial or complete deletion of fljB or a possible deletion of fljA and fljB, although hin was still found to be present (23, 26). In the U.S. isolates a major region containing 76 genes was deleted, but the hin gene at the 3′ end of the deletion was present, which was not the case in the Spanish isolates (23). These and new variants were detected in this study, e.g., isolates with only fljA, only hin, or an insert in fljB. These data clearly indicate that within this region of the S. enterica serovar Typhimurium genome, multiple independent deletions can occur, leading to phenotypically monophasic S. enterica serovar 4,[5],12:i:−.
The relatedness of S. enterica serovar 4,[5],12:i:− to S. enterica serovar Typhimurium has previously been discussed (11, 26). Zamperini et al. (26) observed identical PFGE patterns in S. enterica serovar 4,[5],12:i:− and S. enterica serovar Typhimurium and also some of the typical pathogenicity genes of S. enterica serovar Typhimurium. Another study suggested S. enterica serovar 4,[5],12:i:− as a possible monophasic variant of S. enterica serovar Typhimurium phage type U302 based on comparison of PFGE and resistance profiles (8). Similar conclusions were outlined during investigations on isolates from Thailand (1). DNA microarray-based analyses comparing almost all protein coding regions of S. enterica serovar Typhimurium strain LT2 (genome sequenced) with those of S. enterica serovar 4,[5],12:i:− U302 found only a few differences (13). Both serovars can also share the same multilocus sequence type (23). In this study a comparison of 102 virulence determinants using a comprehensive set of strains clearly showed the close, almost identical, pathogenicity gene repertoire, independently of whether the strains belonged to the monophasic or biphasic S. enterica serovar Typhimurium. All markers indicating fimbrial clusters occurring in S. enterica serovar Typhimurium were also positive in S. enterica serovar 4,[5],12:i:−. It has been previously shown that fimbrial clusters are conserved within a serovar (15, 20). Nevertheless, there were some interesting genetic differences between phage type DT193 isolates of both serovars. Tetracycline resistance was encoded mainly by tet(B) in DT193 S. enterica serovar 4,[5],12:i:− strains, whereas it was encoded by tet(A) in DT193 S. enterica serovar Typhimurium strains. Additionally, the strains clustered in different PFGE clades. Such differences indicate that the S. enterica serovar Typhimurium phage type DT193 lineage is not a direct ancestor of the monophasic phage type DT193. In contrast, S. enterica serovar 4,[5],12:i:− phage type DT120 strains showed more genetic congruence with the S. enterica serovar Typhimurium phage type DT120 strains, suggesting that this biphasic subtype is the recent common ancestor of the monophasic variant.
In conclusion, the typing of isolates received at both German national reference laboratories (NRL-BFR and NRZ-RKI) based on a routine diagnostic indicates that S. enterica serovar 4,[5],12:i:− is a continually emerging pathogen in Germany. Molecular analyses showed that the same genotypes can be isolated from pigs, pork, and humans. Two main lineages are currently spreading in pigs and humans which are characterized by phage type DT193 and DT120, respectively. Both exhibited tetraresistance to AMP, SMX, STR, and TET. Due to the close genetic relatedness to S. enterica serovar Typhimurium, in particular with respect to the pathogenicity gene repertoire, the ongoing control measure programs to eradicate S. enterica serovar Typhimurium in food-producing animals will have to include the monophasic variant, too.
Acknowledgments
This work was funded by the Bundesministerium für Bildung und Forschung, FBI-Zoo (01 KI 07123).
We thank Beatriz Guerra-Roman and Andreas Schroeter for continuous support. We also thank Cornelia Bunge-Croissant for technical assistance and Roberto La Ragione (Veterinary Laboratories Agency, Weybridge, United Kingdom) for his critical revision of the manuscript.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Amavisit, P., W. Boonyawiwat, and A. Bangtrakulnont. 2005. Characterization of Salmonella enterica serovar Typhimurium and monophasic Salmonella serovar 1,4,[5],12:i:− isolates in Thailand. J. Clin. Microbiol. 43:2736-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, E. S., L. R. Ward, M. J. de Saxe, and J. D. H. de Sa. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. (Lond.) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 2006. Commission decision of 29 September 2006 concerning a financial contribution by the Community towards a baseline survey on the prevalence of Salmonella in slaughter pigs to be carried out in the Member States [notified under document number C(2006) 4306 (2006/668/EC)]. Official J. Eur. Union 2006:L275/51-L275/61. [Google Scholar]
- 4.Callow, B. R. 1959. A new phage-typing scheme for Salmonella typhimurium. J. Hyg. (Lond.) 57:346-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2007. Investigation of outbreak of human infections caused by Salmonella serotype I 4,[5],12:i:−. Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/salmonella/4512eyeminus.html.
