Abstract
In acidic forest soils, availability of inorganic nutrients is a tree-growth-limiting factor. A hypothesis to explain sustainable forest development proposes that tree roots select soil microbes involved in central biogeochemical processes, such as mineral weathering, that may contribute to nutrient mobilization and tree nutrition. Here we showed, by combining soil analyses with cultivation-dependent analyses of the culturable bacterial communities associated with the widespread mycorrhizal fungus Scleroderma citrinum, a significant enrichment of bacterial isolates with efficient mineral weathering potentials around the oak and beech mycorrhizal roots compared to bulk soil. Such a difference did not exist in the rhizosphere of Norway spruce. The mineral weathering ability of the bacterial isolates was assessed using a microplaque assay that measures the pH and the amount of iron released from biotite. Using this microplate assay, we demonstrated that the bacterial isolates harboring the most efficient mineral weathering potential belonged to the Burkholderia genus. Notably, previous work revealed that oak and beech harbored very similar pHs in the 5- to 10-cm horizon in both rhizosphere and bulk soil environments. In the spruce rhizosphere, in contrast, the pH was significantly lower than that in bulk soil. Because the production of protons is one of the main mechanisms responsible for mineral weathering, our results suggest that certain tree species have developed indirect strategies for mineral weathering in nutrient-poor soils, which lie in the selection of bacterial communities with efficient mineral weathering potentials.
The mobilization of nutrients via the biotic and abiotic weathering of soil minerals is crucial to satisfying plant nutritional needs (2, 17), especially in acidic forest soils, which are mainly nonfertilized and nutrient poor. Besides the physicochemical weathering reactions, evidence is presently accumulating which indicates that certain soil bacterial strains increase mineral weathering and improve tree nutrition (5, 9, 32, 39-41).
By way of their root exudates, plants alter the structure and activity of microbial communities (6, 25, 51) and selectively favor certain ones that are potentially beneficial to them (15, 16, 21, 45, 46). A hypothesis for sustainable forest development proposes that tree roots select from the soil efficient mineral weathering bacterial communities that may contribute to nutrient mobilization and tree growth (20). In this manner, recent studies (10, 46) have revealed that the oak-Scleroderma citrinum ectomycorrhizal symbiosis selects bacterial communities that are more efficient in mineral weathering than those of the surrounding soil, suggesting that the mycorrhizal symbiosis has an indirect effect on plant nutrition through its selective pressure on the functional diversity of the mycorrhizosphere bacterial communities.
Distinct impacts of the tree species on the soil bacterial community structure have been previously reported (23, 38), suggesting that the composition and activity of soil bacterial communities depend on tree physiology and notably on its impact on the soil physicochemical properties and nutrient cycling (24, 26, 37). However, no study has ever addressed the question of the impact of tree species on the structure of forest soil bacterial communities involved in mineral weathering. This question regarding the impact of tree species on the functional diversity of the bacterial communities remains a major issue in forestry, especially in the context of today's climate change, which will give rise to a shift in the spatial distribution of forest tree species.
To appreciate the effect of tree species on mycorrhizosphere bacterial communities, we focused on a single but ubiquitous mycorrhizal fungus, S. citrinum, which forms mycorrhizae with different tree species. Since no functional genes have been identified to date, a cultivation-dependent analysis was developed in this study. A total of 155 bacterial isolates were randomly chosen among a collection of 400 bacterial isolates from the soil-Scleroderma citrinum mycorrhiza interface (ectomycorrhizosphere), the extramatrical mycelium (hyphosphere), and the surrounding soil (bulk soil) in 28-year-old stands of oak (Quercus sessiliflora Smith), beech (Fagus sylvatica L.), and Norway spruce (Picea abies Karst.). The mineral weathering potential of each bacterial isolate was evaluated by way of an in vitro microplate assay, putting in interaction a calibrated bacterial suspension and the biotite, a mineral widespread in soils (46). The bacterial isolates were genotypically characterized by amplifying and sequencing a portion of the 16S rRNA gene. Their mineral weathering efficiencies and the functional structure of the bacterial communities were compared with the physicochemical characteristics of the surrounding soil.
MATERIALS AND METHODS
Experimental site and soil properties.
The samples were collected at the Breuil-Chenue experimental forest site, established in 1976, in the Morvan (47°18′N, 4°5′E; France). The forest is situated on a plateau at an altitude of 638 m, on a slight, northwest-facing slope. The mean annual air temperature is 9°C, and the annual precipitation averages 1,280 mm year−1. The native forest was clear felled and replaced by monospecific plantations distributed in plots of 0.1 ha of different species, such as oak (Quercus sessiliflora Smith), beech (Fagus sylvatica L.), and Norway spruce (Picea abies Karst.). The soil of the native forest is a “Typic dystrochrept” (49) developed on the “Pierre qui Vire” granite (42). It has a sandy-loam texture (60% sands and less than 20% clays) and is acidic (Table 1). The cation exchange capacity and the base cation saturation are very low and decrease with depth (Table 1). This soil is very sensitive to acidification and nutrient deficiencies. The rhizosphere soil is significantly more acidic than the bulk soil for the Norway spruce, while no pH difference is observed between the bulk and the rhizosphere soils for oak stand (Table 2) (11, 38) and for beech stand (data not shown).
TABLE 1.
Particle size distribution, pH, organic C, and exchangeable cations of the soil profiles of the native forest site of Breuil, France
| Depth (cm) | pHa | % clay | % silt | % sand | CECb (cmolc kg−1) | BSc (%) |
|---|---|---|---|---|---|---|
| 0-8 | 4.3 | 16.0 | 15.0 | 69.0 | 12.2 | 7.7 |
| 10-18 | 4.6 | 13.0 | 26.0 | 61.0 | 10.5 | 5 |
| 20-28 | 4.7 | 14.0 | 24.0 | 62.0 | 8.9 | 3.5 |
Soil pH was measured in H20 with a soil-to-solution ratio of 1:2.
Cation exchange capacity (CEC) = Mg2+ + Ca2+ + Na+ + Fe3+ + Mn2+ + Ba2+ + H+ + Al. cmolc, cmol of charge.
