Abstract
Previously isolated dissimilatory perchlorate-reducing bacteria (DPRB) have been primarily affiliated with the Betaproteobacteria. Enrichments from the cathodic chamber of a bioelectrical reactor (BER) inoculated from creek water in Berkeley, CA, yielded a novel organism most closely related to a previously described strain, WD (99% 16S rRNA gene identity). Strain VDYT has 96% 16S rRNA gene identity to both Magnetospirillum gryphiswaldense and Magnetospirillum magnetotacticum, and along with strain WD, distinguishes a clade of perchlorate-reducing Magnetospirillum species in the Alphaproteobacteria. In spite of the phylogenetic location of VDYT, attempted PCR for the key magnetosome formation genes mamI and mamL was negative. Strain VDYT was motile, non-spore forming, and, in addition to perchlorate, could use oxygen, chlorate, nitrate, nitrite, and nitrous oxide as alternative electron acceptors with acetate as the electron donor. Transient chlorate accumulation occurred during respiration of perchlorate. The organism made use of fermentation end products, such as acetate and ethanol, as carbon sources and electron donors for heterotrophic growth, and in addition, strain VDYT could grow chemolithotrophically with hydrogen serving as the electron donor. VDYT contains a copy of the RuBisCo cbbM gene, which was expressed under autotrophic but not heterotrophic conditions. DNA-DNA hybridization with strain WD confirmed VDYT as a separate species (46.2% identity), and the name Magnetospirillum bellicus sp. nov. (DSM 21662, ATCC BAA-1730) is proposed.
Dissimilatory perchlorate-reducing bacteria (DPRB) use perchlorate as a terminal electron acceptor during respiration, reducing it completely to chloride. As a consequence, bioremediation of perchlorate has been identified as the most effective means of treating this harmful contaminant (10), which, due to historically unregulated release into the environment, has become widespread (13, 20, 41). Fortunately, DPRB are ubiquitous and can be readily isolated from a variety of environments (1, 10, 11, 39, 44), and a key gene in the pathway, the chlorite dismutase (cld) gene, has been broadly detected (6). Much has been revealed about the biochemistry and genetics of microbial perchlorate reduction through the study of several model organisms, including Dechloromonas aromatica and Dechloromonas agitata, by a variety of groups (5, 6, 8, 9, 17, 28, 29, 34, 35, 38, 47, 51, 56, 57).
Less is known about the variation in physiology between these organisms or the evolution of the perchlorate reduction metabolism, highlighting a need for further isolation and characterization of pure cultures. The lack of congruence between phylogenetic trees of cld and the 16S rRNA gene among tested DPRB suggests that the metabolism may be the result of horizontal gene transfer (6). Given that various elements of the pathway may be mobile, it is not unreasonable to expect that organisms with a wide phylogenetic diversity could acquire the ability to reduce perchlorate. As more varied enrichment conditions are tested (2, 39), sometimes as a result of novel bioreactor development for perchlorate treatment (38, 40, 45), the true phylogenetic diversity of DPRB is becoming apparent, supporting the hypothesis that the metabolism may be widespread within the tree of life, similar to other respiratory processes, such as the reduction of sulfate, Fe(III), and nitrate.
Although perchlorate has been primarily regarded as an anthropogenic contaminant, a variety of studies have indicated that perchlorate occurs naturally (29-31, 34), which provides a possible explanation for the selective pressure behind the evolution of perchlorate reduction genes. As more is understood about the chlorine redox cycle on earth, knowledge about the diversity of organisms capable of interacting with the various oxyanions of chlorine is becoming more important. Here, we report the characterization of a unique DPRB in the Alphaproteobacteria. Strain VDYT was isolated from the surface of a working electrode in an active perchlorate-reducing bioelectrical reactor (BER) that was inoculated with water from Strawberry Creek on the University of California, Berkeley, campus (40). This is only the second described DPRB in the Alphaproteobacteria, the other being the closely related strain WD (26), and these strains compose a unique clade of perchlorate-reducing organisms in the genus Magnetospirillum.
MATERIALS AND METHODS
Media and culturing conditions.
Culturing medium was prepared as described previously (8). All cultures were grown and evaluated using freshwater 30 mM bicarbonate-buffered basal medium (pH 6.8) under an N2-CO2 (80%/20%) headspace, except for testing the optimum pH for growth, where a 10 mM phosphate-buffered medium was utilized. NaH2PO4 and Na2HPO4 were added in appropriate concentrations to establish stable pHs at 6.0, 6.5, 6.8, 7.0, 7.2, and 7.5. The medium was boiled, cooled, and dispensed under an N2 headspace. Omitting sodium bicarbonate, all other medium components were identical with bicarbonate-buffered medium. Electron acceptors and donors were added separately from sterile, anoxic aqueous stock solutions. For testing of salinity growth optima, NaCl was added to the culture medium from a 5 M sterile, anoxic aqueous stock solution. Acetate and perchlorate (10 mM) were used for testing of alternative electron donors and acceptors, respectively, and together for testing optimum growth temperature, pH, and salinity.
Scanning electron microscopy (SEM).
Cells for electron microscopy were grown anaerobically in freshwater basal medium amended with acetate (10 mM) and perchlorate (10 mM) and prepared and imaged as described previously (42).
16S rRNA gene sequencing and analysis.
