Abstract
The intestinal microbiota of beef cattle are important for animal health, food safety, and methane emissions. This full-length sequencing survey of 11,171 16S rRNA genes reveals animal-to-animal variation in communities that cannot be attributed to breed, gender, diet, age, or weather. Beef communities differ from those of dairy. Core bovine taxa are identified.
The gastrointestinal tracts (GIT) of beef cattle are colonized by microorganisms that profoundly impact animal physiology, nutrition, health, and productivity (5). The GIT microbiota potentially impact food safety via pathogen shedding (13) by interacting with organisms such as Salmonella and competing for resources in the GIT. Cattle intestinal microbiota also play an important role in methane emissions, with U.S. beef cattle alone contributing an estimated 3.87 million metric tons of methane into the environment each year, both from rumen and large-intestine fermentations (7). Although the bovine fecal microbiota have been well characterized using culture-based methods, these techniques are necessarily limited to characterizing bacteria that can be grown in the laboratory. Culture-independent methods can reveal community members that are recalcitrant to culture. Only a handful of deep-sequencing studies have been done using culture-independent 16S rRNA-based methods (1, 11, 12, 14), all with dairy cattle, which have a fundamentally different diet and metabolism from beef cattle. Despite the potential contributions of the beef cattle GIT microbiota to animal health, food safety, and global warming, these communities remain poorly characterized. With the advent of pyrosequencing technology, researchers now have the tools to characterize these important communities. Pyrosequencing will allow rapid characterization of large-sample data sets (1). However, the taxonomic information generated by rapid sequencing is approximate by necessity (9), and full-length 16S-rRNA sequencing remains the “gold standard” method. Accordingly, we have characterized fecal bacteria from six feedlot cattle by full-length capillary sequence analysis of 11,171 16S rRNA gene clones (Fig. 1).
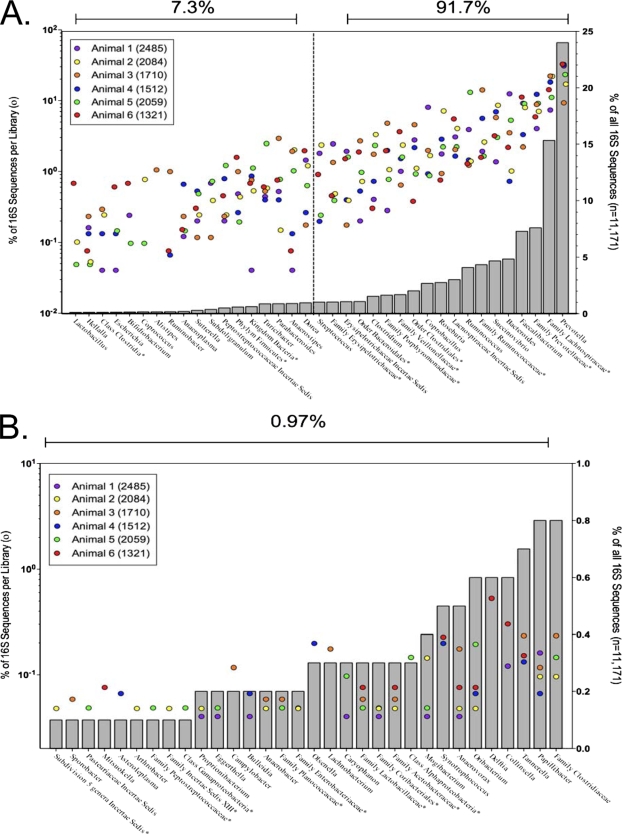
FIG. 1.
Bacterial diversity of six feedlot beef cattle. Gray bars represent the percentages of all 16S sequences that were assigned to each taxonomy. Colored dots represent the percentages of 16S sequences from each library that were assigned to each taxonomic group. Asterisks indicate unclassified members of the named taxon. Panel A shows the data for the first 99% of all the sequences. Panel B shows the data for the remaining 1% of sequences. Note differences in scales for panels A and B.
Rectal grab fecal samples (n = 6) were collected according to institutional animal care guidelines. All animals were female cross-bred MARCIII beef heifers, 6 to 8 months of age, 214 to 241 kg, housed in the same feedlot pen for 2 months prior to fecal collection, and fed the same typical feedlot beef production growing rations consisting of 61.6% corn silage (41.3% dry matter), 15.2% alfalfa hay, 20.9% corn, and 2.3% liquid supplement.