Base saturation (BS) = (Mg2+ + Ca2+ + Na+ + Fe3+ + Mn2+ + Ba2+)/CEC.
TABLE 2.
Characteristics of 5- to 10-cm-depth horizon soil under Norway spruce, oak, and beech at the Breuil-Chenue experimental sitea
| Tree species | Environmentb | pHc | meq 100 g−1d |
g kg soil−1 |
C/N ratio | |||
|---|---|---|---|---|---|---|---|---|
| K | Mg | Ca | N | C | ||||
| Beech | R | 4.47 | 0.79 | 0.29 | 0.30 | 5.52 | 90 | 16.23 |
| BS | 4.48 | 0.36 | 0.20 | 0.23 | 4.48 | 74 | 16.55 | |
| Oak | R | 4.35 | 1.57 | 0.40 | 0.29 | 4.94 | 83 | 16.85 |
| BS | 4.32 | 0.61 | 0.26 | 0.17 | 4.01 | 68 | 16.93 | |
| Spruce | R | 4.14e | 0.68 | 0.41 | 0.28 | 4.92 | 87 | 17.61 |
| BS | 4.30 | 0.42 | 0.21 | 0.15 | 4.12 | 72 | 17.49 | |
Data are from the work of Mareschal (34).
R, rhizosphere; BS, bulk soil.
Soil pH was measured in H2O with a soil-to-solution ratio of 1:2.
Milliequivalent for 100 g of fine earth (dry weight).
The pH of the spruce rhizosphere is significantly more acidic than those of the other soils environments according to a one-factor (tree-species) ANOVA (P = 0.05) and the Bonferroni-Dunn test (adapted from the work of Calvaruso et al., 2009 [11]).
Collection of bacterial strains.
Soil samples and Scleroderma citrinum ectomycorrhizae were collected in autumn under four sporocarps of S. citrinum (approximately 5 m apart) associated with different trees and at 5 to 10 cm in the organo-mineral horizon under oak, beech, and spruce stands. The ectomycorrhizal fungal species S. citrinum was chosen because it is a ubiquitous fungus, widely distributed in the experimental forest site and colonizing all the oak, beech, and spruce stands. Moreover, its mycorrhizae can be unambiguously identified in situ because of its color, morphology, and typical smell. In the experimental forest site, an inventory of the epigeous sporocarps, performed three times each autumn during 5 years, revealed 138 ectomycorrhizal species. Among them, S. citrinum represents the 9th most abundant (8). Metagenomic analysis of the soil fungal communities confirmed that this fungus represented a major species (8). Furthermore, this fungal species was targeted because in vitro assays showed that it was not efficient for mineral weathering (10).
The bacterial strains were isolated from three compartments, as follows: (i) bulk soil, i.e., soil remaining after picking out the roots and the extramatrical mycelium of S. citrinum aggregated in thick white strands; 50 mg of fresh bulk soil was suspended in 1 ml sterile distilled water and shaken manually (five times up and down), and the resulting suspension was called “N”; (ii) hyphosphere soil, i.e., the extramatrical mycelium of S. citrinum; 20 mg of extramatrical mycelium collected only under beech and spruce stands (no extramatrical mycelium was detected under oak stand) were suspended in 1 ml sterile distilled water and manually shaken (five times up and down), and the resulting suspension was called “MY”; (iii) ectomycorrhizosphere soil, i.e., soil adherent to oak/beech/spruce-S. citrinum ectomycorrhizae; 20 mg of fresh ectomycorrhizae were manually shaken in order to remove the nonadherent soil particles, and then the ectomycorrhizae were suspended in 1 ml sterile distilled water and manually shaken (five times up and down); the resulting suspension was called “ML.” One milliliter of each suspension (N, ML, or MY) was sampled, and serial dilutions were spread onto a tryptic soy agar (TSA) medium (10%). Plates were incubated at 25°C for 7 days. The bacterial isolates were purified by three successive platings on TSA (10%). A total of 400 bacterial strains were isolated from the bulk soil (PN), the hyphosphere (PMY), and the soil-mycorrhiza interface (PML). For the present study, 56 to 59 bacterial strains isolated from the beech and spruce stands, respectively, were subsampled at random among the whole collection in the three soil compartments. For the oak bacterial collection, a total of 40 bacterial isolates from the bulk soil (PN) and the ectomycorrhizosphere (PML) were tested. All the bacterial isolates used in this study were cryopreserved at −80°C in 20% glycerol and then cultivated on 1/10-strength TSA medium (tryptic soy broth from Difco, 3 g·liter−1, and agar, 15 g·liter−1) at 25°C.
Mineral weathering potential of bacterial isolates.
The mineral weathering assay was performed as described by Uroz et al. (46). Briefly, bacterial isolates were cultivated overnight in liquid LB medium (50). Two milliliters of each culture was washed three times with sterile ultrapure water. The absorbance of each resulting suspension at 600 nm was then adjusted between 0.8 and 1. Twenty microliters of each suspension was transferred into a well of a sterile MultiScreen microplate (MAGVN22, 0.22-μm pore size; Millipore) containing 10 mg of 200- to 500-μm sterile particles of biotite and 180 μl of sterile Bushnell-Hass medium (BHm) lacking iron [in mg liter−1: KCl, 20; MgSO4·7H2O, 150; NaH2PO4·2H2O, 80; Na2HPO4·2H2O, 90; (NH4)2SO4, 65; KNO3, 100; and CaCl2, 20] buffered at pH 6.5 and supplemented with glucose (2 g liter−1). Biotite was obtained from Bancroft. It is a pure and homogenous mineral, and its composition, in g kg−1, is as follows: SiO2, 410.1; Al2O3, 109; Fe2O3, 22.1; FeO, 100.5; MnO, 2.7; MgO, 189; Na2O, 4.1; K2O, 94.6; TiO2, 22.8; F, 44.2; and Zn, 0.8. This mineral weathers relatively quickly under our experimental conditions and holds K and Mg nutrients (which are indispensable for plants). Biotite and culture media were sterilized by autoclaving (20 min at 120°C). Glucose was sterilized by filtration (0.22-μm pore size; Millipore) and added to Bushnell-Hass medium lacking iron after autoclaving.