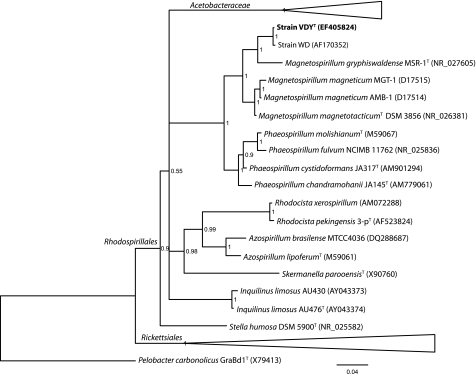
Isolation of genomic DNA, 16S rRNA gene-specific PCR, and sequencing of PCR products were completed as described previously (11, 40). 16S rRNA gene sequences were aligned with Muscle 3.6 (14), and Bayesian analysis of 16S rRNA gene phylogeny was completed with MrBayes 3.2 (19, 33). The program was run until the standard deviation of the split frequencies was stabilized below 0.01, in this case for 241,000 generations, with a sample frequency of 1,000. The first 25% of the samples were discarded for accurate estimation of the posterior probability distribution of the summary tree. The organisms in the Acetobacteraceae that were used were Acetobacter pasteurianusT (X71863), Acetobacter cerevisiaeT (AJ419843), Gluconobacter oxydansT (X73820), Gluconobacter cerinusT (X80775), Paracraurococcus ruberT (D85827), Gluconacetobacter oboediensT (AJ001631), Gluconacetobacter europaeusT (Z21936), Gluconacetobacter hanseniiT (X75620), Roseococcus thiosulfatophilusT (X72908), Acidomonas methanolicaT (D30770), Acidocella facilisT (D30774), Acidiphilium aminolyticaT (D30771), and Acidiphilium cryptumT (D30773); the organisms in the Rickettsiales that were used were Rickettsia honeiT (AF060705), Rickettsia australisT (L36101), Rickettsia aeschlimanniiT (U74757), Wolbachia pipientis (AF179630), and Holospora obtusa (X58198). For all other accession numbers, see Fig. 2.
FIG. 2.
Phylogenetic placement of Dechlorospirillum among the Rhodospirillales in the Alphaproteobacteria according to 16S rRNA gene analysis. The bar represents 0.04 expected change per site; posterior probability values are indicated at the nodes.
DNA analysis.
DNA-DNA hybridization was performed commercially at the DSMZ. DNA was purified and hybridized as described previously (10, 16, 27). The G+C content was determined by high-performance liquid chromatography (HPLC) at the DSMZ as previously outlined (25).
Fatty acid determination.
Cells were grown in freshwater basal medium, harvested by centrifugation, and resuspended in 1 ml buffer. The whole-cell fatty acid content was determined using the Sherlock Microbial Identification System (v. 4.5; MIDI, Newark, DE) according to the manufacturer's protocol.
mamI/mamL PCR.
Genomic DNA was isolated using the Power Soil Kit (MoBio) as directed by the manufacturer's protocol. Primers specific to mamI and mamL (27) were used to amplify these genes (Table 1). The following PCR parameters were used: 95°C for 5 min, then 32 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a final incubation at 72°C for 10 min. Reaction mixtures (50 μl) contained 3 μl (100 ng) genomic DNA as the template, 1 μl (20 μM) each primer, and 1.25 U Taq polymerase (TaKaRa). Genomic DNA from Magnetospirillum magneticum AMB-1 was used as the positive control.
TABLE 1.
Primers used in this study
| Primer name | Gene target | Sequence |
|---|---|---|
| 5EcoI | mamI | GGCGAATTCATGCCAAGCGTGATTTTCGG |
| 3SpeI | mamI | GGCACTAGTTCAACCATCGATGTCAGGG |
| 5EcoL | mamL | GGCGAATTCATGGTAAGATTGATCGGATC |
| 3SpeL | mamL | GGACTAGTTCAGCGCTTGATGACGATG |
| 595f | RuBisCo cbbL | GACTTCACCAAAGACGACGA |
| 1387r | RuBisCo cbbL | TCGAACTTGATTTCTTTCCA |
| cbbMf | RuBisCo cbbM | TCATCAARCCSAARCTSGGCCTGCGTCCC |
| cbbMr | RuBisCo cbbM | MGAGGTGACSGCRCCGTGRCCRGCMCGRTG |
RNA isolation.
Cultures were grown in freshwater basal medium with an N2-CO2 (80%/20%) headspace under autotrophic (H2 as the sole electron donor; CO2 as the carbon source) and heterotrophic (acetate as the electron donor and carbon source) conditions with perchlorate as the sole terminal electron acceptor. Total RNA was isolated by trapping cells on a nylon membrane filter (Nalgene) via vacuum filtration, immersing the filter in 1 ml Trizol reagent (Sigma-Aldrich) in Lysing Matrix E (MP Bio) tubes, and bead beating for 30 s. After a 5-min room temperature incubation, 200 μl chloroform (Fisher Scientific) was added, and the tubes were shaken for 30 s and allowed to incubate at room temperature for a further 10 min. The tubes were then centrifuged (13,000 rpm) at 4°C for 15 min, and the supernatant was transferred to new tubes to which 500 μl diethyl pyrocarbonate (DEPC)-H2O was added. The tubes were mixed, supplemented with 1 ml isopropanol (Sigma-Aldrich), mixed again, and allowed to incubate at room temperature for 10 min. The tubes were then centrifuged (13,000 rpm) at 4°C for 15 min, the supernatant was decanted and replaced with 1 ml 75% ethanol, and the tubes were vortexed and centrifuged (13,000 rpm) again at 4°C for 5 min. After the supernatant was decanted, the pellets were allowed to air dry for 10 min and resuspended in 100 μl DEPC-H2O. The resuspended nucleic acids were then further purified using the AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer's protocol, including the optional DNase I steps (E1 to E4).
cDNA preparation.
