Abstract
The Brazilian Atlantic Forest is one of the 25 biodiversity hot spots in the world. Although the diversity of its fauna and flora has been studied fairly well, little is known of its microbial communities. In this work, we analyzed the Atlantic Forest ecosystem to determine its bacterial biodiversity, using 16S rRNA gene sequencing, and correlated changes in deduced taxonomic profiles with the physicochemical characteristics of the soil. DNAs were purified from soil samples, and the 16S rRNA gene was amplified to construct libraries. Comparison of 754 independent 16S rRNA gene sequences from 10 soil samples collected along a transect in an altitude gradient showed the prevalence of Acidobacteria (63%), followed by Proteobacteria (25.2%), Gemmatimonadetes (1.6%), Actinobacteria (1.2%), Bacteroidetes (1%), Chloroflexi (0.66%), Nitrospira (0.4%), Planctomycetes (0.4%), Firmicutes (0.26%), and OP10 (0.13%). Forty-eight sequences (6.5%) represented unidentified bacteria. The Shannon diversity indices of the samples varied from 4.12 to 3.57, indicating that the soils have a high level of diversity. Statistical analysis showed that the bacterial diversity is influenced by factors such as altitude, Ca2+/Mg2+ ratio, and Al3+ and phosphorus content, which also affected the diversity within the same lineage. In the samples analyzed, pH had no significant impact on diversity.
The Brazilian Atlantic Forest is one of the 25 biodiversity hot spots in the world. Altogether, these hot spots contain more than 60% of the total terrestrial species of the planet (17). The Atlantic Forest is a dense ombrophilous forest with several variations, including coastal (3 to 50 m), submontane (50 to 500 m), montane (500 to 1,200 m), and high montane (1,200 to 1,400 m) forests, creating a vegetation gradient ranging from shrubs to well-developed montane forest (4). The Serra do Mar is a mountainous system that shelters the main remainder of the Atlantic Forest following the Brazilian east coast, from north to south along the coastal line, and it is divided into diverse sections of high and low blocks, which have regional denominations.
The most important law-protected conservation area of the Brazilian Atlantic Forest is located in the Serra do Mar of the southern state of Paraná. This conservation area (∼5,000 km2) shelters 72% of the fauna and flora species that occur in Paraná and was declared a Biosphere Reserve by UNESCO in 1992. Much is known about the diversity of its fauna and flora, but little is known of its microbial diversity, particularly the soil microbial diversity and the soil characteristics that influence it.
The soil microbial diversity is vast, and it is estimated that >99% of species remain unidentified (1, 28). Acidobacteria and Proteobacteria are the most abundant groups in soil (15). However, the Proteobacteria lineage is more diverse and stable than the Acidobacteria lineage, suggesting that the latter group is more susceptible to variation in soil properties and to disturbing factors (33). Seasonal, physical, and physicochemical factors can be relevant to the structure and diversity of microbial communities. For example, seasonal changes in vegetation and temperature led to replacement of dominant groups in a wheat field (25) and in grassland soils (16). The particle size also has an influence on the bacterial diversity of soils. The clay fraction has a more diverse bacterial community than do silt or sand fractions (23). Finally, analyses of communities from North and South American soils showed that pH plays a major role in bacterial diversity, with less diverse communities associated with a lower pH (9).
Human activity can also change the microbial diversity of soils, both qualitatively and quantitatively. Analyses of microbial communities on coral atolls in the central Pacific Ocean under different degrees of human impact showed that the least-impacted atoll had autotrophs and heterotrophs equally distributed in the community, whereas the most-impacted atoll had a dominance of heterotrophs and about 10 times more microbial cells and virus-like particles in the water column, including a large percentage of potential pathogens (7). A comparison between bacterial communities in forest and pasture soil showed that there is a less diverse and more restricted community in pasture soils. The vegetation shift from forest to pasture resulted in changes to G+C% contents of soil bacterial DNA and amplified rRNA gene restriction analysis (ARDRA) profiles (18). Similar changes occurred with communities of soils submitted to agroindustrial treatments and pollutants (3, 30).