For each bacterial strain, eight wells were filled with the same inoculum (four wells for pH determination and four wells for iron measurement). In each experiment, eight wells were used as a control without bacteria: these wells received only 200 ml of Bushnell-Hass medium lacking iron. The two reference strains of Burkholderia, PN3(3) and PML1(4), were used as negative and positive controls, respectively, in each experiment (46).
The MultiScreen microplates were incubated at 25°C for 48 h under constant shaking (orbital; 350 rpm). After incubation, all of the liquid contents in each well were filtered (0.22 μm) by centrifuging the MultiScreen microplates in order to eliminate bacterial cells and biotite particles. To assess biotite weathering, filtrates were recovered in a new microplate containing bromocresol green (1 g liter−1; Sigma) for pH determination or 20 μl of Ferrospectral (Merck) for quantification of total iron (Fe2+ and Fe3+). The iron concentration was used as a weathering indicator because we previously demonstrated that the biotite used has a stoichiometric dissolution under our experimental conditions. The pH and the amount of iron released from biotite in the solution were both determined at A595 with a Bio-Rad model 550 microplate reader, as described by Uroz et al. (46).
Molecular identification of bacterial isolates.
The 16S rRNA gene amplification of the bacterial isolates was performed using the universal primers pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and pH (5′-AAGGAGGTGATCCAGCCGCA-3′), which allowed amplification of almost the entire gene (ca. 1,500 bp) (19). PCRs were performed in 50-μl (total volume) mixtures containing 1× PCR master mix (Eppendorf), 0.1 μM primers, and 1 μl cell extract (7). Cell extract was prepared by adding one colony of each isolate to 100 μl of sterile water. The following temperature cycle was used: initial denaturation for 5 min at 95°C, followed by 30 cycles of 1 min of denaturation at 95°C, 1 min of annealing at 56°C, and 1.5 min of extension at 72°C and then a final extension at 72°C for 10 min. PCR products were purified and concentrated using a Qiaquick gel extraction kit (Qiagen). The sequencing reaction was performed using the multicapillary Beckman CEQ 8000XL automated sequencer system. The sequencing primer used was 518r (5′-ATTACCGCGGATGCTGG-3′) (29). The sequences were compared with the sequences in the GenBank databases (www.ncbi.nlm.nih.gov.BLAST), using the BLAST program (1).
Phylogenetic analysis.
The partial 16S rRNA gene sequences (ca. 450 bp) of the bacterial isolates were aligned with previously described 16S rRNA gene sequences of alpha-, beta-, and gammaproteobacteria and Gram-positive bacteria (Firmicutes and Actinobacteria) using Clustal X (version 1.8) (43). Phylogenetic algorithms were obtained, and tree design (DNA-DIST, NEIGHBOR, and SEQBOOT) was performed using the PHYLIP software package (version 3.65; J. Felsenstein, University of Washington, Seattle, WA [http://evolution.genetics.Washington.edu/phylip.html]). A bootstrap analysis was based on 1,000 replicates.
Statistical analysis.
The effects of the compartment of origin (bulk soil, hyphosphere, or ectomycorrhizosphere) and tree species (oak, beech, and Norway spruce) on the mineral weathering potential (acid compound production and iron mobilization) of the bacterial isolates were determined by analysis of variance (ANOVA) at a threshold level of P = 0.05 using the Superanova software program (Abacus Concepts, Inc., Berkeley, CA). The proportion of bacterial isolates per class, differing in their ability to acidify the medium or to release iron from biotite, were compared between the different isolate origin compartments using a χ2 test (P = 0.05).
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession numbers GU302310 to GU302418.
RESULTS AND DISCUSSION
Understanding the role of tree-microorganism-soil interactions in governing nutrient availability in the ectomycorrhizosphere should allow us to better comprehend forest ecosystem functioning and sustainability. Here we tested the hypothesis that certain tree species develop indirect strategies for nutrient mobilization in nutrient-poor forest soils, consisting in the selection of soil bacterial communities with high mineral weathering potential. To our knowledge, this is the first time that the impact of tree species on the bacterial mineral weathering function has been tested.
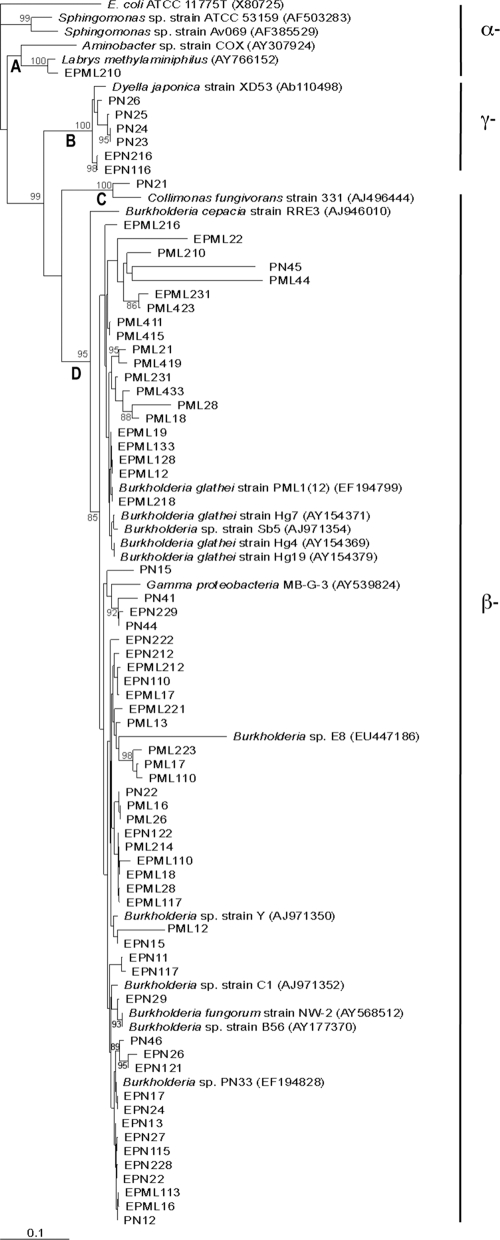