Total RNA (7 μl) from the isolation protocol was added to 1 μl of 3 μM cbbM-specific primers (15, 43) (Table 1), and 4 μl 2.5 mM each deoxynucleoside triphosphate (dNTP) (Invitrogen); heated at 65°C for 5 min; and quickly chilled on ice. To this, 4 μl of 5× First-Strand buffer (Invitrogen), 2 μl 0.1 M dithiothreitol (DTT) (Invitrogen), and 1 μl RNAseOUT (Invitrogen) were added, mixed gently, and incubated at 42°C for 2 min. Superscript II Reverse Transcriptase (0.5 μl; Invitrogen) was added, mixed gently, and incubated at 42°C for 50 min and then again at 70°C for 15 min.
RuBisCo cbbM and cbbL PCR.
Primers designed to amplify form I (cbbL) and form II (cbbM) of the RuBisCo large subunit were utilized as previously described (15, 43) (Table 1). Amplifications involved the following parameters: 95°C for 4 min, then 35 cycles of 95°C for 45 s, 57°C for 45 s, and 72°C for 45 s, followed by a final incubation at 72°C for 10 min. The PCR products were visualized by agarose gel electrophoresis and imaged using a NucleoTech gel-imaging system.
Analytical methods.
All experimental analyses were performed in triplicate to ensure reproducibility, and the results are expressed as the mean of these determinations. The concentrations of perchlorate and chlorate in cultures were determined using ion chromatography as previously described (9). Cell growth in active cultures was monitored by the optical density at 600 nm and by the total direct cell count using phase-contrast microscopy (Petroff-Hausser counting chamber; 0.02-mm depth). Samples collected for counting were immediately fixed in 0.2 μm filter-sterilized formaldehyde (final concentration, 3.7%).
RESULTS
Phylogenetic characterization.
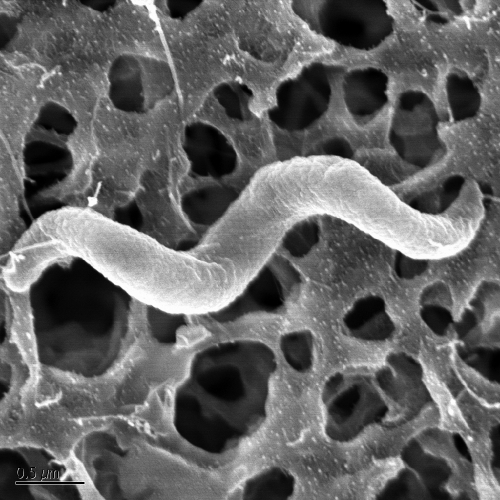
Strain VDYT was spirillum shaped, ∼0.5 μm by ∼3 μm (Fig. 1), typically with two helical rotations, though cells were observed with as few as one or as many as five, and were highly motile. Phylogenetic analysis of the 16S rRNA gene sequence indicated that it was most closely related to the previously described strain WD (99% 16S rRNA gene sequence similarity) (26) and placed it within the family Rhodospirillaceae of the Alphaproteobacteria (Fig. 2). DNA-DNA hybridization between strain VDYT and strain WD showed 46.2% similarity, well below the 70% similarity required to distinguish the two strains as separate species. The next closest organism to strain VDYT based on 16S rRNA gene identity was Magenetospirillum gryphiswaldense MSR-1, with 96% sequence similarity between the two.
FIG. 1.
SEM of strain VDYT. Bar, 0.5 μm.
Magnetosome formation.
To further examine the distinction between strain VDYT and Magnetospirillum species, it was tested for magnetosome production via visual inspection of response to a magnetic field and molecular probing. Strain VDYT was unable to produce magnetosomes in Magnetospirillum-specific medium under microaerophilic conditions (21) or in high ferrous-iron concentrations (5 to 10 mM FeCl2) in its typical culture medium (data not shown). mamI and mamL are genes required for magnetosome formation in Magnetospirillum strains AMB-1, MS-1, and MSR-1 (32). PCR amplification using AMB-1-specific primers for these genes (27) (and predicted by BLAST to also be specific for M. gryphiswaldense mamI and mamL genes) was negative in VDYT compared to AMB-1-positive controls (data not shown).
Fatty acid profiles.
VDYT contained a fatty acid profile very similar to that of WD when grown on perchlorate (10 mM) and acetate (10 mM) (Table 2). The major fatty acid present for both was 18:1ω7c. Secondary fatty acids were the summed feature 16:1ω7c/15 iso 2OH and 16:0. These three fatty acids made up 87.12% and 89.80% of the fatty acid content of strains VDYT and WD, respectively, and were the only fatty acids present above 5% of the total.
TABLE 2.
