Abstract
Sequencing of plasmid pLM33 from the food isolate Listeria monocytogenes Lm1 revealed a molecule of 32,307 bp with a G+C content of 36.2%. The plasmid displays a mosaic pattern of identities common to several closely related L. monocytogenes plasmids isolated from food and clinical sources.
Listeria monocytogenes is a Gram-positive opportunistic pathogen which has received an increasing level of interest in the last few decades due to the severity of the food-borne disease in immunocompromised individuals, mainly characterized by meningitis, encephalitis, and materno-fetal and perinatal infections (3). Dairy products are one of the main vehicles for infection of humans, and they have been responsible for several outbreaks and sporadic cases of listeriosis in Europe and North America (21). In a previous work, we reported a high incidence of L. monocytogenes in artisanal soft-ripened cheeses in Asturias, Spain (18). The molecular typing of 19 strains of L. monocytogenes serogroup 1, isolated from the same type of cheese (Afuega'l Pitu) at different times of the year, revealed the presence of a plasmid of about 33 kb in all the isolates, named pLM33. This plasmid shares large homology regions with other Listeria innocua plasmids isolated from different soft-ripened cheeses (17, 19). In this work, we went deeply into the genetic content of pLM33 and fully sequenced the plasmid, and we performed a bioinformatic analysis of the sequence that has allowed us to conclude that this plasmid or parts of it are widely distributed among different L. monocytogenes strains.
For plasmid sequencing, L. monocytogenes Lm1 (19) was grown overnight in 100 ml of tryptic soy broth (TSB; Scharlau, Barcelona, Spain). A plasmid extraction was carried out with the Qiagen plasmid MidiKit (Qiagen, Inc., Valencia, CA), using a hand-made resuspension buffer containing 20% sucrose, 10 mM Tris-HCl, 10 mM EDTA, 50 mM NaCl, 80 kU/liter mutanolysin, and 1 g/liter lysozyme. All of the other steps were carried out following the manufacturer's instructions. The plasmid was digested with BamHI, and the fragments were cloned in vector pUC19. A BamHI fragment of 6.8 kb cloned in pUC19 (corresponding to the sequence from positions 8796 to 15566 in pLM33) was sequenced, and due to the high homology of this fragment with the plasmids of L. monocytogenes strains FLS J1-194, FSL R2-503, and FSL N1-017 (http://www.broad.mit.edu/annotation/genome/listeria_group), sequence walking on the plasmid (direct sequencing of the plasmid DNA) allowed the sequencing of the full-length plasmid.
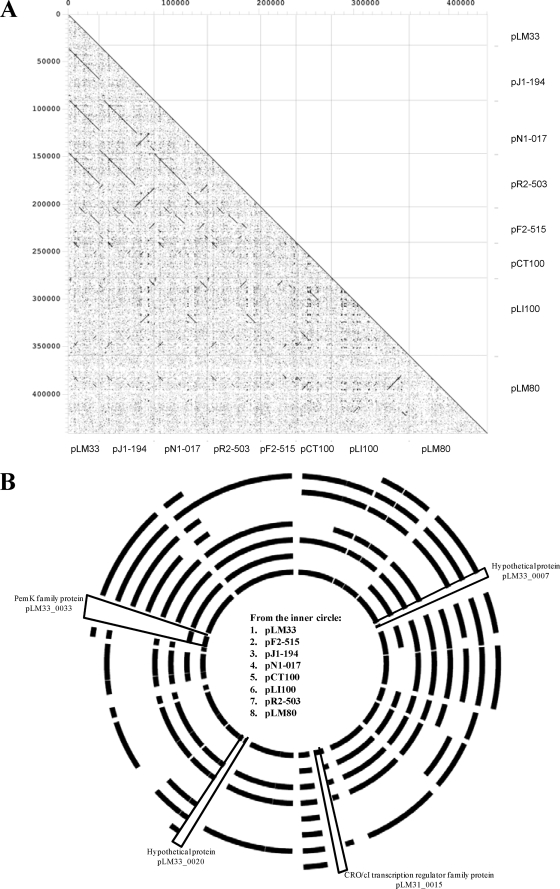