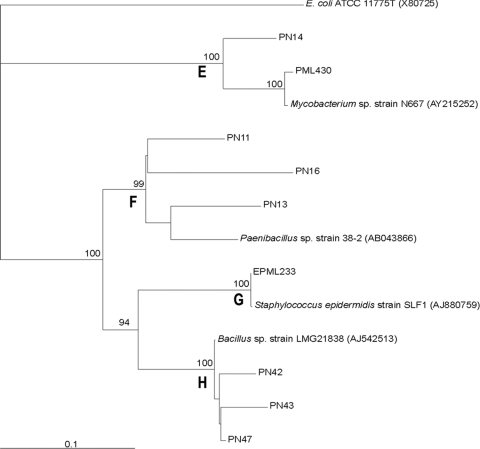
The 16S rRNA gene sequences obtained from the bacterial isolates from the oak, beech and spruce stands revealed that these bacterial isolates belonged to the alpha-, beta-, and gammaproteobacteria and to the Gram-positive (Firmicutes and Actinobacteria) lineages. The 16S rRNA gene sequences exhibited high homology (95 to 100%) with sequences of environmental bacteria belonging to the genera Burkholderia, Bacillus, Collimonas, Dyella, Labrys, Mycobacterium, Nocardia, Paenibacillus, and Staphylococcus (Fig. 1 and 2). Gram-negative and Gram-positive bacteria were both clustered in four groups (Fig. 1 and 2). Gram-positive (clusters E, F, G, and H; Fig. 2) and gammaproteobacterial (cluster B) groups comprised isolates obtained mainly from the bulk soil. On the other hand, cluster A contained one alphaproteobacterium obtained from the mycorrhizosphere (PML). Cluster C contained one sequence strongly related to that of collimonads. Interestingly, collimonads coming from the same experimental site have been noted for their efficient mineral weathering ability (31, 46, 47). A metagenomic analysis performed on the same experimental site showed that the betaproteobacteria, composed mainly of Burkholderia and Collimonas, accounted for about 2 to ca. 5% of the total number of sequences in the bulk soil and rhizosphere environments, respectively (48). The genus occurring most frequently in this study, whatever the tree species, was Burkholderia and corresponds to cluster D. This genus represented 58% of the bulk soil isolates and 92% of the ectomycorrhizosphere isolates. Strains of the Burkholderia genus have been found previously to constitute the dominant cultivated bacterial genus associated with ectomycorrhizal fungi and forest soils (3, 18, 36) and to harbor mineral weathering abilities (46). Similar taxonomic groups were detected for the three tree species.
FIG. 1.
Neighbor-joining tree showing the phylogenetic relationships of the Gram-negative bacterial isolates associated with oak and spruce, based on PCR sequencing of a portion of the 16S rRNA gene with primers pA and pH. A bootstrap analysis was performed with 1,000 repetitions, and only values greater than 80 are indicated at the nodes. The bootstrap analysis identified four clusters: A, B, C, and D. The bacterial isolates used in this study were designated based on their origins. The PN isolates from the bulk soil and the PML isolates from the mycorrhizosphere came from the oak stand. The bacterial isolates preceded by the letter E (EPN and EPML) came from the spruce stand.
FIG. 2.
Neighbor-joining tree showing the phylogenetic relationships of the Gram-positive bacterial isolates associated with oak and spruce, based on PCR sequencing of a portion of the 16S rRNA gene with primers pA and pH. A bootstrap analysis was performed with 1,000 repetitions, and only values greater than 80 are indicated at the nodes. The bootstrap analysis identified four clusters: E, F, G, and H. The bacterial isolates used in this study were designated based on their origins. The PN isolates from the bulk soil and the PML isolates from the mycorrhizosphere came from the oak stand. The bacterial isolates preceded by the letter E (EPN and EPML) came from the spruce stand.
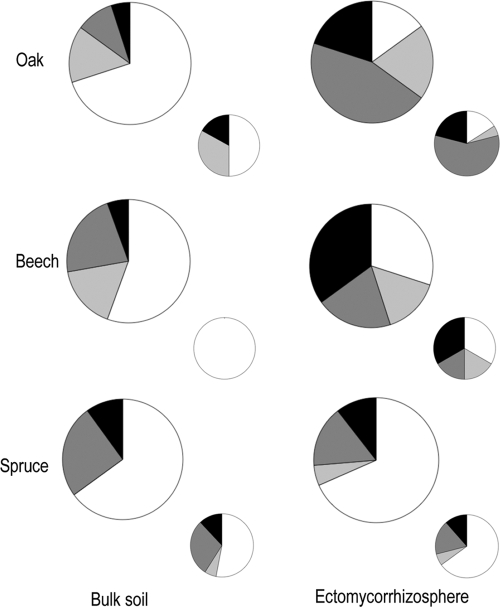
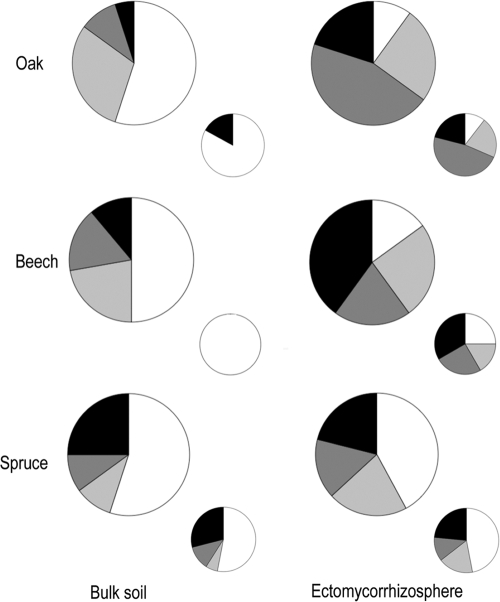
About half of the bacterial isolates tested significantly acidified the microplate solution regardless of the tree species of the stand where they came from (Fig. 3). The most efficient isolates generated a decrease in the pH of the microplate solution from 6.5 to 3.2 in 48 h. It should be kept in mind that production of protons is one of the main factors influencing mineral stability (43). Moreover, Carrillo et al. (12) demonstrated that inoculation of cardon seedlings with a strain of Azospirillum brasilense enhanced acidification of the rhizosphere as well as plant growth in poor desert soil microcosms. About two-thirds of the bacterial isolates were also able to release iron from biotite (Fig. 4). The most efficient isolates released ca. 2 mg liter−1 of iron. With the biotite dissolution being stochiometric under our experimental conditions (46), the iron values obtained are good estimates of the total dissolution of the biotite and of the weathering efficacy of the bacterial strains. These results highlight that bacteria with efficient mineral weathering potentials are widespread in forest soils regardless of the tree species.