Comparative fatty acid compositions of strains VDYT and WD
| Fatty acid type | Contenta |
|
|---|---|---|
| VDYT | WD | |
| 12:0 | 3.25 | 3.14 |
| 14:0 | 0.37 | 0.43 |
| 16:1ω7c | ||
| 16:0 | 12.01 | 9.87 |
| 16:0 3OH | 0.75 | 0.69 |
| 18:1ω9c | ||
| 18:1ω7c | 64.64 | 68.51 |
| 18:1 | ||
| 18:0 | 1.89 | 0.80 |
| 11 methyl 18:1ω7c | 0.83 | 1.36 |
| 18:1 2OH | 1.23 | |
| 18:0 3OH | 0.33 | |
| Summed features | ||
| 14:0 3OH/16:1 iso i | 3.10 | 2.86 |
| 16:1ω7c/15 iso 2OH | 10.47 | 11.42 |
| 18:1c11/t9/t6 | ||
| 19:1 2OH/cy 19:0 | ||
| Unknown | 3.59 | 3.28 |
Values are given as percentages of total membrane content. Boldface indicates values for the major fatty acid.
Growth characteristics.
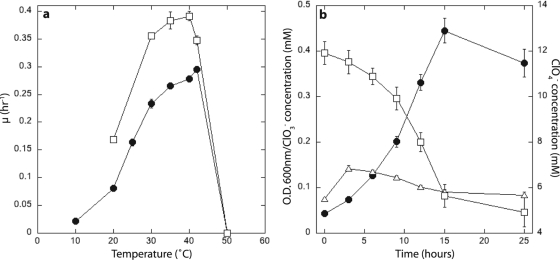
Strain VDYT grew between 10 and 42°C, but not at 50°C, with an optimum growth rate at 42°C (0.29 h−1) at pH 6.8 (Table 3) . It could grow with NaCl concentrations up to 1.5%, but not at 2%, and showed minimal growth defects in medium containing up to 40 mM perchlorate or chlorate. VDYT was non-spore forming, as indicated by the inability of cultures to grow in new medium after incubation at 80°C for 3 min, and was an obligate respiratory organism, being unable to ferment hexoses, Casamino Acids, or yeast extract components. Strain VDYT used electron acceptors similar to those of strain WD and other known DPRB (1, 8, 26), but a wider range than the previously characterized Magnetospirillum species, namely, oxygen, perchlorate, chlorate, and nitrate, with the additional ability to use nitrite and nitrous oxide (Table 3). VDYT had a higher growth rate on nitrate than on perchlorate (Fig. 3 a). Like other Magnetospirillum species, strain VDYT could oxidize a range of fermentation end products, including acetate, lactate, and ethanol, but no sugars (Table 3). VDYT could also utilize the electron donors FeCl2, H2, and the humic substances analog anthrohydroquinone-2,6-disulfonate (AHDS). Chlorate transiently accumulated during growth on perchlorate, but after 4 h, chlorate was removed concomitantly with perchlorate (Fig. 3b).
TABLE 3.
Physiological characteristics of strain VDYT compared to those of closely related Magnetospirillum strainsa
| Characteristic | Valueb |
||||||
|---|---|---|---|---|---|---|---|
| VDYT | WD | M. gryphiswaldense | M. magnetotacticum | M. magneticum | J10 | CC-26 | |
| Cell type | Spirillum | Spirillum | Spirillum | Spirillum | Spirillum | Spirillum | Spirillum |
| Size (μm) | ∼0.5 by ∼3 | ∼0.2 by ∼7 | 0.7 by ∼4 | ∼0.4 by ∼4 | ∼0.5 by >3 | ∼0.5 by ∼4 | ∼0.5 by 2-5 |
| Motility | + | + | + | + | + | + | + |
| Temp range (°C) | <10-42 | 25-37 | ND | 15-37 | ND | ND | ND |
| Temp optima (°C) | 42 | 35 | 28-34 | 30 | 28-34 | ND | ND |
| pH range | 6.0-7.5 | 6.5-7.5 | ND | ND | 5.0-8.2 | ND | ND |
| pH optima | 6.8 | 7.2 | 7.0-7.5 | 7.0-7.5 | 7.0-7.5 | ND | ND |
| NaCl tolerance (%) | 1.5 | 1 | ND | <1 | ND | ND | ND |
| Spore forming | − | − | ND | ND | ND | ND | ND |
| Fermentative | − | − | − | − | − | ND | ND |
| G+C content (%) | 64.8 | 66.1 | 71 | 66.4 | 65.1 | ND | ND |
| RuBisCo cbbM | + | ND | + | + | + | + | ND |
| cld | + | + | − | + | − | ND | ND |
| Magnetotactic | − | − | + | + | + | − | − |
| Electron acceptors | |||||||
| Oxygen | + | + | + | + | + | + | + |
| Perchlorate | + | + | ND | ND | ND | − | ND |
| Chlorate | + | + | ND | ND | ND | − | ND |
| Nitrate | + | + | − | +c | + | + | + |
| Nitrite | + | ND | ND | ND | ND | ND | ND |
| N2O | + | ND | ND | ND | ND | ND | ND |
| Sulfate | − | − | ND | ND | ND | − | ND |
| Thiosulfate | − | − | ND | ND | ND | ND | ND |
| Selenate | − | − | ND | ND | ND | ND | ND |
| Arsenate | − | ND | NA | ND | ND | ND | ND |
| Fumarate | − | − | ND | ND | ND | − | ND |
| Malate | − | − | ND | ND | ND | ND | ND |
| Fe(III) NTAd | − | − | ND | ND | ND | ND | ND |
| AQDS | − | − | ND | ND | ND | ND | ND |
| Electron donors | |||||||
| Acetate | + | + | + | + | + | + | + |
| Propionate | + | + | ND | − | + | ND | + |
| Isobutyrate | + | + | ND | − | ND | ND | + |
| Butyrate | + | + | ND | − | + | ND | + |
| Valerate | + | + | ND | ND | ND | ND | − |
| Formate | − | − | ND | − | ND | ND | − |
