Abstract
A novel PCR primer system that targets a wide range of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase (PAH-RHDα) genes of both Gram-positive and Gram-negative bacteria was developed and used to study their abundance and diversity in two different soils in response to phenanthrene spiking. The specificities and target ranges of the primers predicted in silico were confirmed experimentally by cloning and sequencing of PAH-RHDα gene amplicons from soil DNA. Cloning and sequencing showed the dominance of phnAc genes in the contaminated Luvisol. In contrast, high diversity of PAH-RHDα genes of Gram-positive and Gram-negative bacteria was observed in the phenanthrene-spiked Cambisol. Quantitative real-time PCR based on the same primers revealed that 63 days after phenanthrene spiking, PAH-RHDα genes were 1 order of magnitude more abundant in the Luvisol than in the Cambisol, while they were not detected in both control soils. In conclusion, sequence analysis of the amplicons obtained confirmed the specificity of the novel primer system and revealed a soil type-dependent response of PAH-RHDα gene-carrying soil bacteria to phenanthrene spiking.
Polycyclic aromatic hydrocarbons (PAHs) are hydrophobic compounds composed of two or more fused aromatic rings. Although PAHs are ubiquitous in the environment (from natural oil seeps, brush fires, and plant derivatives), anthropogenic activities, such as disposal of coal-processing waste, mining accidents, petroleum wastes, and vehicle exhaust, have drastically increased their occurrence in the environment. The fate of PAHs in soil is of great interest due to their potential for bioaccumulation, persistence, transport, and toxicity. Microbe-driven aerobic degradation of PAHs is well documented (15-17). The diversity of PAH-degrading genes in soils is assumed to be huge, but the extent of diversity and how it is influenced by different soil types or their history and type of pollution are not yet fully explored. Knowledge of the genes coding for dioxygenase enzymes that catalyze the primary step of PAH degradation by incorporating molecular oxygen into the aromatic nucleus is an essential prerequisite to unraveling the contributions of microbial population networks to transformation, assimilation, and degradation of organic chemicals in soil. Recently, the complete genomes of several PAH-degrading bacteria became available and allowed new insights into degradative pathways (6, 18, 36). Organic pollutants also serve as nutrients for those microbes that have the appropriate genetic makeup to utilize them, resulting in their increased metabolic activity and abundance (4, 14). In the last decade, impressive progress was seen in techniques that allow cultivation-independent analysis of microbial communities and thus overcome the most severe limitations in studying microbial communities in natural habitats, namely, that only a rather small portion of microbes are accessible to standard cultivation conditions (1, 29). For more than a decade, cultivation-independent approaches have also been employed to unravel the responses of microbial communities in soils and sediments to PAH pollution. In all these studies, PCR amplification of PAH-degrading gene fragments from nucleic acids directly extracted from environmental samples was used to explore the abundance and diversity of PAH ring-hydroxylating dioxygenase (PAH-RHDα) genes (4, 8, 9, 13, 14, 22, 34, 37). Despite the known biases of PCR amplification from mixed templates, these techniques allow highly sensitive and specific detection even from minute amounts of nucleic acids. In order to select suitable primer systems, previously published primer systems were analyzed for their ranges of target sequences. The existing primer systems were found to have limitations, as they often target only a rather narrow range of sequences, e.g., nahAc- or phnAc-type sequences (21, 34) or only PAH-RHDα genes from Gram-negative bacteria (3, 13). In other studies, two-primer systems were used to target PAH-RHDα genes of both Gram-positive and Gram-negative bacteria (4, 37). Only one primer system targeting the Rieske gene fragment was described that amplified a small fragment from PAH-RHDα genes from both Gram-negative and Gram-positive bacteria (24). However, the amplicon size was only 78 bp and the primer might also target genes coding for dioxygenases that attack nonpolar aromatic compounds, such as benzene, toluene, and xylene. Therefore, this work aimed to design an improved primer system that targets PAH-RHDα genes from both Gram-positive and Gram-negative bacteria and provides larger amplicon sizes. The novel primer system was tested in silico and validated by sequencing cloned PAH-RHDα genes amplified from total-community (TC) DNA and was used in endpoint and quantitative real-time PCR (qPCR) formats. The primer system was also applied to study the responses of soil microbial communities in two different soils (a Cambisol and a Luvisol representing typical arable soils in Central Europe with different texture compositions) to artificial phenanthrene pollution.