FIG. 3.
Distribution of the total bacterial (large circles) and Burkholderia genus (small circles) isolates based on their compartment of origin (bulk soil [PN isolates] and ectomycorrhizosphere [PML isolates]) and their ability to acidify the microplate solution for the three different tree species. White, pH ≥ 5.30; light gray, pH ≥ 4.30 and < 5.30; dark gray, pH ≥ 3.50 and < 4.30; black, pH < 3.50. The proportion of bacterial isolates in each of these four classes was significantly different between the bulk soil and the ectomycorrhizosphere for oak and beech species only, according to a χ2 test (P < 0.05).
FIG. 4.
Distribution of the total bacterial (large circles) and Burkholderia genus (small circles) isolates based on their compartment of origin (bulk soil [PN isolates] and ectomycorrhizosphere [PML isolates]) and their efficacy to release iron (Fe, in mg liter−1) from the biotite for the three different tree species. White, Fe ≤ 0.1 mg liter−1; light gray, Fe ≥ 0.1 mg liter−1 and < 0.5 mg liter−1; dark gray, Fe ≥ 0.5 mg liter−1 and < 1 mg liter−1; black, Fe ≥ 1 mg liter−1. The proportion of bacterial isolates in each of these four classes was significantly different between the bulk soil and the ectomycorrhizosphere for oak and beech species only, according to a χ2 test (P < 0.05).
We pointed out the strong ability of certain strains belonging to the Burkholderia genus (Fig. 4) to weather biotite. The mineral weathering efficacy of strains of the Burkholderiales was previously reported for other minerals, such as apatite, basalt, and granite (33, 52-53). We also revealed that the weathering potential varies greatly depending on the isolates: about 40% of the isolates belonging to the Burkholderia genus did not weather biotite under our experimental conditions (Fig. 4). These results highlight that within the Burkholderia genus, a certain functional plasticity may exist concerning the weathering ability of these bacteria. Recent genomic advances have made it evident that Burkholderia strains are capable of flexibility, plasticity, and versatility, making them highly adaptable to various niches (14, 44).
The culturable bacterial strains isolated from the S. citrinum ectomycorrhizosphere of oak and beech appeared significantly more efficient in acidifying and releasing iron from biotite than those from the surrounding soil as determined by a one-factor (origin) ANOVA (P = 0.05) and the Bonferroni-Dunn test (Table 3). Moreover, the proportion of culturable bacterial strains able to acidify and to mobilize iron from the biotite was significantly higher in the S. citrinum ectomycorrhizosphere of oak and beech than in the bulk soil (Fig. 3 and 4), suggesting a niche effect. Notably, the culturable bacterial isolates from the S. citrinum hyphosphere of beech appeared significantly more efficient in releasing iron from biotite than those from the surrounding soil as determined by a one-factor (origin) ANOVA (P = 0.05) and the Fisher test, suggesting an impact of the mycorrhizal fungus on the bacterial communities. No significant differences were observed between the bacterial isolates from the S. citrinum hyposphere and those from the ectomycorrhizosphere. As regards the Burkholderia genus, 83% of the strains isolated in the bulk soil did not weather the biotite, while 89% of the strains isolated in the ectomycorrhizosphere weathered the biotite regardless of the tree species (Fig. 4). All together, these results reveal that the S. citrinum mycorrhizal roots associated with oak and beech structure the functional diversity of the soil bacterial community by selecting in their vicinity bacterial strains potentially able to weather the minerals, which may thus contribute to improving tree nutrition and growth. This hypothesis has already been partly confirmed, since Calvaruso et al. (10) demonstrated that a Burkholderia glathei strain from the oak-S. citrinum ectomycorrhizosphere, sampled in 2001 in the same experimental forest site, significantly improved Scots pine seedling nutrition and growth under controlled conditions because of its effects on the weathering of biotite (i.e., mobilization of K and Mg). Similarly, Koele et al. (27) showed that coinoculating S. citrinum and a Collimonas strain significantly improved the pine biomass compared to that of noninoculated pine plants. Finally, Puente et al. (40-41) demonstrated that rhizosplane and endophytic bacteria, by releasing significant amounts of nutrients from rocks, promoted the growth of cactus seedlings.
TABLE 3.
Concentrations of Fe and pH measured in the microplate solutions after 48 h of interactions between bacterial suspensions and biotitea
| Origin of bacterial isolates | Fe (mg liter−1) |
pH |
||||
|---|---|---|---|---|---|---|
| Oak | Beech | Norway spruce | Oak | Beech | Norway spruce | |
| Bulk soil | 0.28 ± 0.08 | 0.34 ± 0.12 | 0.51 ± 0.16 | 4.33 | 4.22 | 4.10 |
| Ectomycorrhizosphere | 0.75* ± 0.10 | 0.88* ± 0.15 | 0.54 ± 0.14 | 3.77* | 3.69* | 4.19 |
Values represent means ± standard deviations for 18 to 20 replicates. In each column, treatments associated with an asterisk are significantly different according to a one-factor (compartment of origin of the bacterial strains) ANOVA (P < 0.05). Under our experimental conditions, no iron was measured in the filtrate of the noninoculated control wells and no modification of the pH was observed.