| Methanol | − | − | ND | − | ND | ND | − |
| Ethanol | + | + | ND | − | ND | ND | − |
| Catechol | − | − | ND | − | ND | ND | − |
| Glycerol | − | − | ND | − | ND | ND | − |
| Benzoate | − | − | ND | − | ND | ND | + |
| Pyruvate | + | ND | + | + | + | ND | + |
| Citrate | − | − | ND | − | ND | ND | − |
| Succinate | + | + | + | + | + | ND | + |
| Lactate | + | + | + | + | + | ND | + |
| Glucose | − | − | ND | − | ND | ND | − |
| Sucrose | − | ND | − | ND | ND | ND | − |
| Fructose | − | ND | ND | − | ND | ND | − |
| Maltose | + | ND | − | − | ND | ND | − |
| Casamino Acids | + | + | ND | ND | ND | ND | ND |
| Fumarate | + | + | ND | + | + | ND | + |
| Malate | + | + | + | + | + | ND | + |
| Hydrogen | + | − | ND | ND | ND | ND | ND |
| FeCl2 | + | + | ND | ND | ND | − | ND |
| AHDS | + | ND | ND | ND | ND | ND | ND |
| H2S | − | ND | + | ND | ND | + | ND |
| Thiosulfate | ND | ND | + | ND | ND | + | ND |
| Methane | − | ND | ND | ND | ND | ND | ND |
| Urea | − | ND | ND | ND | ND | ND | ND |
Data for the table were compiled from references 3, 4, 6, 7, 16-18, 22-24, 26, 35, and 37 and the IMG/M website (for G+C content of M. magnetotacticum and M. magneticum).
+, present or, with regard to electron donors/acceptors, positive for utilization (for VDYT and WD, this was indicated by three successful transfers on the substrate); −, absent or not utilized; ND, not determined.
Nitrate was coreduced with oxygen.
NTA, nitrilotriacetic acid.
FIG. 3.
(a) Comparative temperature-dependent growth optima of VDYT on nitrate (open squares) and perchlorate (closed circles). (b) Transient chlorate accumulation by strain VDYT while growing on perchlorate. Closed circles, optical density at 600 nm (O.D. 600 nm); open squares, ClO4−; open triangles, ClO3−. The error bars represent the standard deviations of triplicate samples.
Chemolithotrophy.
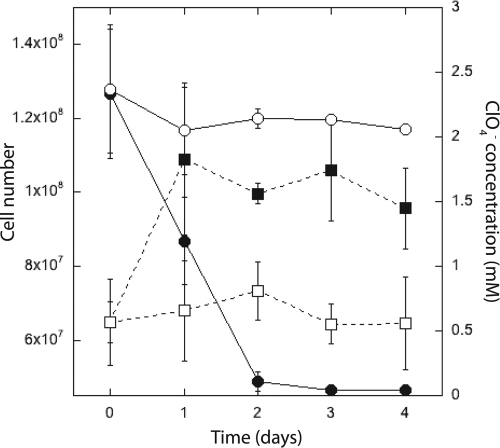
Strain VDYT could grow under both mixotrophic and autotrophic conditions with hydrogen coupled to perchlorate reduction, which has been described for only three other pure cultures (39, 45). Figure 4 shows chemolithotrophic growth of strain VDYT with hydrogen as the electron donor and perchlorate as the sole terminal electron acceptor in the bicarbonate-buffered medium described above. Cells were grown in electron donor-limited medium (10 mM acetate and 15 mM perchlorate) and allowed to reach late stationary phase to reduce the possibility of acetate carryover. Growth was monitored by direct cell counts. Cells reduced 2.2 mM perchlorate over 48 h relative to a no-electron-donor control and increased in number from 6.5 × 107 (±1.2 × 107) to 1.1 × 108 (±2.1 × 107) cells·ml−1 in the first 24 h (Fig. 4). The control, from which the electron donor (H2) was omitted, showed no significant growth. Interestingly, although the cultures entered stationary phase after 24 h of incubation, the organisms continued to respire the perchlorate to completion over 48 h.
FIG. 4.
Chemolithoautotrophic growth of strain VDYT (closed squares) compared to that of a control with no added electron donor (open squares), showing concomitant perchlorate reduction (closed circles, experimental; open circles, control). The error bars represent the standard deviations of triplicate samples.
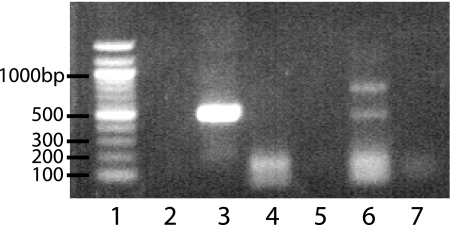
Genetic screening demonstrated the presence of the RuBisCo cbbM large-subunit (form II) gene (Fig. 5, lane 3), but not the cbbL (form I) gene (data not shown), and differential expression of cbbM mRNA under heterotrophic and autotrophic conditions. cbbM was expressed when cells were grown on hydrogen and perchlorate as the sole electron donor and acceptor, respectively, under an N2-CO2 (80%/20%) headspace (Fig. 5, lane 6). In contrast, no cbbM expression (Fig. 5, lane 4) was observed in cells grown under identical conditions with acetate as the electron donor and carbon source. Since upregulation of RuBisCo under autotrophic conditions would be expected if this gene were present and differentially expressed, these data support the involvement of RuBisCo in carbon fixation by strain VDYT.