MATERIALS AND METHODS
Primers.
Primers for PCR amplification of PAH-RHDα genes from both Gram-positive and Gram-negative bacteria were designed based on all 40 PAH-RHDα genes deposited in GenBank (as of 18 December 2007). The criteria for primer design were as follows: (i) no mismatches for the last 7 base pairs at the 3′end of the primer and (ii) two to six mismatches at the 5′ end. The selected primer system consisting of the 20-mer degenerated forward primer PAH-RHDα-396F (5′-ATT GCG CTT AYC AYG GBT GG-3′) and the 21-mer degenerated reverse primer PAH-RHDα-696R (5′-ATA GGT GTC TCC AAC RAA RTT-3′) should allow amplification of an approximately 320-bp fragment from all 40 PAH-RHDα genes. The primer system was tested with Pseudomonas putida KT2442 pNF142 and cloned nahAc, phnAc, ndo-3, ndo-5, and nagAc genes recently described by Gomes et al. (13 and 14) (see Table S1 in the supplemental material). On 26 April 2010, 281 sequences encoding putatively PAH-degradative proteins were obtained using BLAST-N searches and used for in silico analysis of the primers (see Fig. S1 in the supplemental material).
PCR and qPCR conditions.
Optimization of the PCR was done with a gradient of annealing temperatures (46 to 60°C) with DNA from P. putida pNF142 and one environmental sample as a target DNA. PCR amplification was performed in a 25-μl reaction mixture consisting of Stoffel buffer (Applied Biosystems, Foster, CA), 0.2 mM deoxynucleoside triphosphates, 2.5 mM MgCl2, 4% (vol/vol) acetamide, 0.1 μg μl−1 bovine serum albumin, 0.2 μM forward primers, 0.6 μM reverse primers, and 2.5 U Amplitaq DNA polymerase (Stoffel fragment; Applied Biosystems). Denaturation was carried out for 5 min at 94°C with 5 cycles of preamplification, 1 min of denaturation at 94°C, annealing at 46°C for 2 min, and extension at 72°C for 1 min, followed by 30 cycles of 1 min at 95°C, 1 min at 58.5°C, and 1 min at 72°C and 10 min of extension at 72°C. qPCR was performed in 50-μl reaction volumes consisting of Stoffel buffer (Fermentas, St. Leon-Rot, Germany), 0.2 mM deoxynucleoside triphosphates, 2.5 mM MgCl2, 4% (vol/vol) acetamide, 0.1 μg μl−1 bovine serum albumin, 0.2 μM forward primers, 0.6 μM reverse primers, EvaGreen solution (Biotium, Hayward, CA), and 2.5 U TrueStart Hot Start Taq DNA polymerase (Fermentas, St. Leon-Rot, Germany). The amplification was performed as follows. Denaturation was carried out for 5 min at 94°C; 5 cycles of preamplification were done as previously described, followed by 40 cycles of 1 min at 95°C, 45 s at 58.5°C, and 2 min at 60°C and 10 min of extension at 72°C. At the end, dissociation curve analysis was performed in autoincrements from 60°C to 94°C. The standard for quantification of PAH-RHDα genes by qPCR was prepared as follows. The cloned phnAc gene fragment (AF061872) was amplified with the primer pair SP6 and T7. The PCR product was gel purified with a Qiaex II Gel Extraction Kit (Qiagen, Hilden, Germany), and the DNA concentration was measured with a BioPhotometer (Eppendorf, Hamburg, Germany). The copy number of the standard DNA was calculated according to the size (920 bp), assuming a molecular mass of 660 Da for a base pair. The standard stock solution of 1.91 × 1010 copies was prepared. Primers and a TaqMan probe to quantify 16S rRNA genes by qPCR were previously described (30). PCR amplifications were performed in a 50-μl reaction volume containing 1.25 U TrueStart polymerase (Fermentas, St. Leon-Rot, Germany), 0.2 mM each deoxynucleoside triphosphate, 2.5 mM MgCl2, and 0.25 μM primers and probe. Thermocycles were 10 min at 94°C and 40 cycles consisting of 15 s at 95°C, 15 s at 50°C, and 60 s at 60°C. Templates to generate standard curves were prepared by serial dilutions of gel-purified PCR products from Escherichia coli 16S rRNA genes. The amplification efficiency was tested with four serial dilutions of the Sp6/T7 amplicons obtained from five different cloned RHDα genes (AF061872, GQ479194, GQ479076, GQ479118, and GQ479093) used as templates.