Total fecal DNA was isolated from homogenized samples using MoBio UltraClean fecal kit (Carlsbad, CA). PCR was performed using 27F and 1392R primers (11). Amplification consisted of 25 cycles, with an annealing temperature of 55°C. Amplicons from three reactions per sample were pooled (8), cloned using the Invitrogen TOPO TA cloning kit (Carlsbad, CA), and sequenced bidirectionally with M13 primers using an ABI 3700 sequencer (17). Low-quality and chimeric sequences (6) were excluded from further analysis. Distance matrices were compiled from ClustalW alignments (18) in PHYLIP (4). Pairwise estimates of shared richness were calculated using EstimateS, version 8.2 (R. K. Colwell; http://purl.oclc.org/estimates). DOTUR (16) was used to identify operational taxonomic units (OTUs) and to generate rarefaction curves (Fig. 2), richness and evenness estimates, and Shannon's and Simpson's diversity indices (Table 1). A 97% similarity cutoff and an 85% similarity cutoff for estimating OTUs were used to approximate species and class-level designations (15). Taxonomies were assigned to one member of each OTU using the RDP “classifier” tool (19), and the RDP taxonomic information was used for Fig. 1 and 3. Common bovine taxa were identified based on inclusion in all three U.S. culture-independent studies (this study and references 1 and 11).
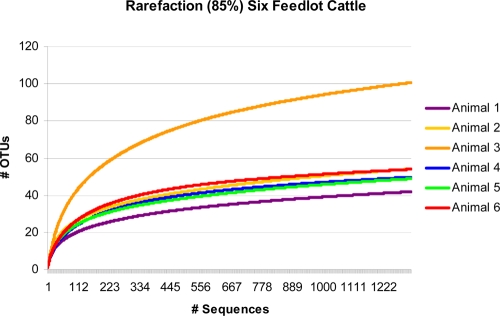
FIG. 2.
Rarefaction curves for six feedlot beef cattle. OTUs were assigned at the 85% DNA sequence similarity level. For comparison purposes, all six curves were truncated after 1,321 sequences.
TABLE 1.
Richness and diversity indices for 6 beef feedlot cattle
| Library and animal (n) | No. of OTUs observed | Species richness (CI)a by: |
Diversity (CI) by: |
||
|---|---|---|---|---|---|
| Chao | ACE | Shannon's index | Simpson's index | ||
| 97% DNA sequence similarity | |||||
| Animal 1 (2,485) | 198 | 372 (294-515) | 329 (280-408) | 3.89 (3.83-3.95) | 0.0422 |
| Animal 2 (2,084) | 416 | 600 (538-694) | 604 (552-675) | 5.40 (5.35-5.45) | 0.0066 |
| Animal 3 (1,710) | 696 | 1,393 (1,224-1,615) | 1,418 (1,327-1,523) | 6.13 (6.08-6.18) | 0.0027 |
| Animal 4 (1,512) | 294 | 526 (439-665) | 483 (425-566) | 4.71 (4.63-4.78) | 0.0237 |
| Animal 5 (2,059) | 314 | 612 (495-805) | 488 (434-566) | 4.93 (4.88-4.99) | 0.0126 |
| Animal 6 (1,321) | 174 | 320 (252-447) | 289 (244-361) | 4.18 (4.11-4.25) | 0.0286 |
| 85% DNA sequence similarity | |||||
| Animal 1 (2,485) | 48 | 61 (51-99) | 62 (52-90) | 2.64 (2.59-2.68) | 0.1056 |
| Animal 2 (2,084) | 77 | 107 (87-165) | 102 (87-139) | 3.38 (3.34-3.43) | 0.0505 |
| Animal 3 (1,710) | 130 | 153 (139-186) | 151 (140-174) | 4.07 (4.02-4.12) | 0.0254 |
| Animal 4 (1,512) | 66 | 75 (68-98) | 77 (70-96) | 2.71 (2.64-2.78) | 0.0931 |
| Animal 5 (2,059) | 69 | 80 (72-109) | 84 (75-110) | 3.31 (3.26-3.36) | 0.0545 |
| Animal 6 (1,321) | 54 | 65 (57-102) | 61 (56-76) | 2.90 (2.83-2.97) | 0.0939 |
CI, confidence interval.