Dot plot comparisons at DNA level gave us an overview of the degree of similarity of pLM33 with respect to the other Listeria plasmids (Fig. 1A). Table 1 includes all of the plasmids used in this report and their main features. In order to view the synteny of the pLM33 genome, its origin region was taken as the corresponding origin region of the other Listeria plasmids. Dot plot analysis showed a close similarity of the genome sequence of pLM33 with the genomes of the plasmids from strains FSL J1-194, FSL N1-017, FSL R2-503, and FSL F2-515 (pJ1-194, pN1-017, pR2-503, and pF2-515, respectively). Sets of contigs corresponding to the DNA sequences of these four plasmids were downloaded in December 2008 from the Listeria monocytogenes database of the Broad Institute (http://www.broad.mit.edu/annotation/genome/listeria_group) (pR2-503, supercontigs of Listeria monocytogenes FSL R2-503 plasmids 2.52, 2.53, and 2.54; pJ1-194, supercontig of Listeria monocytogenes FSL J1-194 plasmid 2.44; pN1-017, supercontigs of Listeria monocytogenes FSL N1-017 plasmids 2.75, 2.76, and 2.77; and pF2-515, supercontigs of Listeria monocytogenes FSL F2-515 plasmids 2.1405, 2.1406, 2.1407, 2.1408, 2.1409, 2.1410, 2.1411, 2.1412, 2.1413, 2.1414, and 2.1415). The contigs were assembled and annotated by us, and the results are shown in the supplemental material. On the other hand, pCT100, pLI100, and pLM80 (accession no. U15554, NC_003383, and AADR01000010/AADR01000058, respectively; and see the supplemental material) displayed only a few homologous regions among them and with the other plasmids. Dot plot analysis also provided evidence that the pLM33 genome sequence is mostly present in pJl-194, pN1-017, and pR2-503, without any large deletion or insertion or, even, any rearrangement. These three plasmid sequences share a region of 31,735 nucleotides (98.23% of pLM33 genome size), which represents an overall identity among them and pLM33 higher than 99.97%; a similar region, more than one-half of its genome sequence, was found in pF2-515, although with some insertion/deletion regions. The high level of similarity among these four plasmids (pLM33, pJ1-194, pN1-017, and pR2-503) supports the idea of a close relationship among and recent origin of this group of plasmids. Finally, the genomic region shared by pN1-017 and pR2-503, but not with pLM33, contains a large inverted region. Furthermore, comparative genomic analyses between the pLM33-encoded predicted proteome and those of the other seven plasmids revealed pLM33-specific sequences encompassing hypothetical encoded proteins or transcriptional regulators (Fig. 1B). Each of the seven plasmid proteomes was compared with the pLM33 predicted proteome using BLAST (1). Proteins of pLM33 have been used as a database, and a Blastp search has been performed for every one of the seven sets of plasmid proteins (cutoff, E value of 1 × 10−6 and 30% identity over at least 80% of both protein sequences).
FIG. 1.
Comparative analyses of Listeria plasmid genomes. Panel A shows the dot plot matrix calculated for the complete sequences of pLM33, pJ1-194, pN1-017, pR2-503, pF2-515, pCT100, pLI100, and pLM80. Panel B depicts a circular plot of genome diversity in Listeria plasmids. Each ring indicates a Listeria plasmid; the order is shown within the inner circle of the figure.
TABLE 1.