- 6.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7, 7th ed., vol. 23. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Dauga, C., A. Zabrovskaia, and P. A. Grimont. 1998. Restriction fragment length polymorphism analysis of some flagellin genes of Salmonella enterica. J. Clin. Microbiol. 36:2835-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Torre, E., D. Zapata, M. Tello, W. Mejia, N. Frias, F. J. Garcia Pena, E. M. Mateu, and E. Torre. 2003. Several Salmonella enterica subsp. enterica serotype 4,5,12:i:− phage types isolated from swine samples originate from serotype Typhimurium DT U302. J. Clin. Microbiol. 41:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dionisi, A. M., C. Graziani, C. Lucarelli, E. Filetici, L. Villa, S. Owczarek, A. Caprioli, and I. Luzzi. 2009. Molecular characterization of multidrug-resistant strains of Salmonella enterica serotype Typhimurium and monophasic variant (S. 4,[5],12:i:−) isolated from human infections in Italy. Foodborne Pathog. Dis. 6:711-717. [DOI] [PubMed] [Google Scholar]
- 10.Echeita, M. A., A. Aladuena, S. Cruchaga, and M. A. Usera. 1999. Emergence and spread of an atypical Salmonella enterica subsp. enterica serotype 4,5,12:i:− strain in Spain. J. Clin. Microbiol. 37:3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Echeita, M. A., S. Herrera, and M. A. Usera. 2001. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:− appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 39:2981-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Food Safety Authority. 2008. Report of the Task Force on Zoonoses Data Collection on the analysis of the baseline survey on the prevalence of Salmonella in slaughter pigs, in the EU, 2006-2007. EFSA J. 135:1-111. [Google Scholar]
- 13.Garaizar, J., S. Porwollik, A. Echeita, A. Rementeria, S. Herrera, R. M. Wong, J. Frye, M. A. Usera, and M. McClelland. 2002. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40:2074-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimont, P. A., and F. X. Weill. 2007. Antigenic formulae of the Salmonella serovars, 9th ed. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
- 15.Huehn, S., C. Bunge, E. Junker, R. Helmuth, and B. Malorny. 2009. Poultry associated Salmonella enterica subsp. enterica serovar 4,12:d:− reveals high clonality and a distinct pathogenicity gene repertoire. Appl. Environ. Microbiol. 75:1011-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacterial. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsson, J. T., M. Torpdahl, R. F. Petersen, G. Sørensen, B. A. Lindsted, and E. M. Nielsen. 2009. Development of a new nomenclature for Salmonella Typhimurium multilocus variable number of tandem repeats analysis (MLVA). Euro Surveill. 14:pii19174. [PubMed] [Google Scholar]
- 17.Lim, Y. H., K. Hirose, H. Izumiya, E. Arakawa, H. Takahashi, J. Terajima, K. Itoh, K. Tamura, S. I. Kim, and H. Watanabe. 2003. Multiplex polymerase chain reaction assay for selective detection of Salmonella enterica serovar Typhimurium. Jpn. J. Infect. Dis. 56:151-155. [PubMed] [Google Scholar]
- 18.Lindstedt, B. A., T. Vardund, L. Aas, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59:163-172. [DOI] [PubMed] [Google Scholar]
- 19.Mossong, J., P. Marques, C. Ragimbeau, P. Huberty-Krau, S. Losch, G. Meyer, G. Moris, C. Strottner, W. Rabsch, and F. Schneider. 2007. Outbreaks of monophasic Salmonella enterica serovar 4,[5],12:i:− in Luxembourg, 2006. Euro Surveill. 12:E11-E12. [DOI] [PubMed] [Google Scholar]
- 20.Porwollik, S., E. F. Boyd, C. Choy, P. Cheng, L. Florea, E. Proctor, and M. McClelland. 2004. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J. Bacteriol. 186:5883-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 22.Schroeter, A., B. Hoog, and R. Helmuth. 2004. Resistance of Salmonella isolates in Germany. J. Vet. Med. B 51:389-392. [DOI] [PubMed] [Google Scholar]
- 23.Soyer, Y., S. A. Moreno, M. A. Davis, J. Maurer, P. L. McDonough, D. J. Schoonmaker-Bopp, N. B. Dumas, T. Root, L. D. Warnick, Y. T. Grohn, and M. Wiedmann. 2009. Salmonella enterica serotype 4,5,12:i:−, an emerging Salmonella serotype that represents multiple distinct clones. J. Clin. Microbiol. 47:3546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Switt, A. I., Y. Soyer, L. D. Warnick, and M. Wiedmann. 2009. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i:−. Foodborne Pathog. Dis. 6:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavechio, A. T., A. C. Ghilardi, and S. A. Fernand es. 2004. “Multiplex PCR” identification of the atypical and monophasic Salmonella enterica subsp. enterica serotype 1,4,[5],12:i:− in Sao Paulo State, Brazil: frequency and antibiotic resistance patterns. Rev. Inst. Med. Trop. Sao Paulo 46:115-117. [DOI] [PubMed] [Google Scholar]
- 26.Zamperini, K., V. Soni, D. Waltman, S. Sanchez, E. C. Theriault, J. Bray, and J. J. Maurer. 2007. Molecular characterization reveals Salmonella enterica serovar 4,[5],12:i:− from poultry is a variant Typhimurium serovar. Avian Dis. 51:958-964. [DOI] [PubMed] [Google Scholar]