Soil samples.
The Eutric Cambisol (10) was collected from a long-term field experiment near Ultuna (60°N, 17°E) in central Sweden. The physicochemical characteristics, as well as previous treatments, have been described (12). The Luvisol (48°N, 11°E) was collected from a farm in Scheyern located north of Munich, Germany, without a known contamination history. The physicochemical parameters determined in this study for both soils are given in Table 1. Briefly, all soils were passed through a 2-mm mesh sieve to remove stones and plant debris and were stored at 4°C for less than 1 month before the batch experiment. The water contents of the soils were determined by leaving the soil in the oven at 110°C overnight. The carbon and nitrogen content of the soils was determined with a Euro EA elemental analyzer, and the pH was measured in 0.01 M CaCl2. Texture was determined after H2O2 oxidation of organic matter by sieving and by measuring the X-ray absorption of the soil-water suspension during sedimentation of the soil particles with a Micromeritics Sedigraph 5100 (Micromeritics, Norcross, GA).
TABLE 1.
Properties of soils in this study
| Soil parameter | Value |
|
|---|---|---|
| Cambisol | Luvisol | |
| Soil type | ||
| Sand (%) | 19 | 16 |
| Silt (%) | 44 | 70 |
| Clay (%) | 37 | 14 |
| pH | 5.8 ± 0.1 | 5.5 ± 0.1 |
| Total organic C (mg g−1) | 14.6 ± 0.2 | 14.9 ± 0.2 |
| Total N (mg g−1) | 1.51 ± 0.03 | 1.59 ± 0.03 |
| C/N ratio | 11.3 | 10.9 |
| Depth (cm) | 0-20 | 0-20 |
In addition, phenanthrene sorption was investigated in batch sorption experiments according to the guidelines of the Organization for Economic Cooperation and Development (OECD) (25). The phenanthrene concentrations of the liquid phase were determined by gas chromatography-mass spectrometry (GC-MS), and sorption isotherms were calculated using the solubility-normalized Freundlich isotherm (see the supplemental material).
Experimental design and TC DNA extraction.
Prior to the experiment, the soils were kept at room temperature for 1 week to equilibrate. Contaminating the soils with phenanthrene was done as follows. Twenty grams of each replicate was used as seeding soil, which was first contaminated by spiking it with phenanthrene dissolved in acetone (200 mg ml−1) to reach a final concentration of 20 mg g soil−1. Soil that received only acetone was used as seeding soil for the control. The seeding soil was kept in a chemical hood overnight to allow evaporation of the acetone. The adjusted weight of the seed soil was added to 150 g natural soil (final phenanthrene concentration, 2 mg g of soil−1), mixed thoroughly, and used to fill 250-ml glass flasks that were covered with lids and incubated in the dark at room temperature (23°C). The moisture of the soil was adjusted to ca. 60% of the maximum water-holding capacity by adding sterilized Milli-Q water. At days 0, 21, and 63, samples were taken from the four soil microcosms per treatment and kept at −20°C before DNA extraction. The following abbreviations are used in the text for the treatments: CA, Cambisol control; CP, phenanthrene-spiked Cambisol; LA, Luvisol control; and LP, phenanthrene-spiked Luvisol, taken at day 21 (T21) and day 63 (T63) of the incubation experiment. TC DNA was extracted from 0.5 g of soil after a harsh cell lysis step using the FastPrep FP120 bead-beating system (QBiogene, Carlsbad, CA) for 30 s at high speed (this step was repeated twice) by means of the Bio-101 DNA spin kit for soil (QBiogene, Heidelberg, Germany) according to the instructions of the manufacturer.
Cloning, sequencing, and sequence analysis.
