Abstract
Endoglucanase C5614-1 comprises a catalytic module (CM) and an X module (XM). The XM showed no significant homology with known carbohydrate-binding modules (CBMs). Recombinant full-length endoglucanase could bind Avicel, whereas the CM could not. The XM could bind various polysaccharides. The results demonstrated that the XM was a new CBM.
Most cellulases are modular proteins that comprise two or more discrete modules, such as catalytic modules (CMs) and carbohydrate-binding modules (CBMs), each of which can function independently (9). CBMs are classified into 59 families based on their amino acid similarity in the CAZY database (http://www.cazy.org/fam/acc_CBM.html). The main functions of CBMs are to recognize and bind polysaccharides and to increase the hydrolytic activities of the enzymes against insoluble and soluble substrates (3). Endoglucanase C5614-1 (GenBank accession no. ACA61140), which was identified from the metagenome of the contents of buffalo rumen (5), is a modular enzyme comprising an N-terminal signal peptide (amino acids [aa] 1 to 20), a CM belonging to the glycosyl hydrolase family 5 (aa 40 to 334), and a C-terminal X module (XM) of unknown function (aa 335 to 537) (Fig. 1A). No linker region rich in Ser/Pro/Thr was found between the CM and the XM. In this study, we aimed to ascertain the function of the XM in the endoglucanase C5614-1.
FIG. 1.
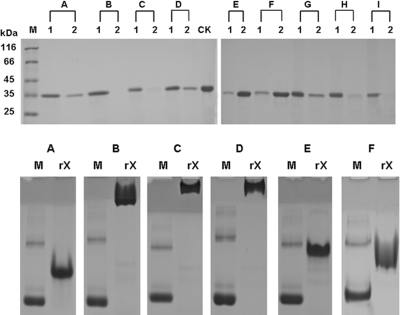
Endoglucanase C5614-1 and its derivatives. (A) Modular organization of C5614-1 and its truncated derivatives. Abbreviations: SP, signal peptide; GHF5, GHF5 catalytic module; X, the X module (XM). (B) SDS-PAGE of purified recombinant proteins. Protein samples were analyzed on a 10% gel. Lane 1, protein molecular mass standard (molecular masses are shown on the left); lane 2, rC5614-1; lane 3, rGHF5; lane 4, rX.
The XM showed no significant homology to known CBMs. The XM shared 25% to 33% identities and 41% to 45% similarities with about 200 amino acids at the C terminus of a xylanase (GenBank accession no. AAC36862) from the ruminal bacterium Prevotella ruminicola, uncultured ruminal microbial cellulases (ABX76045, ACA61132, ACA61135, ACA61137, and ABB46200), and an uncultured bacterial bifunctional mannanase-xyloglucanase (ADA62505). None of these homologous polypeptides was confirmed to show carbohydrate-binding activity. An alignment of XM with these homologous sequences using ClustalW (http://www.ebi.ac.uk/Tools/clustalw) is shown in Fig. 2. Seven conserved aromatic amino acid residues were found in the respective sequences. Aromatic amino acid residues in CBMs play critical roles in recognizing and binding polysaccharides (4).
FIG. 2.
Multiple sequence alignment of the XM (203 aa) in endoglucanase C5614-1 (aa 335 to 537), with its homologous sequences. The identical and similar amino acid residues are indicated by asterisks and dots, respectively, below the alignment. The conserved aromatic amino acid residues are indicated by arrows. GenBank accession no. AAC36862, Prevotella ruminicola xylanase (aa 376 to 584); ABB46200, ruminal uncultured bacterium endoglycosidase precursor protein (aa 719 to 917); ABX76045, ruminal uncultured microorganism endo-1,4-beta-d-glucanase (aa 342 to 552); ADA62505, ruminal uncultured bacterium bifunctional mannanase-xyloglucanase (aa 721 to 919); ACA61132, ruminal uncultured microorganism cellulase C29-2 (aa 341 to 553); ACA61135, ruminal uncultured microorganism cellulase C35-2 (aa 337 to 552); ACA61137, ruminal uncultured microorganism cellulase C67-1 (aa 335 to 546). The amino acid numbers in each of the parentheses above define the range of the homologous region in each sequence.