In this work, we used a culture-independent approach based on 16S rRNA gene sequences to survey the bacterial community of the Atlantic Forest soils and determined the physicochemical factors affecting its bacterial biodiversity.
MATERIALS AND METHODS
Soil sampling.
Atlantic Forest soil samples were collected along the PR 410 highway in the State of Paraná, Brazil, which transverses 28.5 km of an area of Atlantic Forest. GPS coordinates (Gardin) of the collection point for each sample were recorded (see Table S1 in the supplemental material). For sample collection, the site was cleaned superficially to remove plants and decomposing organic matter. The soil in a circle of approximately 50 cm in diameter, from 0 to 20 cm in depth, was thoroughly mixed, and soil samples (approximately 500 g) were then collected, transferred to sterilized Falcon tubes, and stored on ice. Collection tools were washed in water, followed by disinfection with 70% alcohol and 2% sodium hypochlorite and, finally, were washed thoroughly with sterile water. A total of 10 soil samples were collected from sites in the submontane (50 to 500 m of altitude) and montane (500 to 1,200 m) forest (4) (Table 1). The following physicochemical parameters of the collected soil were determined: pH, Al3+, H+ + Al3+, Ca2+, Mg2+, K+, total bases (SB; = Ca2+ + Mg2+ + K+), effective cation exchange capability (T; = SB + H+ + Al3+), phosphorus level, carbon content, base saturation (V), aluminum saturation (m), Ca2+/Mg2+ ratio, and clay content (see Table S2 in the supplemental material). Soil analyses were performed by the Laboratory of Soil Analyses of the Department of Soils of the Universidade Federal do Paraná, using standard methods (27).
TABLE 1.
Bacterial diversity of the Brazilian Atlantic Forest soila
| Library | Altitude (m)b | No. of reads | No. of OTUs | H′ | E |
|---|---|---|---|---|---|
| MA01 | 874 | 77 | 64 | 4.08 | 0.94 |
| MA02 | 900 | 78 | 66 | 4.12 | 0.95 |
| MA03 | 896 | 70 | 48 | 3.79 | 0.97 |
| MA04 | 810 | 74 | 58 | 3.96 | 0.92 |
| MA05 | 604 | 83 | 59 | 3.87 | 0.87 |
| MA06 | 375 | 80 | 54 | 3.84 | 0.88 |
| MA07 | 161 | 71 | 51 | 3.81 | 0.96 |
| MA08 | 95 | 69 | 43 | 3.57 | 0.95 |
| MA09 | 44 | 81 | 51 | 3.60 | 0.92 |
| MA10 | 29 | 70 | 47 | 3.67 | 0.95 |
Sequences with identities of ≥95% were assumed to belong to the same OTU. Indices were calculated from the number and abundance of species in each soil sample by using DOTUR (22). H′, Shannon index; E, evenness index.
Referenced to the average level of the sea.
Soil DNA extraction, 16S rRNA gene amplification, and cloning.
After collection, the soil samples were stored on ice for no more than 4 h before DNA extraction. Soil DNA was extracted using an UltraClean soil DNA kit (MoBio Laboratories) following the manufacturer's instructions. Briefly, soil (0.5 g) was added to a tube containing 2 ml of bead suspension and vigorously mixed. The mixture was treated with an inhibitor removal solution, and then the DNA was purified on silica columns. 16S rRNA gene amplification was performed using the universal primers for the Bacteria domain: 27F (5′-AGAGTTTGATCCTGGCTCAG) and 1492R (5′-ACGGCTACCTTGTTACGACTT) (31). The PCR mixture (20 μl) contained 2 U of Taq DNA polymerase, 4 pmol of each primer, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), approximately 10 ng of extracted soil DNA, and PCR buffer (200 mM Tris-HCl, pH 8.4, 500 mM KCl). The thermocycler program was as follows: 1 cycle at 95°C for 5 min, followed by 20 sequential cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min and a final step at 72°C for 5 min. The PCR products were cloned using the pGEM-T Easy vector system (Promega) according to the manufacturer's instructions.