The impact of the S. citrinum ectomycorrhizosphere on the composition of the soil bacterial communities observed in our study is in accordance with results from previous works which compared the functional diversity of bacterial communities isolated from bulk soil and oak-S. citrinum ectomycorrhizosphere in the same experimental forest site and during the same autumn season but 4 years earlier (10, 46). Interestingly, the Burkholderia genus was also the main taxonomic group detected at this sampling time (46). Contrary to the studies of Berardesco et al. (4) and Grayston et al. (22), which have revealed a high temporal variation in the functional and metabolic diversity of bacterial communities, here we have revealed that the functional structure of culturable bacterial communities in soil in relation to the mineral weathering process is stable during a 2-year period, suggesting a temporal stability of the bacterial communities in the forest ecosystem considered. Similarly, Krave et al. (28) previously demonstrated a seasonal stability of bacterial diversity in the mineral soil horizon in nutrient-poor soils intensively reforested with Pinus merkusii, even though the soil moisture level greatly varied during the year. To explain the interannual temporal stability we observed, we propose that the structure effect of S. citrinum ectomycorrhizal symbiosis on the functional diversity of the culturable bacterial communities in soil is a functional requirement strongly linked to the strategy that certain tree species may have developed for improving their mineral nutrition. This hypothesis is supported by previous results of Frey-Klett et al. (20) with another species of mycorrhizal fungus. These authors demonstrated in a different context (9-month seedlings of Douglas fir-Laccaria bicolor S238N) that the Douglas fir-Laccaria bicolor ectomycorrhizal symbiosis also selects Pseudomonas fluorescens isolates likely to be beneficial to the nutrition of Douglas fir seedlings due to their efficacy in mobilizing phosphorus and iron.
Contrary to the case with oak and beech stands, in the Norway spruce stand no significant difference in terms of acidification and iron mobilization potentials was observed between bacterial strains isolated from the S. citrinum ectomycorrhizosphere, hyposphere, and bulk soil (Table 3; Fig. 2 and 3). Regarding the Burkholderia genus, the percentage of strains able to weather biotite is about 50% in the bulk soil and in the ectomycorrhizosphere (Fig. 3). This reveals for the first time that the structuring effect of the S. citrinum ectomycorrhizal symbiosis on the functional diversity of soil bacterial communities depends on the tree species. In a previous study carried out in the same experimental site as detailed in this study, Lejon et al. (30) demonstrated differences in bacterial density and community composition between oak and Norway spruce. According to these authors, these differences result mainly from soil physicochemical characteristics linked to tree functioning, which are very distinct between oak and Norway spruce. Notably, Colin-Belgrand et al. (13) and Zeller et al. (54), who investigated the effect of tree species on nitrogen transformations in the Breuil site, clearly demonstrated that the amounts of nitrate produced during mineralization were lower in the Norway spruce stand than those measured in oak and beech stands. The presence of large amounts of ammonium in the vicinity of Norway spruce roots, resulting from a strong ammonification, allows this species to consume mainly nitrogen in this form. To balance this uptake of positive charges, Norway spruce roots thus exudate large amounts of protons (35), resulting in a decrease in the soil pH in the rhizosphere of Norway spruce, as observed by Calvaruso et al. (11) (Tables 2 and 3). These authors demonstrated that due to this massive production of protons by the roots, the dissolution of clay-size minerals, the soil fraction that is the most reactive and the richest in nutrients, is higher in the rhizosphere of Norway spruce than in that of oak in the Breuil site (Table 2). All together, these results suggest that differential impacts of tree species on the structure of culturable mineral weathering bacterial communities could be linked to the nutrient availability rate in the soils under the different tree species.
To conclude, our study highlights the presence of bacterial strains harboring efficient mineral weathering potential in the soil surrounding the trees regardless of the tree species considered. However, an enrichment of efficient mineral weathering bacteria was observed in the S. citrinum ectomycorrhizosphere of oak and beech but not of Norway spruce. A detailed analysis showed that the Burkholderia isolates were the most abundant and among the most efficient mineral weathering bacteria regardless of the tree species considered, suggesting that they are well adapted to nutrient-poor and acidic environments. Finally, our study demonstrates the temporal stability of the functional structure of culturable soil bacterial communities under oak stands. All these observations suggest that certain forest tree species have at their disposal a very effective strategy to balance low-nutrient availability in soils through the selection of efficient mineral weathering bacterial communities in their vicinity. Complementary studies will be necessary to determine by which mechanism(s) trees can select bacterial strains favorable to their nutrition and to study the impact of other ectomycorrhizal species on the structure of the mineral weathering bacterial communities.
Acknowledgments
This work was supported by the French Research Ministry (ACI quantitative ecology project), the Lorraine Region, Andra (Agence Nationale pour la Gestion des Déchets Radioactifs), and the INRA (Institut National de la Recherche Agronomique) Center of Nancy. Sequencing facilities were supported by IFR110, the Lorraine Region, and INRA.
We thank J. Weber, B. Palin, C. Delaruelle, and G. Nourrisson for technical assistance and K. Bateman for English language review.
Footnotes
Published ahead of print on 28 May 2010.
REFERENCES
- 1.Altschul, C., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber, S. A. 1995. Soil nutrient bioavailability: a mechanistic approach, 2nd ed. J. B. Wiley & Sons, New York, NY.