A PCR-based approach was used to produce constructs expressing C5614-1 derivatives (Fig. 1A). Plasmid C5614 (5), carrying the endoglucanase gene C5614-1, was used as a PCR template. The primer pairs used to amplify the portions of C5614-1 encoding C5614-1 amino acids 20 to 537 (recombinant C5614-1 [rC5614-1], comprising the CM and the XM), 20 to 348 (rGHF5, containing the CM only), and 349 to 537 (rX, containing the XM only) are, respectively, as follows: C5614-1F (5′CAGCCATGGAGGCACAAGATTTTGAGACTGCTACCGAA-3′) and C5614-1R (5′-GACCTCGAGTTGTGCTATGTATTTTTTGCCGTTCTGG-3′), C5614-1F and C5614-1CM (5′-GGGCTCGAGGGTTAATGTCTCAGCCAGGTCAGGCTG-3′), and C5614-1X (5′-GGGCCATGGCCAAAGCCTATCATGGCAGCGCGTTC-3′) and C5614-1R. The underlined sequences in the primers are NcoI and XhoI restriction sites.
The digested PCR products were ligated into the same digested expression vector, pET-30a(+) (Novagen, Madison, WI). The recombinant plasmids were transformed into Escherichia coli Rosetta(DE3)pLysS (Novagen), and the cloned fragments were expressed as proteins with 6×His tags at both the N and C termini. The recombinant C5614-1 derivatives were purified by affinity chromatography with Cobalt immobilized metal chromatography resin (Clontech, Palo Alto, CA), according to the user manual. Each of the purified proteins produced a single band on an SDS-PAGE gel (8), and their molecular sizes were in agreement with those of the deduced polypeptides (Fig. 1B).
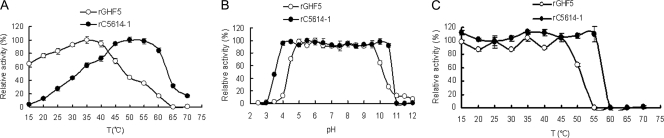
The hydrolytic activities of rC5614-1 and rGHF5 toward carboxymethyl cellulose (CMC) were determined essentially as described by Duan et al. (5). The pH profiles of the enzymatic reactions of rC5614-1 and rGHF5 were similar, and both showed maximum activities at pH 5.0 (data not shown). However, rC5614-1 and rGHF5 showed different temperature profiles (Fig. 3A). The rGHF5 showed narrower pH stability and lower temperature stability profiles than those of rC5614-1 (Fig. 3B and C). These results indicated that the XM is required for the stability of rC5614-1.
FIG. 3.
Effects of pH and temperature on the activities and stability of rC5614-1 and rGHF5. (A) Influence of temperature on the activities of rC5614-1 and rGHF5. Cellulase activity was measured at pH 5.0 in citrate-phosphate buffer at the indicated temperatures for 10 min. Values are expressed as percentages of maximal activity at 50°C and 35°C for rC5614-1 and rGHF5, respectively. (B) Influence of pH on rC5614-1 and rGHF5 stability. Purified enzyme was first incubated in citrate-phosphate buffer (pH 2.5 to 7.0), 0.1 M Tris-HCl buffer (pH 7.5 to 8.5), or 0.1 M glycine-NaOH buffer (pH 9.0 to 12) at 4°C for 24 h, and activity was measured under optimal conditions for 10 min. Values are expressed as percentages of maximal activity when the sample was incubated under pH 5.5 and pH 6.0 for rC5614-1 and rGHF5, respectively. (C) Thermal stability of recombinant rC5614-1 and rGHF5. Purified enzymes were first incubated at the indicated temperatures for 1 h; activity was then measured under optimal conditions for 10 min. Values are expressed as percentages of untreated-sample activity.
The hydrolytic activities of rC5614-1 toward particle substrates, including birch wood xylan, lichenan, and acid-swollen cellulose (ASC), were about three times greater than those of rGHF5; however, rC5614-1 and rGHF5 showed similar hydrolytic activities toward soluble substrates (Table 1). This result indicated that the presence of the XM enhanced the hydrolytic activity of the enzyme toward insoluble substrates but not toward soluble substrates. Similar phenomena were reported for Irpex lacteus exocellulase I (7), Clostridium thermocellum Xyn10C (1), and Clostridium stercorarium Xyn10B (2).