Plasmid DNA extraction and sequencing.
Plasmid DNA was purified in 96-well plates by the alkaline lysis method (20). The V1-V2 region of cloned 16S rRNA genes (∼300 bp at the 5′ end of the 16S rRNA gene) was sequenced with the forward primer Y1 (5′-TGGCTCAGAACGAACGCTGGCGGC) and the reverse primer Y2 (5′-CCCACTGCTGCCTCCCGTAGGAGT) (32) in a Megabace 1000 automatic sequencer, using a DYEnamic ET dye terminator cycle sequencing kit (GE Healthcare).
Sequence assembly and analysis.
The Phred program was used for base calling (8). The Phrap program was used to assemble the reads into the 16S rRNA partial gene sequence. Finally, the Consed program (11) was used to view and edit the sequence assembly. The final sequences were compared with the Ribosomal Database Project II (6), using the SeqMatch tool. Partial 16S rRNA gene sequences were aligned using ClustalW (26), and the alignment was used to construct distance matrices with the DNAdist program (J. Felsenstein, University of Washington [http://evolution.genetics.washington.edu/phylip.html]). Distance matrices were used as input for the DOTUR program (22), which was used to cluster sequences into operational taxonomic units (OTUs) (identities of ≥95%).
Biodiversity evaluation.
Sequences with identities of ≥95% were assumed to belong to the same OTU (5, 19). The bacterial diversity was evaluated (13) by the Shannon diversity index (H′), calculated by the DOTUR program (22). Rarefaction curves, ACE estimators, and Shannon indices for high- and low-altitude groups were also calculated using DOTUR. Evenness (E) was calculated by the equation E = H′/ln S, where S (species richness) is the total number of OTUs.
The similarities in the compositions of the clone libraries were examined by using the S-LibShuff program (21). Graphical analyses were done using the LibShuff program (24). The LibShuff program generates homologous and heterologous coverage curves (CX and CXY, respectively), at any level of sequence similarity or evolutionary distance (D), from two 16S rRNA gene clone libraries (X and Y). To determine if the coverage curves CX(D) and CXY(D) are significantly different, the distances between the two curves are first calculated by using the Cramér-von Mises test. The two libraries were considered significantly different when the P value was <0.05.
Statistical methods.
Statistical analyses of the biological diversity indices and physicochemical characteristics of soil were performed for samples with high (MA01 to MA04) and low (MA07 to MA10) levels of diversity. An independent two-sample Student t test and the Mann-Whitney test were performed to screen for variables with statistically significant differences between the two groups of samples. The Hodges-Lehman (HL) estimator of the difference in central tendency between the two groups was calculated for all biological and physicochemical variables. Principal component analysis (PCA) was carried out on the mean, centered with unit variance scaled data by a matlab routine developed in-house. Data were visualized in the form of the principal component score plots and loading plots. Partial least-square discriminant analysis (PLS-DA) was performed to determine which variables were correlated with the biodiversity and to validate the results obtained with the unsupervised PCA model. Validation of statistical data was performed using jackknifing and cross-validation tests. The model predictive value was assessed by the Q2 parameter (10), indicating how well the model predicts new data by using leave-one-out cross-validation.
Nucleotide sequence accession numbers.
The obtained 16S rRNA gene sequences were deposited in the GenBank database under accession no. EF135620 to EF136358 and GU071058 to GU071072.
RESULTS
Atlantic Forest soil physical and chemical properties.
The physicochemical properties of the soil samples are shown in Table S2 in the supplemental material. All of the samples had a low pH (≤4.50) and high aluminum saturation level (>50%). The base saturation (V%) was low (<50%), and thus the soil was classified as infertile or dystrophic. The organic matter content (C) was high only in sample MA01 (>50 g/dm3). The other samples had low organic matter contents (<50 g/dm3). The amount of clay was also determined and varied from 150 to 500 g per kg of soil.
Sequence identification and diversity characterization.