- 3.Bending, G. D., E. J. Poole, J. M. Whipps, and D. J. Read. 2002. Characterization of bacteria from Pinus sylvestris-Suillus luteus mycorrhizas and their effects on root-fungus interactions and plant growth. FEMS Microbiol. Ecol. 39:219-227. [DOI] [PubMed] [Google Scholar]
- 4.Berardesco, G., S. Dyhrman, E. Gallagher, and M. P. Shiaris. 1998. Spatial and temporal variation of phenanthrene-degrading bacteria in intertidal sediments. Appl. Environ. Microbiol. 64:2560-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle, J. R., and G. K. Voigt. 1973. Biological weathering of silicate materials. Implications for tree nutrition and soil genesis. Plant Soil 38:191-201. [Google Scholar]
- 6.Brant, J. B., D. D. Myrold, and E. W. Sulzman. 2006. Root controls on soil microbial community structure in forest soils. Oecologia 148:650-659. [DOI] [PubMed] [Google Scholar]
- 7.Bruce, K. D., W. D. Hiorns, J. L. Hobman, A. M. Osborn, P. Strike, and D. A. Ritchie. 1992. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3413-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buée, M., M. Reich, C. Murat, E. Morin, R. H. Nilsson, S. Uroz, and F. Martin. 2009. 454 pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 184:449-456. [DOI] [PubMed] [Google Scholar]
- 9.Calvaruso, C., M. P. Turpault, and P. Frey-Klett. 2006. Root-associated bacteria contribute to mineral weathering and to mineral nutrition in trees: a budgeting analysis. Appl. Environ. Microbiol. 72:1258-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvaruso, C., M. P. Turpault, E. Leclerc, and P. Frey-Klett. 2007. Impact of ectomycorrhizosphere on the functional diversity of soil bacterial and fungal communities from a forest stand in relation to nutrient mobilization processes. Microb. Ecol. 54:567-577. [DOI] [PubMed] [Google Scholar]
- 11.Calvaruso, C., L. Mareschal, M. P. Turpault, and E. Leclerc. 2009. Rapid clay weathering in the rhizosphere of Norway spruce and oak in an acid forest ecosystem. Soil Sci. Soc. Am. J. 73:331-338. [Google Scholar]
- 12.Carrillo, A. E., C. Y. Li, and Y. Bashan. 2002. Increased acidification in the rhizosphere of cactus seedlings induced by Azospirillum brasilense. Naturwissenschaften 89:428-432. [DOI] [PubMed] [Google Scholar]
- 13.Colin-Belgrand, M., E. Dambrine, S. Bienaimé, C. Nys, and M. P. Turpault. 2003. Influence of tree roots on nitrogen mineralization. Scand. J. Forest Res. 18:260-268. [Google Scholar]
- 14.Compant, S., J. Nowak, T. Coenye, C. Clément, and E. Ait Barka. 2008. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32:607-626. [DOI] [PubMed] [Google Scholar]
- 15.Cook, R. J., L. Thomashow, M. D. Weller, D. Fujimoto, M. Mazzola, G. Bangera, and D. S. Kim. 1995. Molecular mechanism of defence by rhizobacteria against root disease. Proc. Natl. Acad. Sci. U. S. A. 92:4197-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dakora, D. F., and D. A. Phillips. 2002. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 245:35-47. [Google Scholar]
- 17.Drever, J. I. 2005. Surface and ground water, weathering, and soils. Treatise on geochemistry. Elsevier, Amsterdam, Netherlands.
- 18.Duponnois, R., and J. Garbaye. 1991. Mycorrhization helper bacteria associated with the Douglas fir-Laccaria laccata symbiosis: effects in aseptic and in glasshouse conditions. Ann. Sci. For. 48:239-251. [Google Scholar]
- 19.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frey-Klett, P., M. Chavatte, M. L. Clausse, S. Courrier, C. Le Roux, J. Raaijmakers, M. Martinotti, J. C. Pierrat, and J. Garbaye. 2005. Ectomycorrhizal symbiosis affects functional diversity of rhizosphere fluorescent pseudomonads. New Phytol. 165:317-328. [DOI] [PubMed] [Google Scholar]
- 21.Garbeva, P., J. A. Van Veen, and J. D. Van Elsas. 2004. Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for suppressiveness. Annu. Rev. Phytopathol. 42:243-270. [DOI] [PubMed] [Google Scholar]
- 22.Grayston, S. J., G. S. Griffith, J. L. Mawdsley, C. D. Campbell, and R. D. Bardgett. 2001. Accounting for variability in soil microbial communities of temperate upland grassland ecosystems. Soil Biol. Biochem. 33:533-551. [Google Scholar]
- 23.Grayston, S. J., and C. E. Prescott. 2005. Microbial communities in forest floors under four tree species in coastal British Columbia. Soil Biol. Biochem. 37:1157-1167. [Google Scholar]
- 24.Hackl, E., S. Zechmeister-Boltenstern, L. Bodrossy, and A. Sessitsch. 2004. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 70:5057-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haichar, F. Z., C. Marol, O. Berge, J. I. Rangel-Castro, J. I. Prosser, J. Balesdent, T. Heulin, and W. Achouak. 2008. Plant host habitat and root exudates shape soil bacterial community structure. Intern. Soc. Microb. Ecol. 2:1221-1230. [DOI] [PubMed] [Google Scholar]
- 26.Högberg, M. N., P. Högberg, and D. D. Myrold. 2007. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia 150:590-601. [DOI] [PubMed] [Google Scholar]
- 27.Koele, N., M. P. Turpault, E. E. Hildebrand, S. Uroz, and P. Frey-Klett. 2009. Interactions between mycorrhizal fungi and mycorrhizosphere bacteria during mineral weathering: budget analysis and bacterial quantification. Soil Biol. Biochem. 41:1935-1942. [Google Scholar]
- 28.Krave, A. S., B. Lin, M. Braster, A. M. Laverman, N. M. van Straalen, and W. F. Röling. 2002. Stratification and seasonal stability of diverse bacterial communities in a Pinus merkusii (pine) forest soil in central Java, Indonesia. Environ. Microbiol. 4:361-373. [DOI] [PubMed] [Google Scholar]
- 29.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-176. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. B. Wiley & Sons, Chichester, United Kingdom.