FIG. 5.
Reverse transcription (RT)-PCR of the RuBisCO cbbM gene. Strain VDYT contains a genomic copy of the cbbM gene (lane 3), that is differentially expressed during chemolithoautotrophic growth (lane 6), but not chemoorganoheterotrophic growth (lane 4). Lane 1, 100-bp ladder; lane 2, negative control (water). Lanes 5 and 7 are RNA controls to discount genomic DNA contamination.
DISCUSSION
These studies examined the phylogeny and physiology of strain VDYT and included molecular probing to assess the presence of magnetosome-forming genes and those responsible for autotrophic growth. PCR for the mamL and mamI genes (essential for magnetosome formation in other Magnetospirillum species) was negative, although this does not rule out the possibility that homologs undetected by these primers exist. However, strain VDYT was unable to form magnetosomes in the typical medium used for testing Magnetospirillum species, nor was it able to form magnetosomes in other medium with ferrous iron present. The inability of an organism closely related to Magnetospirillum species to form magnetosomes is not unprecedented. Strain WD and four other nonmagnetotactic Magnetospirillum strains, including J10 and CC-26 (Table 3), have been reported (17, 26, 37), and a non-magnetosome-forming strain of M. gryphiswaldense was described which contained a large genomic lesion corresponding to loss of the magnetosome island (36). Similarly, it is also not unprecedented for a member of the genus Magnetospirillum to contain perchlorate reduction genes. M. magnetotacticum contains a homolog of the cld gene (6), but it lacks all other perchlorate reduction genes. The cld gene sequences of strains VDY, WD, and MS-1 are more similar to each other than those of other strains containing cld, but this gene is not found in other sequenced Magnetospirillum species (6; J. C. Thrash, K. Byrne-Bailey, and J. D. Coates, unpublished results). These data and those presented in this study suggest that magnetosome-related and perchlorate reduction genes are poor taxonomic markers.
VDYT had some other notable physiological and genetic features. The temporary chlorate accumulation in strain VDYT indicates a differentiation between the initial perchlorate reduction step in strain VDYT and other DPRB. A recent study quantified the kinetic difference in another chlorate-accumulating DPRB, Azospira strain PCC (12), and found that the chlorate accumulation by strain PCC was consistent with competitive inhibition of chlorate reduction by perchlorate. Two other recently reported strains also showed similar chlorate accumulation (39), generally around 20% of the initial perchlorate concentration, whereas in strain VDYT, the accumulation was closer to 1%. VDYT, therefore, represents a third class of perchlorate reduction physiology intermediate between no accumulation (in strains like D. agitata [8]) and the considerable accumulation of chlorate seen in strains PCC, MP, and CR. Comparative analysis of the pcr genes and enzymes between organisms from these three classes could yield important information about the evolution of the perchlorate reduction pathway. The presence and differential expression of RuBisCo is enticing evidence for this autotrophy pathway in strain VDYT and is the first evidence for a specific mechanism of carbon fixation in DPRB. However, given that cells continued to respire after growth was completed, some form of energy storage by the organism cannot be ruled out.
Taxonomic conclusions.
The taxonomic characterization of strain VDYT has demonstrated that the strain shares many characteristics with strain WD and other Magnetospirillum species but is nevertheless distinct. Like all described Magnetospirillum species, VDYT is strictly respiratory, motile, and Gram negative and did not utilize the sugars tested. However, like strain J10, CC-26, or WD, strain VDYT did not form magnetosomes under the conditions tested and, furthermore, does not contain copies of the necessary magnetosome formation genes mamI and mamL with close homology to those found in AMB-1. Although these results are negative data and not sufficient to prove that magnetosome formation is impossible, they add to the physiological and genetic distinction between strain VDYT and other Magnetospirillum species. VDYT uses a wide range of electron acceptors, similarly to strain WD and in contrast to other Magnetosprillum species. Since it is only 4% divergent at the 16S rRNA gene from M. gryphiswaldense, VDYT may not be considered a novel genus, but this divergence is enough for it to be considered a novel species. The low DNA-DNA hybridization value between this organism and its closest phylogenetic relative, strain WD, also distinguishes strain VDYT as a separate species from strain WD. Thus, based on physiological and phylogenetic characteristics, strain VDYT should be considered a distinct species of Magnetospirillum.
Description of Magnetospirillum bellicus sp. nov.
Magnetospirillum bellicus (bel′li.cus. L. adj. bellicus, martial, warlike, referring to the propensity of the organism to degrade perchlorate, a principal component of munitions and solid rocket fuel).