PCR products were obtained for all DNA samples, using primers 27F and 1492R, and were used to construct 10 libraries of soil organism 16S rRNA gene amplicons in pGEM-T Easy (Promega). Ninety-six clones of each library were isolated and used as templates for sequencing reactions. Among the 960 templates, 754 complete sequences of the V1-V2 region were obtained, and they varied from 234 to 341 bp in length. All of the reads used in the assembly of the contigs had a Phred quality index of at least 30.
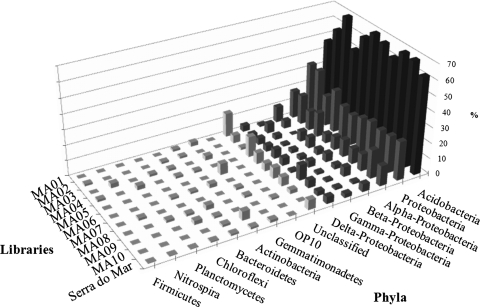
The partial 16S rRNA gene sequences were compared to the RDP II database through the RDPquery program (Fig. 1). Approximately 63% (473 sequences) of the sequences were grouped in the phylum Acidobacteria. The Proteobacteria phylum was ranked second, with 25.2% (190 sequences) of the sequences, which were distributed as follows: Alphaproteobacteria (52.1%), Betaproteobacteria (20%), Deltaproteobacteria (16.3%), and Gammaproteobacteria (11.5%). Other phyla found were Actinobacteria (1.2%), Bacteroidetes (1%), Chloroflexi (0.66%), Firmicutes (0.26%), Gemmatimonadetes (1.6%), Nitrospira (0.4%), Planctomycetes (0.4%), and OP10, a thermophilic bacterium phylum (0.13%). Forty-eight sequences (6.5%) matched the 16S rRNA genes of unclassified, usually uncultured, bacteria and could not be grouped with sequences of known bacteria phyla.
FIG. 1.
Bacterial phyla in Brazilian Atlantic Forest soil.
The number of OTUs (sequences with identities of ≥95%) differed from sample to sample. The MA01 and MA02 samples had the highest species richness (S), with 64 OTUs in 77 sequences and 66 OTUs in 78 sequences, respectively. These two samples also showed the highest Shannon indices, of 4.02 and 4.12, respectively (Table 1). The other samples had lower species richness and Shannon indices. The evenness index varied from 0.97 to 0.87, suggesting that the species were equally represented in the analyzed samples, without dominance of specific bacterial phylotypes (Table 1).
Sequences from the 10 libraries were compared using the S-LibShuff program to evaluate their degrees of similarity. Analyses of homologous coverage curves (see Table S3 in the supplemental material) indicated that libraries for samples MA01 to MA05 had similar bacterial communities (P > 0.05). These libraries were grouped in a cluster and were different from the libraries for samples MA06 to MA10. Similarly, libraries for samples MA07 to MA10 also seemed to have similar communities. On the other hand, the MA06 library was different from all the others (P < 0.05).