The PAH-RHDα gene amplicons from four replicates of each soil were combined, gel purified, ligated into pGEM-T vectors, and transformed into competent cells (E. coli JM109; Promega, Mannheim, Germany) according to the instructions of the manufacturer. After the primer sequences were removed, the sequences were analyzed by BLAST-N and TBLASTX (http://www.ncbi.nlm.nih.gov/BLAST/) to identify PAH-RHDα gene sequences (ca. 280 bp). All sequences obtained were submitted to GenBank (see Table S2 in the supplemental material). Translated amino acid sequences of cloned PAH-RHDα genes, as well as of most known PAH-RHDα groups, were aligned, and the tree was calculated according to the neighbor-joining method and tested by bootstrap analysis using Molecular Evolutionary Genetics Analysis (MEGA4) (20) integrated software. Based on a DNA distance matrix created with Phylip, the program DOTUR (distance-based operational taxonomic unit [OTU] and richness determination) (27) was used to assign sequences to operational units (OUs) (sequences that have less than 5% DNA distance) and for rarefaction analyses. Rarefaction curves were plotted and further analyzed with three regression equations: (regression 1 [R1]) y = a × (1 − exp−bx), (R2) y = a × (1 − exp−bx^c) (31), and (R3) y = (a × x)/(b + x), where x is the sample size, y is the observed number of OUs, a is the number of OUs expected with infinite sample size, and b and c are constants to fit the model. SigmaPlot v. 11.0 was used for regression analysis.
Phenanthrene analysis.
Quantitation of phenanthrene was done according to the method of Baran and Oleszczuk (2) with the following modifications. Briefly, 1 gram soil was extracted using an ultrasonic bath and overhead shaker. The final extract was separated with an acetonitrile-water gradient on a C18 column (Luna C18 [2]; 100 A, 150 by 2.0 mm, 3 μm; Phenomenex, Aschaffenburg, Germany) and the phenanthrene was detected at 254 nm (UVD 340 S UV detector; Dionex). External quantitation was done with the multicomponent standard solution SRM 1647D (National Institute of Standards and Technology, Promochem, Wesel, Germany) at concentrations of 1.0, 2.5, 5.0, and 7.5 μg ml−1. Linearity was excellent (R2 = 0.996).
RESULTS
Design of a new PAH-RHDα primer set.
In silico analysis indicated that the primer set designed on the basis of all 40 different PAH-RHDα sequences deposited in GenBank (as of December 2007) would amplify a wide spectrum of PAH-RHDα genes, such as nahAc, phnAc, nagAc, bphA1, ndo-3, and ndo-5 from Gram-negative bacteria and phdAc, narAa, nidA, and other RHDα genes from Gram-positive bacteria, as no mismatches were detected for at least 7 base pairs from the 3′ ends of both the forward and reverse primers (see Fig. S2 in the supplemental material). Recently, 281 sequences putatively coding for PAH-degradative genes from GenBank (as of 26 April 2010) were obtained using BLAST-N searches and used for in silico analysis of the primers. Of the 281 sequences, only 143 were long enough to be checked for the specificity of both primers; 138 sequences had 7-bp perfect matches at the 3′ ends of both primers, while 2 to 6 mismatches were observed at the 5′ ends of the primers. However, 130 out of 281 sequences in GenBank were too short for in silico analysis of both primers, and thus, no prediction for amplification was possible. However, the annealing site that could be analyzed met the criteria described above (see Fig. S1 in the supplemental material). In order to amplify diverse PAH-RHDα sequences from TC DNA despite mismatches of the primers at the 5′ ends, a preamplification with five cycles using an annealing temperature of only 46°C for 2 min was done, followed by 30 cycles with an annealing temperature of 58.5°C. Furthermore, amplicons of the expected size (approximately 320 bp) were obtained from the genomic DNA of P. putida pNF142 and of eight cloned ndo genes (see Table S1 in the supplemental material).
The same primer set was also used for qPCR. Testing the amplification efficiencies of five different RHDα genes revealed that the amplification efficiencies ranged from 73.3% to 97.4% (see Table S3 in the supplemental material). The cloned phnAc gene used for the standard curve had the lowest amplification efficiency.
Detection of PAH-RHDα genes in soils.