TABLE 1.
Specific activities of rC5614-1 and rGHF5 toward various substrates
| Test substrate | Sp act (U/mg protein)a |
|
|---|---|---|
| rC5614-1 | rGHF5 | |
| Barley glucan | 126.1 ± 4.5 | 319.8 ± 19.9 |
| Carboxymethyl cellulose | 72.5 ± 2.1 | 62.8 ± 4.4 |
| Lichenan | 57.2 ± 3.1 | 20.6 ± 0.8 |
| 2-Hydroxyethyl cellulose | 24.6 ± 0.8 | 20.0 ± 0.1 |
| Methyl cellulose | 8.8 ± 0.7 | 13.6 ± 0.03 |
| Birch wood xylan | 6.7 ± 0.7 | 1.9 ± 0.16 |
| Acid-swollen cellulose | 1.7 ± 0.05 | 0.5 ± 0.005 |
| p-Nitrophenyl-d-cellobioside | <0.005 | <0.002 |
| Avicel (β-1,4-glucan) | 0 | 0 |
| Laminarin | 0 | 0 |
| p-Nitrophenyl-d-glucopyranoside | 0 | 0 |
One unit of enzyme activity was defined as the amount of enzyme releasing 1 μmol of glucose equivalent or p-nitrophenol per min from substrates.
Binding of rC5614-1 and rGHF5 to Avicel was determined quantitatively, as described in the supplemental material. As shown in Table 2, rC5614-1 bound to Avicel, and the binding capability of rC5614-1 increased with increasing Avicel concentration and with prolonged incubation time. Its binding capability was also influenced by the pH of the mixture solution. Approximately 90% of rC5614-1 bound to 4% cellulose in the mixture solution at pH 5.0 after incubation for 5 h, whereas the proportion of the enzyme bound to cellulose dropped slightly to 83% at pH 4.0 and dropped significantly to less than 65% at pHs ≥6. Bovine serum albumin (BSA) in the mixture solution only slightly affected the binding of rC5614-1 to Avicel, suggesting that its binding to Avicel was specific. However, 100% of rGHF5 remained in the supernatant of the binding mixture, showing that rGHF5 could not bind Avicel. These results demonstrated that the XM is absolutely required for the binding of rC5614-1 to Avicel, suggesting that the XM is a CBM.
TABLE 2.
Adsorption properties of rC5614-1 to Avicel
| Additive(s) (pH)a | Binding to Avicel (%) after incubation for: |
|
|---|---|---|
| 1 hb | 5 h | |
| 1% Avicel | 25.0 ± 1.3 | 47.8 ± 1.8 |
| 2% Avicel | 38.1 ± 2.8 | 72.7 ± 1.2 |
| 4% Avicel | 55.3 ± 1.7 | 92.1 ± 1.2 |
| 8% Avicel | 65.1 ± 3.8 | 96.2 ± 1.2 |
| 4% Avicel + 0.08% BSA | ND | 79.8 ± 1.1 |
| 4% Avicel + 0.01% BSA | ND | 81.5 ± 1.9 |
| 4% Avicel (4.0) | ND | 83.6 ± 0.4 |
| 4% Avicel (6.0) | ND | 63.5 ± 3.2 |
| 4% Avicel (7.0) | ND | 62.0 ± 3.5 |
Unless otherwise stated, the mixture solution was at pH 5.0.
ND, not determined.
To further confirm the XM as a CBM, the binding of purified rX to insoluble polysaccharides was investigated by incubating the polypeptide with various polysaccharides and comparing the proteins in the supernatant fraction (unbound protein) and in the precipitated fraction (bound protein) by SDS-PAGE, as described in the supplemental material. rX could bind to the insoluble polysaccharides Avicel, ASC, chitin, lichenan, xylan from sugarcane bagasse, powder of sugarcane bagasse, and raw cassava starch. It bound slightly to agarose and Sephadex G-100 (Fig. 4, top). The affinities of rX for soluble polysaccharides were examined by native-affinity PAGE. The migration of rX was strongly retarded by inclusion of methylcellulose, 2-hydroxyethylcellulose, and barley glucan in gels, whereas it was only slightly affected by the presence of birch wood xylan and CMC (Fig. 4, bottom) and not affected by the inclusion of laminarin and soluble starch (data not shown). The affinities of the XM in C5614-1 to insoluble substrates were similar to those of CBM37 from Ruminococcus albus (10) and CBM54 from Clostridium thermocellum (6), which could also bind various insoluble substrates. However, the binding of CBM54 and CBM37 to soluble substrates was either negative or not tested. We propose that the X module in endoglucanase C5614-1 is a novel CBM.