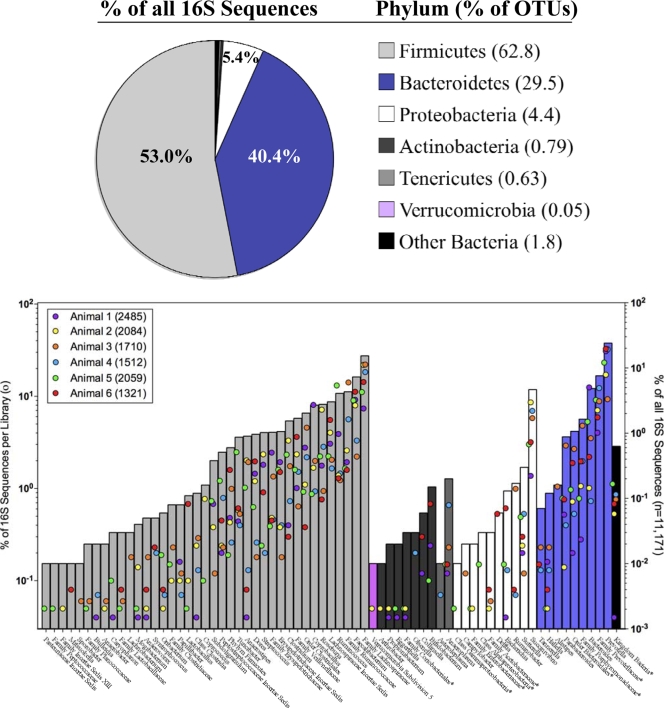
FIG. 3.
Phylum-level distribution of bacterial sequences from six beef feedlot cattle. Asterisks indicate unclassified members of the named taxon.
The GIT community of beef feedlot cattle characterized in this study was found to share many taxa with the bovine GIT community described for dairy cattle (1, 11, 14), although the relative abundances of the major bacterial groups differed considerably. The fecal microbiota of beef cattle were dominated by members of the Firmicutes, with 62.8% of the OTUs belonging to this taxonomic group (Fig. 3). Bacteroidetes (29.5% of the OTUs) and Proteobacteria (4.4% of the OTUs) were also represented in feces (Fig. 3). A total of seven phyla were found in our six animals.
Total estimated species richness values (Chao) for each of the six animals were 372, 600, 1,393, 526, 612, and 320 (Table 1). These cattle richness numbers are higher than those observed for three human subjects (164, 332, and 297) (2). The mean of Chao pairwise estimates of shared richness between any two of the six cattle fecal libraries was 230.
Our findings, in addition to those from pyrosequencing studies (1), identify a core set of bovine GIT bacterial taxa, including the Bacteroidetes Prevotella and Bacteroides; the Firmicutes Faecalibacterium, Ruminococcus, Roseburia, and Clostridium; and the proteobacterium Succinovibrio (Fig. 1). These genera are consistently identified in bovine feces and likely compose part of the bovine resident microbiota. Although the potential exists for culture-independent methods to reveal minority microbial community members, 16S rRNA gene sequencing in dairy (1, 11) and beef cattle supports the list of core taxa identified using culture-based methods.
Comparisons between our data set and recent studies done with dairy cattle (1, 11, 12) suggest that although beef and dairy cattle share many of the same major bacterial groups, the relative abundances of these groups in beef and dairy cattle may differ, and there may be differences between the two groups in the compositions of minority community members. The most common genus in beef cattle from our study was Prevotella, representing 24% of the total number of sequences evaluated. In comparison, Dowd et al. (1) found that Prevotella spp. represented only 5.5% of the total 16S genes sequenced from 20 dairy cattle, and Prevotella was not listed in the top 10 most frequently occurring OTUs in either of the studies from McGarvey et al. (11, 12). Likewise, Clostridium represented only 1.5% of the total beef sequences but 19% of the dairy pyrosequences (1). There were a number of bacterial sequences present in the beef cattle sequences but not reported in the dairy sequences, including Arthrobacter, Asteroleplasma, Bifidobacterium, Collinsella, Delftia, Eggerthella, Lactobacillus, Mitsuokella, Olsenella, and Propionibacterium (1, 11), although a number of these genera have been cultured from dairy animals in the past. It must be noted that all of these sequencing studies examined only a small number of animals, and each method has limitations which affect interpretation of the results. The full-length sequencing performed as part of this beef cattle study and two dairy studies (11, 12) relies on a PCR step which can potentially affect the relative numbers of each taxon observed due to PCR bias, while the pyrosequeincg method used in the 20-animal dairy study suffers from artifacts that potentially affect taxonomic assignment and richness estimates due to short read lengths and potential biases in evenness (how many of each group) due to primer and template mismatches (3). Nonetheless, these studies indicate that there may be fundamental differences between the gastrointestinal communities of beef and dairy cattle, they provide a comprehensive examination of the communities present in the specific animals tested, and they serve to provide important baseline information for further studies examining various factors which can impact cattle gastrointestinal communities.