Spirillum-shaped cells, 0.5 by 3 μm, non-spore forming, nonfermentative, facultatively anaerobic. Cells are motile and may occur singly or in chains. Strictly respiring, complete oxidizer that oxidizes acetate with O2, ClO4−, ClO3−, NO3−, or N2O as the electron acceptor. Perchlorate and chlorate are completely reduced to Cl− with transient chlorate accumulation during growth on perchlorate. Cells can use substrates such as carboxylic acids—including acetate and propionate—and ethanol for chemoorganotrophic growth, as well as hydrogen for lithotrophic growth (Table 3 shows a complete list). Cells contain the RuBisCo cbbM gene. Optimum growth occurs at 42°C, pH 6.8, in freshwater basal medium with 0% NaCl. The G+C content of the genomic DNA of the type strain is 64.8 mol% (determined by HPLC). The type strain is VDYT (DSM 21662, ATCC BAA-1730) and was isolated from the cathodic chamber of an active perchlorate-reducing BER enrichment.
Acknowledgments
Support to J.D.C. for this research was from grant DACA72-00-C-0016 from the U.S. Department of Defense SERDP program.
We gratefully acknowledge Dennis A. Bazylinski for supplying M. gryphiswaldense; Arash Komeili, Meghan E. Byrne, and Dorothee Murat for assistance with physiological and genetic testing of magnetosome formation; and Jason E. Stajich and Thomas J. Sharpton for assistance with MrBayes.
Footnotes
Published ahead of print on 21 May 2010.
REFERENCES
- 1.Achenbach, L. A., R. A. Bruce, U. Michaelidou, and J. D. Coates. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 51:527-533. [DOI] [PubMed] [Google Scholar]
- 2.Balk, M., T. van Gelder, S. A. Weelink, and A. J. M. Stams. 2008. (Per)chlorate reduction by the thermophilic bacterium Moorella perchloratireducens sp. nov., isolated from underground gas storage. Appl. Environ. Microbiol. 74:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazylinski, D. A., and R. P. Blakemore. 1983. Denitrification and assimilatory nitrate reduction in Aquaspirillum magnetotacticum. Appl. Environ. Microbiol. 46:1118-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 5.Bender, K. S., S. M. O'Connor, R. Chakraborty, J. D. Coates, and L. A. Achenbach. 2002. Sequencing and transcriptional analysis of the chlorite dismutase gene of Dechloromonas agitata and its use as a metabolic probe. Appl. Environ. Microbiol. 68:4820-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender, K. S., M. R. Rice, W. H. Fugate, J. D. Coates, and L. A. Achenbach. 2004. Metabolic primers for detection of (per)chlorate-reducing bacteria in the environment and phylogenetic analysis of cld gene sequences. Appl. Environ. Microbiol. 70:5651-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blakemore, R. P., D. Maratea, and R. S. Wolfe. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 140:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruce, R. A., L. A. Achenbach, and J. D. Coates. 1999. Reduction of (per)chlorate by a novel organism isolated from a paper mill waste. Environ. Microbiol. 1:319-331. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri, S. K., S. M. O'Connor, R. L. Gustavson, L. A. Achenbach, and J. D. Coates. 2002. Environmental factors that control microbial perchlorate reduction. Appl. Environ. Microbiol. 68:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coates, J. D., and L. A. Achenbach. 2004. Microbial perchlorate reduction: rocket-fuelled metabolism. Nat. Rev. Microbiol. 2:569-580. [DOI] [PubMed] [Google Scholar]
- 11.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudley, M., A. Salmone, and R. Nerenberg. 2008. Kinetics of a chlorate-accumulating, perchlorate-reducing bacterium. Water Res. 42:2403-2410. [DOI] [PubMed] [Google Scholar]
- 13.Dyke, J. V., K. Ito, T. Obitsu, Y. Hisamatsu, P. K. Dasgupta, and B. C. Blount. 2007. Perchlorate in dairy milk. Comparison of Japan versus the United States. Environ. Sci. Technol. 41:88-92. [DOI] [PubMed] [Google Scholar]
- 14.Edgar, R. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elsaied, H., and T. Naganuma. 2001. Phylogenetic diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from deep-sea microorganisms. Appl. Environ. Microbiol. 67:1751-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geelhoed, J., R. Kleerebezem, D. Sorokin, A. Stams, and M. Van Loosdrecht. 2010. Reduced inorganic sulfur oxidation supports autotrophic and mixotrophic growth of Magnetospirillum strain J10 and Magnetospirillum gryphiswaldense. Environ. Microbiol. 12:1031-1040. [DOI] [PubMed] [Google Scholar]
- 17.Geelhoed, J., D. Sorokin, E. Epping, T. Tourova, H. Banciu, G. Muyzer, A. Stams, and M. Van Loosdrecht. 2009. Microbial sulfide oxidation in the oxic-anoxic transition zone of freshwater sediment: involvement of lithoautotrophic Magnetospirillum strain J10. FEMS Microbiol. Ecol. 70:54-65. [DOI] [PubMed] [Google Scholar]
- 18.Heyen, U., and D. Schuler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 19.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 20.Kirk, A. B., P. K. Martinelango, K. Tian, A. Dutta, E. E. Smith, and P. K. Dasgupta. 2005. Perchlorate and iodide in dairy and breast milk. Environ. Sci. Technol. 39:2011-2017. [DOI] [PubMed] [Google Scholar]
- 21.Komeili, A., H. Vali, T. J. Beveridge, and D. K. Newman. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl. Acad. Sci. U. S. A. 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maratea, D., and R. P. Blakemore. 2008. Aquaspirillum magnetotacticum sp. nov., a magnetic spirillum. Int. J. Syst. Evol. Microbiol. 31:452-455. [Google Scholar]
- 23.Matsunaga, T., T. Sakaguchi, and F. Tadokoro. 1991. Magnetite formation by a magnetic bacterium capable of growing aerobically. Appl. Microbiol. Biotechnol. 35:651-655. [Google Scholar]
- 24.Matsunaga, T., and N. Tsujimura. 1993. Respiratory inhibitors of a magnetic bacterium Magnetospirillum sp. AMB-1 capable of growing aerobically. Appl. Microbiol. Biotechnol. 39:368-371. [Google Scholar]
- 25.Mesbah, M., U. Premachandran, and W. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 26.Michaelidou, U., L. A. Achenbach, and J. D. Coates. 2000. Isolation and characterization of two novel (per)chlorate-reducing bacteria from swine waste lagoons, vol. 57. Kluwer Academic/Plenum Publishers, New York, NY.