FIG. 4.
Binding of rX to insoluble (top) and soluble (bottom) polysaccharides. In the experiment whose results are shown in the top panel, purified rX was incubated with insoluble polysaccharides, including Avicel (lanes A), ASC (lanes B), chitin (lanes C), lichenan (lanes D), agarose (lanes E), Sephadex G-100 (lanes F), xylan from sugarcane bagasse (lanes G), the powder of sugarcane bagasse (lanes H), and raw cassava starch (lanes I). CK is a control (the amount of protein used in the binding assay). After centrifugation, proteins in the precipitate (lanes 1) and the supernatant (lanes 2) were analyzed by SDS-PAGE. In the experiment whose results are shown in the bottom panel, purified rX and bovine serum albumin (BSA) were separated in nondenaturing polyacrylamide gels containing 0.1% (wt/vol) soluble polysaccharides, including methylcellulose (B), 2-hydroxyethylcellulose (C), barley glucan (D), birch wood xylan (E), and CMC (F). A gel without polysaccharide served as a reference (A). Lanes M contained BSA as a control.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30960094, 30960013), a grant from the Ministry of Science and Technology of China (2009DFA30700), a grant from the National Hi-Tech Research and Development Program of China (863 program 2007AA021307), and a grant from the Development Program of Guangxi Key Laboratory of Bioindustry Technology (07-109-001-2A).
Footnotes
Published ahead of print on 14 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ali, E., G. Zhao, M. Sakka, T. Kimura, K. Ohmiya, and K. Sakka. 2005. Functions of family-22 carbohydrate-binding module in Clostridium thermocellum Xyn10C. Biosci. Biotechnol. Biochem. 69:160-165. [DOI] [PubMed] [Google Scholar]
- 2.Ali, M. K., H. Hayashi, S. Karita, M. Goto, T. Kimura, K. Sakka, and K. Ohmiya. 2001. Importance of the carbohydrate-binding module of Clostridium stercorarium Xyn10B to xylan hydrolysis. Biosci. Biotechnol. Biochem. 65:41-47. [DOI] [PubMed] [Google Scholar]
- 3.Beguin, P., and J. P. Aubert. 1994. The biological degradation of cellulose. FEMS Microbiol. Rev. 13:25-58. [DOI] [PubMed] [Google Scholar]
- 4.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan, C. J., L. Xian, G. C. Zhao, Y. Feng, H. Pang, X. L. Bai, J. L. Tang, Q. S. Ma, and J. X. Feng. 2009. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J. Appl. Microbiol. 107:245-256. [DOI] [PubMed] [Google Scholar]
- 6.Dvortsov, I. A., N. A. Lunina, L. A. Chekanovskaya, W. H. Schwarz, V. V. Zverlov, and G. A. Velikodvorskaya. 2009. Carbohydrate-binding properties of a separately folding protein module from beta-1,3-glucanase Lic16A of Clostridium thermocellum. Microbiology 155:2442-2449. [DOI] [PubMed] [Google Scholar]
- 7.Hamada, N., R. Kodaira, M. Nogawa, K. Shinji, R. Ito, Y. Amano, M. Shimosaka, T. Kanda, and M. Okazaki. 2001. Role of cellulose-binding domain of exocellulase I from white rot basidiomycete Irpex lacteus. J. Biosci. Bioeng. 91:359-362. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 9.Shoseyov, O., Z. Shani, and I. Levy. 2006. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 70:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu, Q., M. Morrison, K. E. Nelson, E. A. Bayer, N. Atamna, and R. Lamed. 2004. A novel family of carbohydrate-binding modules identified with Ruminococcus albus proteins. FEBS Lett. 566:11-16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.