- 30.Lejon, D. P. H., R. Chaussod, J. Ranger, and L. Ranjard. 2005. Microbial community structure and density under different tree species in an acid forest soil (Morvan, France). Microb. Ecol. 50:614-625. [DOI] [PubMed] [Google Scholar]
- 31.Leveau, J. H. J., S. Uroz, and W. de Boer. 2009. The bacterial genus Collimonas: mycophagy, weathering, and other adaptive solutions to life in oligotrophic environments. Environ. Microbiol. doi: 10.1111/j.1462-2920.2009.02010.x. [DOI] [PubMed]
- 32.Leyval, C., and J. Berthelin. 1991. Weathering of a mica by roots and rhizospheric micro-organisms of pine. Soil Sci. Soc. Am. J. 55:1009-1016. [Google Scholar]
- 33.Mailloux, B. J., E. Alexandrova, A. R. Keimowitz, K. Wovkulich, G. A. Freyer, M. Herron, J. F. Stolz, T. C. Kenna, T. Pichler, M. L. Polizzotto, H. Dong, M. Bishop, and P. S. Knappett. 2009. Microbial mineral weathering for nutrient acquisition releases arsenic. Appl. Environ. Microbiol. 75:2558-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mareschal, L. 2008. Effet des substitutions d'essences forestières sur l'évolution des sols et de leur mineralogy. Ph.D. thesis. Université Nancy, Nancy, France.
- 35.Marschner, H., M. Haussling, and E. George. 1991. Ammonium and nitrate uptake rates and rhizosphere pH in non-mycorrhizal roots of Norway spruce [Picea abies (L.) Karst]. Trees 5:14-21. [Google Scholar]
- 36.Poole, E. J., G. D. Bending, J. M. Whipps, and D. J. Read. 2001. Bacteria associated with Pinus sylvestris-Lactarius rufus ectomycorrhizas and their effects on mycorrhiza formation in vitro. New Phytol. 151:553-766. [DOI] [PubMed] [Google Scholar]
- 37.Priha, O., S. J. Grayston, T. Pennanen, and A. Smolander. 1999. Microbial activities related to C and N cycling and microbial community structure in the rhizospheres of Pinus sylvestris, Picea abies and Betula pendula seedlings in an organic and mineral soil. FEMS Microbiol. Ecol. 30:187-199. [DOI] [PubMed] [Google Scholar]
- 38.Priha, O., S. J. Grayston, R. Hiukka, T. Pennanen, and A. Smolander. 2001. Microbial community structure and characteristics of the organic matter in soils under Pinus sylvestris, Picea abies and Betula pendula at two forest sites. Biol. Fertil. Soils 33:17-24. [Google Scholar]
- 39.Puente, M. E., Y. Bashan, C. Y. Li, and V. K. Lebsky. 2004. Microbial populations and activities in the rhizoplane of rock-weathering desert plants. I. Root colonization and weathering of igneous rocks. Plant Biol. 6:629-642. [DOI] [PubMed] [Google Scholar]
- 40.Puente, M. E., C. Y. Li, and Y. Bashan. 2004. Microbial populations and activities in the rhizoplane of rock-weathering desert plants. II. Growth promotion of cactus seedlings. Plant Biol. 6:643-650. [DOI] [PubMed] [Google Scholar]
- 41.Puente, M. E., C. Y. Li, and Y. Bashan. 2009. Endophytic bacteria in cacti seeds can improve the development of cactus seedlings. Environ. Exp. Bot. 66:402-408. [Google Scholar]
- 42.Seddoh, F. K. 1973. Altération des roches cristallines du Morvan (granite, granophyres, rhyolites). Etude minéralogique, géochimique et micro-morphologique. Ph.D thesis. University of Dijon, Dijon, France.
- 43.Thompson, J. D., D. J. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumapa, S., M. T. Holden, M. Vesaratchavest, V. Wuthiekanun, D. Limmathurotsakul, W. Chierakul, E. J. Feil, B. J. Currie, N. P. Day, W. C. Nierman, and S. J. Peacock. 2008. Burkholderia pseudomallei genome plasticity associated with genomic island variation. BMC Genomics 25:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ullman, W. J., D. L. Kirchman, S. A. Welch, and P. Vandevivere. 1996. Laboratory evidence for microbially mediated silicate mineral dissolution in nature. Chem. Geol. 132:11-17. [Google Scholar]
- 46.Uroz, S., C. Calvaruso, M. P. Turpault, J. C. Pierrat, C. Mustin, and P. Frey-Klett. 2007. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the soil bacterial communities involved in mineral weathering in a forest soil. Appl. Environ. Microbiol. 73:3019-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uroz, S., C. Calvaruso, M. P. Turpault, A. Sarniguet, W. de Boer, J. H. J. Leveau, and P. Frey-Klett. 2009. Efficient mineral weathering is a distinctive functional trait of the bacterial genus Collimonas. Soil Biol. Biochem. 41:2178-2186. [Google Scholar]
- 48.Uroz, S., M. Buée, C. Murat, P. Frey-Klett, and F. Martin. 2010. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2:281-288. [DOI] [PubMed] [Google Scholar]
- 49.USDA. 1999. Soil taxonomy: a basic system of soil classification for making and interpreting soil surveys, 2nd ed. U.S. Government Printing Office, Washington, DC.
- 50.Vaudequin-Dransart, V., A. Petit, C. Poncet, C. Ponsonnet, X. Nesme, J. B. Jones, H. Bouzar, W. S. Chilton, and Y. Dessaux. 1995. Novel Ti plasmids in Agrobacterium strains isolated from fig tree and chrysanthemum tumors and their opinelike molecules. Mol. Plant Microbe Interact. 8:311-321. [DOI] [PubMed] [Google Scholar]
- 51.Walker, T. S., H. P. Bais, E. Grotewold, and J. M. Vivanco. 2003. Root exudation and rhizosphere biology. Plant Physiol. 132:44-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, L., A. D. Jacobson, H. C. Chen, and M. Hausner. 2007. Characterization of elemental release during microbe-basalt interactions at T = 28°C. Geochim. Cosmochim. Acta 71:2224-2239. [Google Scholar]
- 53.Wu, L., A. D. Jacobson, H. C. Chen, and M. Hausner. 2008. Characterization of elemental release during microbe-granite interactions at T = 28°C. Geochim. Cosmochim. Acta 72:1076-1095. [Google Scholar]
- 54.Zeller, B., S. Recous, M. Kunze, J. Moukoumi, M. Colin-Belgrand, S. Bienaimé, J. Ranger, and E. Dambrine. 2007. Influence of tree species on gross and net N transformation in forest soils. Ann. For. Sci. 64:151-158. [Google Scholar]