Plasmids analyzed in this study
| Plasmid | No. of bases | No. of predicted ORFs | Accession no. (original DNA sequence source) | Host strain, origin (serotype) | Reference(s) |
|---|---|---|---|---|---|
| pLM33 | 32,307 | 37 | GU244485 | L. monocytogenes Lm1, cheese (1/2b) | 19 |
| pJ1-194 | 57,536 | 76 | NZ_AARJ00000000 (www.broad.mit.edu) | L. monocytogenes FSL J1-194, human (1/2b) | 6 |
| pR2-503 | 56,540 | 99 | NZ_AARR00000000 (www.broad.mit.edu) | L. monocytogenes FSL R2-503, human (1/2b) | 6 |
| pN1-017 | 56,037 | 65 | NZ_AARP00000000 (www.broad.mit.edu) | L. monocytogenes FSL N1-017, trout (4b) | 6 |
| pF2-515 | 37,163 | 71 | NZ_AARI00000000 (www.broad.mit.edu) | L. monocytogenes FSL F2-515, meat (1/2a) | 6, 23 |
| pCT100 | 37,279 | 43 | U15554 | L. monocytogenes DRDC8, milk (4) | 4 |
| pLI100 | 81,905 | 86 | NC_003383 | L. innocua CLIP 11262, cheese (6a) | 5 |
| pLM80 | 81,588 | 96 | AADR01000010/AADR01000058 | L. monocytogenes H7858, meat (4b) | 22 |
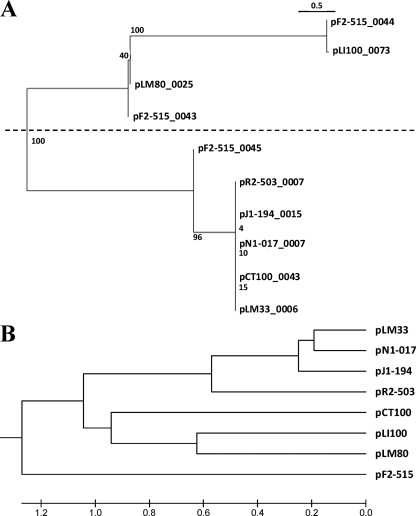
Due to the mobile nature of plasmids and the lack of the most common molecular markers for phylogeny, reconstruction of the phylogenetic history of plasmids in Listeria proved to be a difficult task to achieve. However, further information regarding their relationships can be inferred from the sequence analysis of the replication proteins. Phylogenetic analysis of replication proteins present in Listeria plasmids, using maximum likelihood, supports the idea of two main plasmid groups (Fig. 2A). Nevertheless, the position of pF2-515 is not clear, since it contains three putative replication genes, while the other plasmids contain just one. Furthermore, despite the close relationship of pR2-503, pJ1-194, pN1-107, pCT100, and pLM33 in the tree, dot plot comparisons of their DNA sequences contradict the presence of pCT100 in this group. Sequence data comparison of the replication gene sequences alone is not informative enough to solve this discrepancy, and neither is the position of pF2-515. A tree based on the gene content of these eight plasmids (Fig. 2B) provides further information about the degree of relatedness of these plasmid sequences. In addition, a comparison matrix (Table 2) was built that compares the proteomes of Listeria plasmids at the protein level. Comparisons were done by counting reciprocal hits sharing more than 30% sequence identity using Blastp over more than 80% of total sequence length, so that sets of pairwise hits were obtained. Average values of the sequence identity based on the pairwise comparisons were also obtained. Relationships observed in this analysis are in agreement with the dot plot matrix: pLM33 is much closer to pN1-017, pJl-194, and pR2-503 than to pCT100, pLI100, and pLM80. The position of pF2-215 on the unweighted-pair group method using average linkages (UPGMA) tree based on pairwise protein comparisons as an independent branch might contradict the sequence relationships shown in Fig. 1A and 3B, despite the observation of high-identity values of shared proteins with other plasmids closely related to pLM33 (Table 2). A deeper analysis of the region of pF2-515 homologous to the pLM33 group reveals frameshift and nonsense mutations which produce shorter predicted genes (data not shown). These changes, perhaps due to higher selective pressures suffered by this plasmid sequence, reduce the length of many proteins in pF2-515, which in turn reduces dramatically the number of orthologs obtained accordingly. This would explain the external branch position of pF2-515 in the UPGMA tree with respect to the other plasmids.
FIG. 2.
Phylogenetic relationship analysis of Listeria plasmids. The trees were constructed by maximum likelihood (program PHYML, bootstrap 100, HKY +G model) using DNA sequences of putative replication proteins in the plasmids (A) and UPGMA of pairwise distances calculated as the logarithm of shared orthologs (cutoffs, 30% identity, E value of 1 × 10−6, and a match of over 80% of both proteins) between two plasmid genomes, normalized by the weighted mean of the total number of plasmid genes (12) (B). The tree was built using MEGA4 software (28).
TABLE 2.