- 27.Murat, D., A. Quinlan, H. Vali, and A. Komeili. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. U. S. A. 107:5593-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connor, S. M., and J. D. Coates. 2002. A universal immuno-probe for (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 68:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajagopalan, S., T. Anderson, S. Cox, G. Harvey, Q. Cheng, and W. A. Jackson. 2009. Perchlorate in wet deposition across North America. Environ. Sci. Technol. 43:616-622. [DOI] [PubMed] [Google Scholar]
- 30.Rajagopalan, S., T. A. Anderson, L. Fahlquist, K. A. Rainwater, M. Ridley, and W. A. Jackson. 2006. Widespread presence of naturally occurring perchlorate in high plains of Texas and New Mexico. Environ. Sci. Technol. 40:3156-3162. [DOI] [PubMed] [Google Scholar]
- 31.Rao, B., T. A. Anderson, G. J. Orris, K. A. Rainwater, S. Rajagopalan, R. M. Sandvig, B. R. Scanlon, D. A. Stonestrom, M. A. Walvoord, and W. A. Jackson. 2007. Widespread natural perchlorate in unsaturated zones of the southwest United States. Environ. Sci. Technol. 41:4522-4528. [DOI] [PubMed] [Google Scholar]
- 32.Richter, M., M. Kube, D. A. Bazylinski, T. Lombardot, F. O. Glockner, R. Reinhardt, and D. Schuler. 2007. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J. Bacteriol. 189:4899-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 34.Scanlon, B. R., R. C. Reedy, W. A. Jackson, and B. Rao. 2008. Mobilization of naturally occurring perchlorate related to land-use change in the southern high plains, Texas. Environ. Sci. Technol. 42:8648-8653. [DOI] [PubMed] [Google Scholar]
- 35.Schleifer, K.-H., D. Schuler, S. Spring, M. Weizenegger, R. Amann, W. Ludwig, and M. Kohler. 1991. The genus Magnetospirillum gen. nov. Description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst. Appl. Microbiol. 14:379-385. [Google Scholar]
- 36.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. H. Madkour, F. Mayer, R. Reinhardt, and D. Schuler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinoda, Y., Y. Sakai, M. Ue, A. Hiraishi, and N. Kato. 2000. Isolation and characterization of a new denitrifying spirillum capable of anaerobic degradation of phenol. Appl. Environ. Microbiol. 66:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thrash, J. C., and J. D. Coates. 2008. Review: direct and indirect electrical stimulation of microbial metabolism. Environ. Sci. Technol. 42:3921-3931. [DOI] [PubMed] [Google Scholar]
- 39.Thrash, J. C., J. Pollock, T. Torok, and J. D. Coates. 2010. Description of the novel perchlorate-reducing bacteria Dechlorobacter hydrogenophilus gen. nov., sp. nov., and Propionivibrio militaris, sp. nov. Appl. Microbiol. Biotechnol. 86:335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thrash, J. C., J. I. VanTrump, K. A. Weber, E. Miller, L. A. Achenbach, and J. D. Coates. 2007. Electrochemical stimulation of microbial perchlorate reduction. Environ. Sci. Technol. 41:1740-1746. [DOI] [PubMed] [Google Scholar]
- 41.Urbansky, E. T. 1998. Perchlorate chemistry: implications for analysis and remediation. Bioremed. J. 2:81-95. [Google Scholar]
- 42.Weber, K. A., D. B. Hedrick, A. D. Peacock, J. C. Thrash, D. C. White, L. A. Achenbach, and J. D. Coates. 2009. Physiological and taxonomic description of the novl autotrophic, metal oxidizing bacterium, Pseudogulbenkiania sp. strain 2002. Appl. Microbiol. Biotechnol. 83:555-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber, K. A., J. Pollock, K. A. Cole, S. M. O'Connor, L. A. Achenbach, and J. D. Coates. 2006. Anaerobic nitrate-dependent iron(II) bio-oxidation by a novel lithoautotrophic betaproteobacterium, strain 2002. Appl. Environ. Microbiol. 72:686-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolterink, A., S. Kim, M. Muusse, I. S. Kim, P. J. M. Roholl, C. G. van Ginkel, A. J. M. Stams, and S. W. M. Kengen. 2005. Dechloromonas hortensis sp. nov. and strain ASK-1, two novel (per)chlorate-reducing bacteria, and taxonomic description of strain GR-1. Int. J. Syst. Evol. Microbiol. 55:2063-2068. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, H. S., M. A. Bruns, and B. E. Logan. 2002. Chemolithoautotrophic perchlorate reduction by a novel hydrogen-oxidizing bacterium. Environ. Microbiol. 4:570-576. [DOI] [PubMed] [Google Scholar]