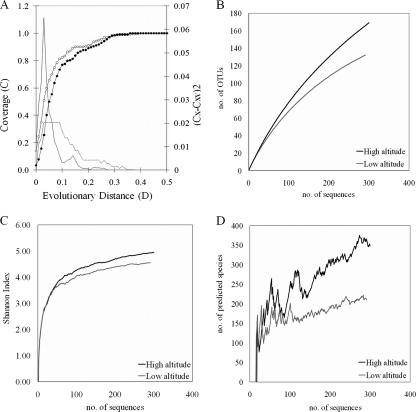
A linear regression plot considering the Shannon index of each library versus the altitude of the sampling site (see Fig. S1 in the supplemental material) revealed that sample clustering may be influenced by the altitude of the collection points and can be divided into three groups: those with a high level of diversity (MA01 to MA04), those with an intermediate level of diversity (MA05 and MA06), and those with a low level of diversity (MA07 to MA10). To evaluate this separation, we grouped sequences from libraries according to altitude, i.e., high altitude (MA01 to MA04 [between 900 and 800 m above sea level]) and low altitude (MA07 to MA10 [between 160 and 30 m above sea level]), and compared them using the LibShuff program. Graphic analyses of homologous and heterologous coverage curves generated by LibShuff (Fig. 2 A) indicated that the bacterial community in the first group was different from that in the second group in the interval of evolutionary distances from 0.0 (100% of identity and 0% of differences) to 0.3 (70% of identity and 30% of differences). This result suggests that the genetic diversity between these two groups occurs not only at lower taxonomic ranks but also at higher taxonomic levels (Fig. 2A). The separation in the two groups was also evident when we analyzed the tendency curves for rarefaction (Fig. 2B), Shannon indices (Fig. 2C), and ACE estimators (Fig. 2D) on DOTUR plots. The high-altitude group had higher Shannon indices and OTU numbers (95% 16S rRNA gene sequence similarity) than the low-altitude group. Also, the rarefaction curve for the high-altitude group is less saturated than that for the low-altitude group, indicating that more phylotypes could be recovered from the first than from the second group of libraries. The LibShuff and DOTUR results suggest that the high-altitude group had a different, more diverse, richer microbial community than that of the low-altitude group.
FIG. 2.
High-altitude samples have more diverse microbial communities than low-altitude samples. Sequences from the MA01 to MA04 libraries were grouped in the high-altitude, high-diversity cluster, and sequences from MA07 to MA10 were grouped in the low-altitude, low-diversity cluster. (A) Phylogenetic diversity in the high- and low-altitude clusters was compared using the LibShuff program. Homologous (○) and heterologous (•) coverage curves for 16S rRNA gene sequence libraries are shown. Solid lines indicate the value of (CX − CXY)2 for the original samples at each value of D. D is equal to the Jukes-Cantor evolutionary distance, determined by the DNADIST program of PHYLIP. Broken lines indicate the 950th value (or P = 0.05) of (CX - CXY)2 for the randomized samples. (B, C, and D) DOTUR graphic analyses comparing the groups according to rarefaction curves (B), Shannon indices (C), and ACE estimators (D).
Microbial diversity is significantly different in high- and low-altitude soil samples.
To understand the impact of altitude and the physicochemical characteristics of soil on microbial biodiversity, the groups were compared for differences in mean and central tendency, using an independent two-sample Student t test and the Mann-Whitney test, respectively. Table S4 in the supplemental material shows the results for the biological diversity indices (richness [S], evenness [E], and Shannon index [H]), and Table S5 in the supplemental material shows the results for the physicochemical characteristics of the soil samples. Microbial diversity of soil samples at low and high altitudes was also compared using the Wilcoxon-Mann-Whitney two-sample rank sum test. The effect of the altitude (difference between groups) was quantified using the HL estimator, which is consistent with the Wilcoxon test. The results showed an association between the altitude level and soil microbial diversity. On the other hand, there was no statistically significant effect of a particular soil parameter. Hence, the results suggest that the difference found in the biodiversity between groups may be explained by interactions between the physicochemical soil characteristics. A PCA model was developed to explore this hypothesis.
PCA reveals a perfect separation between soil samples with high and low levels of microbial biodiversity.