Average protein identities and number of shared proteins
| Protein | No. of shared proteins or % identitya |
|||||||
|---|---|---|---|---|---|---|---|---|
| pLM33 | pN1-107 | pR2-503 | pF2-515 | pJ1-194 | pCT100 | pLI100 | pLM80 | |
| pLM33 | 99.93 | 95.35 | 98.54 | 99.99 | 82.59 | 78.20 | 59.68 | |
| pN1-107 | 31 | 95.57 | 97.25 | 99.81 | 68.14 | 86.24 | 55.22 | |
| pR2-503 | 14 | 25 | 97.25 | 96.94 | 60.78 | 80.88 | 55.08 | |
| pF2-515 | 4 | 6 | 8 | 96.35 | 77.12 | 83.86 | 76.98 | |
| pJ1-194 | 27 | 45 | 30 | 10 | 64.57 | 89.69 | 54.92 | |
| pCT100 | 9 | 11 | 2 | 2 | 8 | 89.33 | 97.95 | |
| pLI100 | 7 | 17 | 7 | 3 | 15 | 10 | 91.01 | |
| pLM80 | 8 | 10 | 4 | 2 | 10 | 7 | 26 | |
Pairwise protein comparisons are shown. Numbers of shared proteins are shown in the lower triangle, and corresponding protein identity averages (expressed in percentage) are shown in the upper triangle. Pairwise protein comparisons were calculated using Blastp with cutoff values represented by an E value of 1 × 10−6 and sequence identity of 30% over 80% of both protein sequences.
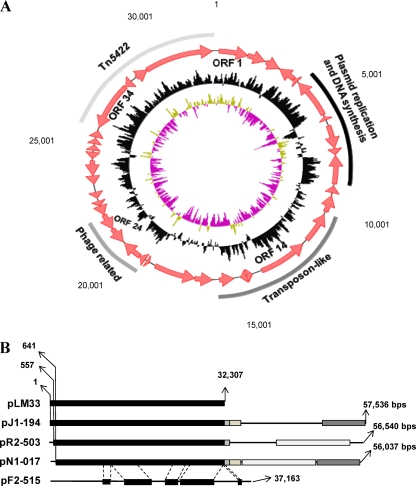
FIG. 3.
Genetic structure of pLM33 (A) and its homology with sequences of other L. monocytogenes plasmids (B). In panel A, the black circle represents the GC content and the green and purple circles represent the GC skew. In panel B, the same filling patterns indicate homologous sequences with an identity of >98%.
pLM33 contains 37 putative open reading frames (ORFs) (Table 3). It is worth noting that 8 ORFs code for putative proteins involved in genetic mobilization events (transposases or transposase-like proteins, integrases, and resolvases), suggesting that this plasmid could have undergone insertion and/or deletion events during evolution. Nowadays, only scarce genetic information is available for Listeria plasmids. Plasmid profiling has been used for L. monocytogenes typing (7, 9, 10, 19, 20, 24), and the presence of plasmids has mainly been linked to resistance phenotypes for cadmium (13, 14), antibiotics (2, 15, 25, 26), and biocides (26). pLM33 contains the transposon Tn5422, a transposon of the Tn3 family that contains a cadmium efflux pump, a transposase, and a resolvase (13). Lebrun and coworkers suggested that the transposition of Tn5422 could be one of the reasons for the size diversity of L. monocytogenes plasmids (13). Apart from Tn5422, pLM33 seems to be structured in different blocks. ORFs 22, 23, and 24 seem to represent the outcome of a prophage integration event. ORF 24 codes for a putative Cas (CRISPR-associated) protein, which allows, in conjunction with CRISPR (clustered regularly interspaced short palindromic repeats) sequences, a microbial population to survive phage predation and provides a historical perspective on phage exposure (8). ORF 23 is a homolog of the major prohead scaffolding core protein of Listeria phage A006 (16) and ORF 22 codes for a putative catalytic region of a viral integrase (11). On the other hand, pLM33 seems to have acquired a Clp protein from some lactic acid bacteria, since ORF 14 displayed a high homology with Clp proteases from Enterococcus, Lactobacillus, and Lactococcus. These proteins are involved in the response to several environmental stresses, such as heat shock, in some Lactobacillus species. Interestingly, a homolog of ORF 14 was found to be surrounded by transposase genes in Lactobacillus rhamnosus and was shown to be mobilized during prolonged cultivation at elevated temperature (27). This suggests that ClpL was acquired via horizontal gene transfer. Finally, another module is constituted by ORFs 6 to 10, which code for proteins involved in plasmid replication and DNA synthesis (Fig. 3A).