The taxonomic information generated by deep sequencing of beef cattle feces revealed considerable animal-to-animal variation in the operational taxonomic unit (OTU) composition of the individual libraries (Fig. 1). The OTU designation facilitates an analysis of the community data without forcing the assignment of sequences into an incomplete and imperfect bacterial taxonomic system. It relies on DNA sequence similarity to assign sequences to a particular OTU defined by the level of DNA sequence similarity. In total, 1,906 OTUs (97% OTU designation) were identified in the six libraries. Of these, only 24 OTUs (1.2%) (comprising 1,253 [11.2%] of sequences) were present in all six libraries, while 1,348 OTUs (69%) were found only in individual libraries. Of these, 1,064 OTUs (77%) were unique, represented by a solitary clone (range of 3% to 29% of the total clones from each individual animal). These data hint at considerable animal-to-animal variation in bacterial community structure at the species level that cannot be readily attributed to breed, gender, age, macroecologic factors such as weather conditions, or diet, given that the animals in this study were controlled for these variables, and support the conclusions of Manter et al. (10) that pooling samples can obscure rare phylotypes.
Our results from beef cattle suggest that there may be differences in the bacterial community members present in the GIT of each individual animal that cannot be attributed to diet, breed, gender, age, or macroecologic factors such as weather and suggest the need for the high-resolution community sequencing of much larger numbers of animals before “core” minority community members can be identified. Considering the limited nature of the community surveys to date and all of the genetic, management, geographic, and temporal factors that can contribute to the composition of GIT microbiota, much work remains before we are able to understand and predict the community composition of any individual animal.
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA clones used in this study may be found under GenBank accession no. FJ672948 to FJ674268 and FJ675665 to FJ685516.
Acknowledgments
We thank R. Mlejnek, S. Simcox, R. Lee, and S. Fryda-Bradley for technical assistance; J. Rosch for secretarial assistance; J. McGarvey and J. Wells for critical reading; Randy Bradley, Phil Anderson, and William Dailey for IT support; and the MARC cattle crew for animal care.
Support for this study was provided by the USDA, ARS, National Program 108.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Dowd, S. E., T. R. Callaway, R. D. Wolcott, Y. Sun, T. McKeehan, R. G. Hagevoort, and T. S. Edrington. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelbrektson, A., V. Kunin, K. C. Wrighton, N. Zvenigorodsky, F. Chen, H. Ochman, and P. Hugenholtz. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and eveness. ISME J. 4:642-647. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 5.Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 6.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 7.Johnson, K. A., and D. E. Johnson. 1995. Methane emissions from cattle. J. Anim. Sci. 73:2483-2492. [DOI] [PubMed] [Google Scholar]
- 8.Kanagawa, T. 2003. Bias and artifacts in multitemplate polymerase chain reaction. J. Biosci. Bioeng. 96:317-323. [DOI] [PubMed] [Google Scholar]
- 9.Liu, Z., T. Z. DeSantis, G. L. Andersen, and R. Knight. 2008. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 36:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manter, D. K., T. L. Weir, and J. M. Vivanco. 2010. Negative effects of sample pooling on PCR-based estimates of soil microbial richness and community structure. Appl. Environ. Microbiol. 76:2086-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGarvey, J. A., W. G. Miller, S. Sanchez, and L. Stanker. 2004. Identification of bacterial populations in dairy wastewaters by use of 16S rRNA gene sequences and other genetic markers. Appl. Environ. Microbiol. 70:4267-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGarvey, J. A., S. W. Hamilton, E. J. DePeters, and F. M. Mitlehner. 2010. Effect of dietary monensin on the bacterial population structure of dairy cattle colonic contents. Appl. Microbiol. Biotechnol. 85:1947-1952. [DOI] [PubMed] [Google Scholar]
- 13.Nurmi, E., and M. Rantala. 1973. New aspects in Salmonella infections in broiler production. Nature 241:210-211. [DOI] [PubMed] [Google Scholar]
- 14.Ozutsumi, Y., H. Hayashi, M. Sakamoto, H. Itabashi, and Y. Benno. 2005. Culture-independent analysis of fecal microbiota in cattle. Biosci. Biotechnol. Biochem. 69:1793-1797. [DOI] [PubMed] [Google Scholar]
- 15.Schloss, P. D., and J. Handelsman. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, T. P. L., R. A. Godtel, and R. T. Lee. 2000. PCR-based setup for high-throughput cDNA library sequencing on the ABI 3700 automated DNA sequencer. Biotechniques 29:698-700. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]