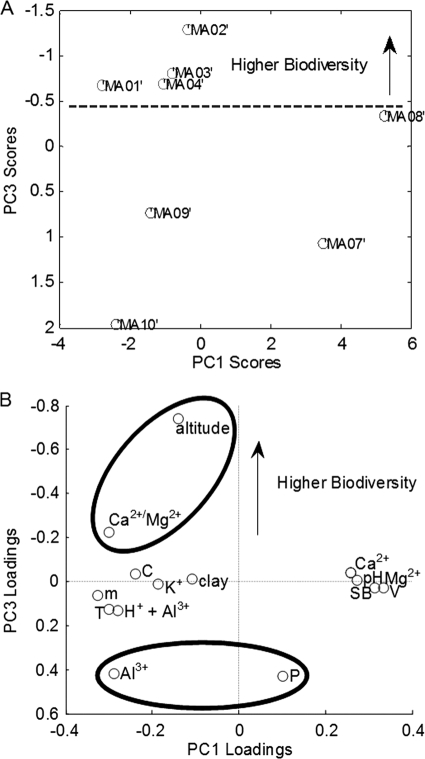
PCA was performed to visualize the interdependence between the variables that could explain the differences between the groups of high- and low-biodiversity soil samples. The score plots of the first and third principal components show a perfect separation between samples of each group (Fig. 3 A). A PCA model with only three components captures over 90% of the variance of the soil samples. The third principal component of the PCA model perfectly discriminates samples with low levels of biodiversity from those with high levels of biodiversity. In order to determine which variables are important for discriminating between groups, the loadings of the third principal component were plotted (Fig. 3B). The variables associated with higher biodiversity levels have larger magnitudes in the same direction as the high-biodiversity samples in the score plots. Higher altitudes and Ca2+/Mg2+ ratios were found to be associated with higher levels of biodiversity, while higher levels of Al3+ and phosphorus were associated with lower levels of biodiversity.
FIG. 3.
PCA. (A) First and third principal component scores showing complete class separation between high and low levels of soil bacterial diversity. (B) First and third principal component loadings. Loadings with higher magnitudes have more impact on the model. The variables that significantly increase biodiversity are altitude and the Ca2+/Mg2+ ratio. The variables Al3+ and phosphorus significantly decrease biodiversity.
To identify the physicochemical characteristics that play a major role in discriminating between low- and high-biodiversity soil samples, PLS-DA was performed. The PLS-DA model achieved a very high predictive value (Q2Y = 0.8) and attained an out-of-sample prediction accuracy of 100%. The significance of the PLS regression coefficients was estimated using the one-sample Student t test on all variables (see Table S6 in the supplemental material). The samples with higher levels of biodiversity were confined to a very small and dense cluster, while the low-biodiversity samples were spread over the space defined by the scores of the first three latent variables (see Fig. S2A in the supplemental material). There are also other variables contributing to reduce the biodiversity in the discriminant model; for example, a similar decrease in biodiversity can be achieved by increasing any of the variables Al3+, clay, and phosphorus because they have very similar contributions to the PLS regression coefficients (22%, 20%, and 17%, respectively) (see Fig. S2B in the supplemental material). On the other hand, an identical increase in the altitude increases the biodiversity indicator variable by 40%, while the Ca2+/Mg2+ ratio increases the biodiversity indicator by only 13.5%. These results show a perfect separation between the low- and high-biodiversity soil samples and provide evidence to support the hypothesis that interdependencies between soil characteristics are associated with the biodiversity in soil samples.
DISCUSSION
In this work, we investigated the microbial biodiversity in Atlantic Forest soil and the factors that influence it. The dominant phylum in Atlantic Forest soil samples was Acidobacteria (63%), followed by the Proteobacteria (25.2%). These two groups are frequently the most numerous in soil samples. In a meta-analysis of 16S rRNA gene sequences from distinct soils, Janssen (15) determined that the most abundant bacterial phyla were Proteobacteria (39%) and Acidobacteria (19%), followed by Verrucomicrobia, Bacteroidetes, Chloroflexi, Planctomycetes, Gemmatimonadetes, and Firmicutes (15). Except for Verrucomicrobia, all of these phyla were represented in Atlantic forest soils, although in different proportions.
The profile of the bacterial community found in Atlantic Forest soils is similar to that found in European forests in eastern Austria (12). In a spruce-fir-beech forest, the Acidobacteria phylum was dominant (35%), followed by the Alphaproteobacteria (27%) and Verrucomicrobia (10%) phyla. In the Kolmberg oak-hornbeam forest, the Acidobacteria were also dominant (28%), followed by the Verrucomicrobia (24%) and Bacteroidetes (11%) phyla. While these similarities occur at the phylum level, it is very unlikely that they also occur at the species level. The dominance of Acidobacteria is common in forest soils, while a dominance of Proteobacteria occurs in disturbed soils (18), possibly because Acidobacteria species are slow-growing bacteria fit to nutrient-limited environments such as pristine forest soils (29). When the soil nutrient content is altered, Acidobacteria organisms are replaced by fast-growing bacteria. The main difference between the Brazilian Atlantic Forest and European forests was the apparent absence of Verrucomicrobia phylum sequences in the Atlantic Forest soils, suggesting that this group is much less represented or absent in the latter environment.