TABLE 3.
Putative open reading frames present in pLM33 and their putative functions
| CDSa | % GC content | No. of amino acids | BLAST-based putative function (E value) |
|---|---|---|---|
| pLM33_0001 | 36.35 | 408 | Transposase element IS712A, Listeria grayi DSM 20601 (0; 100% identity) |
| pLM33_0002 | 37.02 | 252 | Putative transposase helper protein for IS712A, Lactococcus lactis subsp. cremoris MG1363 (3E−79; 98.0159% identity) |
| pLM33_0003 | 35.49 | 200 | Transposase, Listeria monocytogenes FSL J1-194 (1E−111; 100% identity) |
| pLM33_0004 | 41.67 | 251 | Hypothetical protein |
| pLM33_0005 | 38.79 | 54 | Hypothetical protein |
| pLM33_0006 | 42.59 | 586 | Plasmid replication protein RepR, Listeria grayi DSM 20601 (0; 94.0199% identity) |
| pLM33_0007 | 28.93 | 52 | Hypothetical protein |
| pLM33_0008 | 30.09 | 348 | Rep63B, Listeria monocytogenes FSL J1-194 (0; 100% identity) |
| pLM33_0009 | 29.63 | 98 | Hypothetical protein |
| pLM33_0010 | 42.15 | 426 | Possible DNA-directed DNA polymerase, Listeria grayi DSM 20601 (0; 100% identity) |
| pLM33_0011 | 41.16 | 114 | Hypothetical protein |
| pLM33_0012 | 32.55 | 254 | Hypothetical protein |
| pLM33_0013 | 35.11 | 449 | Putative transposase for insertion sequence element IS3 family protein, Listeria monocytogenes FSL J (0; 100% identity) |
| pLM33_0014 | 40.05 | 704 | ATP-dependent Clp protease, ATP-binding subunit ClpL, Listeria monocytogenes FSL J1-194 (0; 100% identity) |
| pLM33_0015 | 34.6 | 78 | Cro/CI family phage transcriptional regulator, Listeria monocytogenes FSL J1-194 (1E−37; 100% identity) |
| pLM33_0016 | 36.92 | 92 | Hypothetical protein |
| pLM33_0017 | 38.42 | 202 | Putative resolvase, N-terminal domain protein, Listeria monocytogenes FSL J1-194 (1E−104; 93.9698% identity) |
| pLM33_0018 | 29.82 | 246 | Hypothetical protein |
| pLM33_0019 | 29.32 | 588 | Hypothetical protein |
| pLM33_0020 | 36.3 | 44 | Hypothetical protein |
| pLM33_0021 | 40.78 | 84 | Hypothetical protein |
| pLM33_0022 | 38.05 | 289 | Integrase catalytic region, Listeria monocytogenes FSL J1-194 (1E−170; 100% identity) |
| pLM33_0023 | 33.61 | 238 | Hypothetical protein |
| pLM33_0024 | 33.75 | 240 | CRISPR-associated Cas5 family protein, Listeria monocytogenes FSL J1-194 (1E−128; 100% identity) |
| pLM33_0025 | 34.74 | 141 | Hypothetical protein |
| pLM33_0026 | 32.89 | 75 | Hypothetical protein |
| pLM33_0027 | 35.84 | 92 | Hypothetical protein |
| pLM33_0028 | 38.62 | 125 | Hypothetical protein |
| pLM33_0029 | 39.97 | 220 | Hypothetical protein |
| pLM33_0030 | 37.92 | 108 | Hypothetical protein |
| pLM33_0031 | 34.6 | 78 | Hypothetical protein |
| pLM33_0032 | 41.15 | 80 | Putative PemI-like inhibitor, Listeria monocytogenes FSL J1-194 (5E−39; 100% identity) |
| pLM33_0033 | 41.23 | 113 | Putative PemK-like protein, Listeria monocytogenes FSL J1-194 (8E−47; 100% identity) |
| pLM33_0034 | 39.98 | 711 | Cadmium resistance protein B, Listeria monocytogenes FSL J1-194 (0; 100% identity) |
| pLM33_0035 | 35.28 | 119 | Putative cadmium efflux system accessory protein, Bacillus sp. strain NRRL B-14911 (4E−29; 92.562% identity) |
| pLM33_0036 | 38.02 | 184 | Resolvase, Listeria monocytogenes FSL J1-194 (1E−102; 100% identity) |
| pLM33_0037 | 36.83 | 971 | Transposase, Listeria monocytogenes FSL J1-194 (0; 100% identity) |
Database coding sequence designation.