The new primer set was used to study the occurrence and diversity of PAH-RHDα genes in two different soils (Luvisol and Cambisol) and variants that were spiked with 2 g phenanthrene per kg soil. At day 21, amplicons were detected only in all four replicates of the phenanthrene-contaminated Luvisol. At day 63, amplicons of putative PAH-RHDα genes were amplified from TC DNA of all replicates of both contaminated soils, but not from both control soils (data not shown).
Diversity of PAH-RHDα genes in two phenanthrene-contaminated soils.
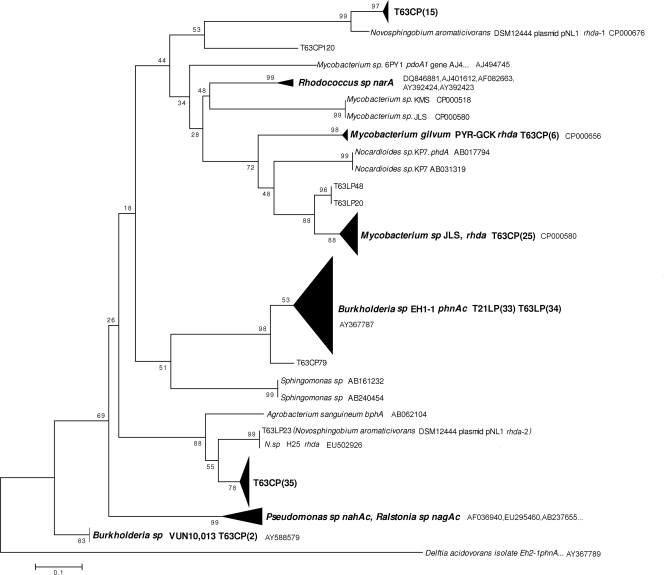
To experimentally determine the specificity of the novel primers and the diversity of PAH-RHDα genes amplified with this system from soil DNA, cloning of the pooled PCR products and sequencing were done. BLAST-N and TBLASTX (http://www.ncbi.nlm.nih.gov/BLAST/) were used to identify the PAH-RHDα gene sequences (ca. 280 bp). Thirty-three and 37 sequences analyzed from the clone library from phenanthrene-contaminated Luvisol at day 21 (T21LP) and day 63 (T63LP), respectively, were identified as PAH-RHDα gene sequences corresponding to 61.1% and 94.9% of the clones with sufficient read length (>233 bp). All 33 sequences from clone library T21LP and 34 out of 37 sequences from clone library T63LP shared 98% of the phnAc gene sequence of Burkholderia sp. strain Eh1-1 (Fig. 1; see also Table S4 in the supplemental material). Two sequences were affiliated with the PAH-RHDα genes of Mycobacterium sp. strain JLS. These genes might be novel, as they shared only 87% nucleotide sequence similarity. One of the 37 sequences of the T63LP library had 100% similarity to one PAH-RHDα gene of the Novosphingobium aromaticivorans DSM 12444 plasmid pNL1.
FIG. 1.
Neighbor-joining rooted phylogenetic tree based on the multiple alignment of PAH-RHDα gene sequences from translated amino acid sequences. The triangles represent compressed branches containing various sequences from different treatments as indicated; the vertical length of a triangle reflects the number of sequences, and the horizontal length reflects the largest distance between sequences. Value at each node = (bootstrap value/500) × 100.
The diversity of PAH-RHDα genes in the phenanthrene-spiked Cambisol could not be determined for day 21, as no amplicon was obtained. The vast majority (85/94) of T63CP sequences with sufficient read length were identified as PAH-RHDα genes. Many of the PAH-RHDα gene sequences determined (62/85) are likely to be novel, as they shared only 72 to 90% similarity (BLAST-N) with their closest corresponding genes in GenBank. However, 18 out of 87 sequences were affiliated with different PAH-RHDα genes from mycobacteria with more than 97% nucleotide sequence identity. Two sequences of clones showed high similarity to PAH-RHDα genes of Burkholderia spp. The phylogenetic analysis of the PAH-RHDα genes amplified from T63CP revealed high diversity, with a Chao1 value of 30.2 (Table 2), and confirmed that the novel primer set amplified PAH-RHDα genes from both Gram-positive and Gram-negative bacteria. The majority of PAH-RHDα genes amplified were affiliated with PAH-RHDα genes from Mycobacterium spp. (31/85) and two PAH-RHDα genes from N. aromaticivorans DSM 12444 plasmid pNL1 (50/85) (Fig. 1; see also Table S4 in the supplemental material).