A similar study of the Brazilian Amazon Rainforest (2) revealed a different bacterial community from that found in the Atlantic Forest. The dominant bacterial phylum in the Amazon Rainforest soil (pH 5.0) was the Firmicutes/Clostridium phylum (22%), followed by Acidobacteria/Fibrobacterium (18%), Planctomycetes (16%), and Proteobacteria (12%). In contrast, in the Brazilian Atlantic Forest, the Firmicutes/Clostridium phylum was much less represented. Similar to the case for the Atlantic Forest soil, sequences from the thermophilic OP10 phylum were also found in the Amazon Rainforest soil. This phylum, initially found in the Obsidian Pool, a 75 to 95°C hot spring at the Yellowstone Caldera (14), has frequently been identified in soil 16S rRNA gene libraries (15), but little is known about its role in soil. One hypothesis to be explored is the presence of nonthermophilic species in this group.
Statistical analyses showed that physicochemical characteristics have specific contributions to soil biodiversity. The variability in samples with a high level of biodiversity in the PLS score space was relatively small, and there were more variables contributing significantly to reducing biodiversity. This suggests that a decrease in microbial biodiversity of the soil samples is associated with a complex interaction of multiple factors, while an increase in biodiversity is associated mainly with altitude and, to a lesser extent, the Ca2+/Mg2+ ratio. The influence of abiotic factors was also evident for the dominant lineages. The LibShuff analysis of high- and low-altitude samples indicated that the communities are different at evolutionary distances of 0% (species level) to 30% (phylum level). Since Acidobacteria and Proteobacteria are the dominant groups, this result suggests that there is variation within lineages between high- and low-altitude groups. This intralineage variation is probably related to physicochemical characteristics of the soil (33). Considering that there is not a large variation in pH, other physicochemical (Ca2+/Mg2+ ratio and phosphorus and Al3+ content) and spatial (altitude) factors must act on biodiversity.
Altitude is relevant to variables that affect the ecosystem, such as temperature and oxygen availability. The results show that altitude is statistically correlated with the Shannon index (r = 0.77; α = 0.05) and is also significantly different between the high- and low-diversity groups of samples. The effect of altitude may be related to a vegetation change and/or to human activity at low altitudes, which are at the limits of the conservation area, in contrast to the high-altitude levels of the Serra do Mar. These factors may result in complex alterations of the soil physicochemical properties and, consequently, the bacterial diversity.
To correctly evaluate a microbial ecosystem, it is necessary to integrate the influences of biotic and abiotic factors on the community structure and biodiversity. Recently, an analysis of soil samples from different ecosystems across North and South America showed that bacterial diversity could be predicted by a single variable, the soil pH (9). However, we show here that in the acidic soils of the Brazilian Atlantic Forest the bacterial diversity is influenced by additional factors, such as the Ca2+/Mg2+ ratio, altitude, and Al3+ and phosphorus content, which also affected the diversity within the same lineage. Thus, characterization of abiotic properties is important to understanding the factors that affect bacterial diversity and providing a clearer view of how microbial communities change.
Supplementary Material
Acknowledgments
We thank the Brazilian Research Council (CNPq/MCT Programa Instituto do Milênio) and Fundação Araucária of the State of Paraná, Brazil, for financial support.
We thank Julieta Pie and Roseli Prado for technical support.
Footnotes
Published ahead of print on 21 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-L. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borneman, J., and E. W. Triplet. 1997. Molecular microbial diversity in soils from Eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckley, D. H., and T. M. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 4.Câmara, I. G. 2003. Brief history of conservation in the Atlantic Forest, p. 31-42. In C. Galindo-Leal and I. G. Câmara (ed.), The Atlantic Forest of South America: biodiversity status, threats and outlook. Island Press, Washington, DC.