It is highly probable that the large amount of transposases present in this plasmid could be responsible for DNA sequence rearrangements (i.e., insertions and deletions), leading to the mosaic-like structure of pLM33. The full plasmid is contained in other larger plasmids (Fig. 3B), suggesting that pLM33 could act as a stable structural block. In relation to this, one of the most striking findings of this work is that pLM33 is fully contained within the plasmid pJ1-194 from strain FSL J1-194 (match of >99.8%), a serotype 1/2b strain isolated from a sporadic case of human listeriosis in the United States (6) (Fig. 3B). Furthermore, almost the complete plasmid pLM33 was contained within the plasmids of strains FSL R2-503 (match of >99.8% starting at position 557 of pLM33) and FSL N1-017 (match of >99.9% starting at position 641 of pLM33). FSL R2-503 is a serotype 1/2b strain responsible for a gastrointestinal listeriosis outbreak in the United States in 1994, and FSL N1-017 is a serotype 4b strain with a ribotype that was exclusively associated with foods and not with any human disease cases (6). On the other hand, pLM33 displays several homology zones with plasmid pF2-515 of strain FSL F2-515, a serotype 1/2a1 strain bearing ribotype DUP-1062A, one of the most common subtypes found among food isolates and rarely causing human disease (6). These findings indicate that pLM33 is widely distributed among L. monocytogenes isolates from different geographic areas in Europe and North America, alone or as part of larger plasmids. The findings also suggest that pLM33 could act as a single mobilizable unit. In this respect, it is worthwhile mentioning that some authors have pointed out the presence of similar DNA electrophoretic patterns shared by plasmids of different L. monocytogenes strains (14), as well as common DNA/DNA hybridization bands shared by L. monocytogenes and L. innocua plasmids (17), suggesting the presence of common homology zones among the different plasmids.
In summary, we have fully sequenced plasmid pLM33 of an L. monocytogenes strain isolated from a soft-ripened cheese and have shown that this plasmid, alone or as part of larger DNA molecules, is present in several L. monocytogenes isolates of either food or human origin. This suggests a wide distribution of this plasmid and indicates that pLM33 can be mobilized as a single genetic unit.
Nucleotide sequence accession number.
The sequence of the full-length pLM33 plasmid has been submitted to GenBank under accession no. GU244485.
Supplementary Material
Footnotes
Published ahead of print on 28 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Charpentier, E., G. Gerbaud, and P. Courvalin. 1999. Conjugative mobilization of the rolling-circle plasmid pIP823 from Listeria monocytogenes BM4293 among gram-positive and gram-negative bacteria. J. Bacteriol. 181:3368-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cossart, P., and A. Toledo-Arana. 2008. Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10:1041-1050. [DOI] [PubMed] [Google Scholar]
- 4.Francis, M. S., and C. J. Thomas. 1997. The Listeria monocytogenes gene ctpA encodes a putative P-type ATPase involved in copper transport. Mol. Gen. Genet. 253:484-491. [DOI] [PubMed] [Google Scholar]
- 5.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. García-del Portillo, P. Garrido, W. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 6.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey, J., and A. Gilmour. 2001. Characterization of recurrent and sporadic Listeria monocytogenes isolates from raw milk and nondairy foods by pulsed-field gel electrophoresis, monocin typing, plasmid profiling, and cadmium and antibiotic resistance determination. Appl. Environ. Microbiol. 67:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horvath, P., A. C. Coûté-Monvoisin, D. A. Romero, P. Boyaval, C. Fremaux, and R. Barrangou. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 131:62-70. [DOI] [PubMed] [Google Scholar]