- 5.Coenye, T., D. Gevers, Y. Van de Peer, P. Vandamme, and J. Swings. 2005. Towards a prokaryotic genomic taxonomy. FEMS Microbiol. Rev. 29:147-167. [DOI] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. Mcgarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:294-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinsdale, E. A., O. Pantos, S. Smriga, R. A. Edwards, F. Angly, L. Wegley, M. Hatay, D. Hall, E. Brown, M. Haynes, L. Krause, E. Sala, S. A. Sandin, R. V. Thurber, B. L. Willis, F. Azam, N. Knowlton, and F. Rohwer. 2008. Microbial ecology of four coral atolls in the Northern Line islands. PLoS One 3:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing, B., L. Hililier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 9.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Perez, I., A. Couto Alves, S. Angulo, J. V. Li, J. Utzinger, T. E. Ebbels, C. Legido-Quigley, J. K. Nicholson, E. Holmes, and C. Barbas. 2010. Bidirectional correlation of NMR and capillary electrophoresis fingerprints: a new approach to investigating Schistosoma mansoni infection in a mouse model. Anal. Chem. 82:203-210. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 12.Hackl, E., S. Zechmeister-Boltenstern, L. Bodrossy, and A. Sessitsch. 2004. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 70:5057-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 14.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipson, D. A., and S. K. Schimidt. 2004. Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Appl. Environ. Microbiol. 70:2867-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers, N., R. A. Mittermeier, C. G. Mittermeier, G. A. B. da Fonseca, and J. Kent. 2000. Biodiversity hotspots for conservation priorities. Nature 403:853-858. [DOI] [PubMed] [Google Scholar]
- 18.Nusslein, K., and J. M. Tiedje. 1999. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl. Environ. Microbiol. 65:3622-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomé, J. B., Jr. 1997. Manual para interpretação de análise de solo. Agropecuária, Guaíba, Brazil.
- 28.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward, N. L., J. F. Challacombe, P. H. Janssen, B. Henrissat, P. M. Coutinho, M. Wu, G. Xie, D. H. Haft, M. Sait, J. Badger, R. D. Barabote, B. Bradley, T. S. Brettin, L. M. Brinkac, D. Bruce, T. Creasy, S. C. Daugherty, T. M. Davidsen, R. T. DeBoy, J. C. Detter, R. J. Dodson, A. S. Durkin, A. Ganapathy, M. Gwinn-Giglio, C. S. Han, H. Khouri, H. Kiss, S. P. Kothari, R. Madupu, K. E. Nelson, W. C. Nelson, I. Paulsen, K. Penn, Q. Ren, M. J. Rosovitz, J. D. Selengut, S. Shrivastava, S. A. Sullivan, R. Tapia, L. S. Thompson, K. L. Watkins, Q. Yang, C. Yu, N. Zafar, L. Zhou, and C. R. Kuske. 2009. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75:2046-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, Y.-H., J. Yao, S. Hu, and Y. Qi. 2000. Effects of agricultural chemicals on DNA sequence diversity of soil microbial community: a study with RAPD marker. Microb. Ecol. 39:72-79. [DOI] [PubMed] [Google Scholar]
- 31.Yoon, J.-H., S. T. Lee, and Y.-H. Park. 1998. Inter- and intraspecific phylogenetic analysis of the genus Nocardioides and related taxa based on 16S rDNA sequences. Int. J. Syst. Bacteriol. 48:187-194. [DOI] [PubMed] [Google Scholar]
- 32.Young, J. P. W., H. L. Downer, and B. D. Eardly. 1991. Phylogeny of the phototrophic Rhizobium strain BTAi by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J. Bacteriol. 173:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youssef, N. H., and M. S. Elshahed. 2009. Diversity rankings among bacterial lineages in soil. ISME J. 3:305-313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.