- 9.Kolstad, J., D. A. Caugant, and L. M. Rørvik. 1992. Differentiation of Listeria monocytogenes isolates by using plasmid profiling and multilocus enzyme electrophoresis. Int. J. Food Microbiol. 16:247-260. [DOI] [PubMed] [Google Scholar]
- 10.Kolstad, J., L. M. Rørvik, and P. E. Granum. 1991. Characterization of plasmids from Listeria sp. Int. J. Food Microbiol. 12:123-131. [DOI] [PubMed] [Google Scholar]
- 11.Konsavage, W. M., Jr., M. Sudol, and M. Katzman. 2008. Effects of varying the spacing within the D, D-35-E motif in the catalytic region of retroviral integrase. Virology 379:223-233. [DOI] [PubMed] [Google Scholar]
- 12.Korbel, J. O., B. Snel, M. A. Huynen, and P. Bork. 2002. SHOT: a web server for the construction of genome phylogenies. Trends Genet. 18:158-162. [DOI] [PubMed] [Google Scholar]
- 13.Lebrun, M., A. Audurier, and P. Cossart. 1994. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917. J. Bacteriol. 176:3049-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebrun, M., J. Loulergue, E. Chaslus-Dancla, and A. Audurier. 1992. Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl. Environ. Microbiol. 58:3183-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemaître, J. P., H. Echchannaoui, G. Michaut, C. Divies, and A. Rousset. 1998. Plasmid-mediated resistance to antimicrobial agents among listeriae. J. Food Prot. 61:1459-1464. [DOI] [PubMed] [Google Scholar]
- 16.Loessner, M. J., I. B. Krause, T. Henle, and S. Scherer. 1994. Structural proteins and DNA characteristics of 14 Listeria typing bacteriophages. J. Gen. Virol. 75:701-710. [DOI] [PubMed] [Google Scholar]
- 17.Margolles, A., and C. G. de los Reyes-Gavilán. 1998. Characterization of plasmids from Listeria monocytogenes and Listeria innocua strains isolated from short-ripened cheeses. Int. J. Food Microbiol. 39:231-236. [DOI] [PubMed] [Google Scholar]
- 18.Margolles, A., A. Rodríguez, and C. G. de los Reyes-Gavilán. 1996. Some chemical and bacteriological characteristics of regional cheeses from Asturias, Spain. J. Food Prot. 59:509-515. [DOI] [PubMed] [Google Scholar]
- 19.Margolles, A., B. Mayo, and C. G. de los Reyes-Gavilán. 1998. Polymorphism of Listeria monocytogenes and Listeria innocua strains isolated from short-ripened cheeses. J. Appl. Microbiol. 84:255-262. [DOI] [PubMed] [Google Scholar]
- 20.McLauchlin, J., M. D. Hampton, S. Shah, E. J. Threlfall, A. A. Wieneke, and G. D. Curtis. 1997. Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J. Appl. Microbiol. 83:381-388. [DOI] [PubMed] [Google Scholar]
- 21.McLauchlin, J., R. T. Mitchell, W. J. Smerdon, and K. Jewell. 2004. Listeria monocytogenes and listeriosis: a review of hazard characterisation for use in microbiological risk assessment of foods. Int. J. Food Microbiol. 92:15-33. [DOI] [PubMed] [Google Scholar]
- 22.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orsi, R. H., Q. Sun, and M. Wiedmann. 2008. Genome-wide analyses reveal lineage specific contributions of positive selection and recombination to the evolution of Listeria monocytogenes. BMC Evol. Biol. 12:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterkin, P. I., M. A. Gardiner, N. Malik, and E. S. Idziak. 1992. Plasmids in Listeria monocytogenes and other Listeria species. Can. J. Microbiol. 38:161-164. [DOI] [PubMed] [Google Scholar]
- 25.Poyart-Salmeron, C., C. Carlier, P. Trieu-Cuot, A. L. Courtieu, and P. Courvalin. 1990. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 335:1422-1426. [DOI] [PubMed] [Google Scholar]
- 26.Romanova, N., S. Favrin, and M. W. Griffiths. 2002. Sensitivity of Listeria monocytogenes to sanitizers used in the meat processing industry. Appl. Environ. Microbiol. 68:6405-6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suokko, A., K. Savijoki, E. Malinen, A. Palva, and P. Varmanen. 2005. Characterization of a mobile clpL gene from Lactobacillus rhamnosus. Appl. Environ. Microbiol. 71:2061-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.