TABLE 2.
Observed and estimated total OUs for phenanthrene-contaminated samplesa
| Library | nb | No. of OUs | Rarefaction analysisc |
Chao1d | |||||
|---|---|---|---|---|---|---|---|---|---|
| R1 |
R2 |
R3 |
|||||||
| a1 | r12 | a2 | r22 | a3 | r32 | ||||
| T63LP | 37 | 4 | 4.01 + 0.22 | 0.941 | 4.63 + 0.19 | 0.977 | 5.16 + 0.21 | 0.963 | 4 |
| T63CP | 85 | 23 | 23.22 + 0.16 | 0.991 | 27.72 + 0.37 | 0.999 | 30.96 + 0.16 | 0.998 | 30.2 |
Rarefaction analysis and the nonparametric richness estimator Chao1 were used for diversity estimation.
n, number of clones in the clone library.
a, asymptote of the regression equation giving the estimated total diversity; r, correlation coefficient.
Chao1, estimated total diversity.
Observed OUs and regression analysis revealed that a higher diversity of PAH-RHDα genes was enriched in the contaminated Cambisol than in the Luvisol. A total of 23 OUs were identified in the clone library of the Cambisol (T63CP; 85 sequences analyzed) but only four OUs in the clone library of the Luvisol (T63LP; 37 sequences analyzed). To compare the richnesses of the two clone libraries, 37 sequences were sampled randomly 1,000 times from the clone library of T63CP, and an average of 16.4 OUs was reached (see Fig. S3 in the supplemental material). Regression analyses done with three equations suggested a significantly higher number of OUs from T63CP than from T63LP was expected with infinite sample size. According to regression analysis, the two clone libraries generated from amplicons obtained from Luvisol and Cambisol at day 63 after phenanthrene pollution covered 80 to 100% and 74 to 100% of PAH-RHDα gene diversity, respectively. Clone library T63LP had a higher coverage of diversity than T63CP, as suggested by regression 2 and regression 3 (Table 2).
Quantification of PAH-RHDα genes.
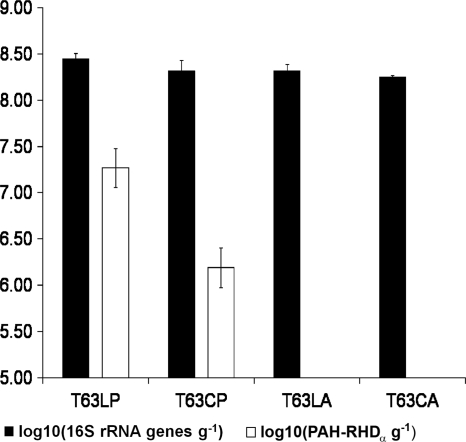
Total bacterial 16S rRNA gene copy numbers and copies of PAH-RHDα genes were determined by qPCR for all samples collected at day 63. In the Luvisol, a significantly higher 16S rRNA gene copy number per gram soil was detected in phenanthrene-contaminated soil than in the control soil. Also, the phenanthrene-contaminated Cambisol showed a slightly higher 16S rRNA gene copy number than the control. Between the two control soils, there was no significant difference in 16S rRNA gene copy numbers, while the contaminated Luvisol had significantly higher copy numbers than the contaminated Cambisol. PAH-RHDα genes were detected only in all replicates of both contaminated soil samples, with 2.03 × 107 ± 1.05 × 107 copies per gram soil and 1.68 × 1066 ± 0.66 × 106 copies per gram soil for T63LP and T63CP, respectively, but not in the corresponding control soils (Fig. 2).
FIG. 2.
Determination of 16S rRNA gene and PAH-RHDα gene copy numbers for samples collected after 63 days of incubation by qPCR. The error bars indicate standard deviations of the four replicates of each treatment.
Detection of phenanthrene in the Luvisol and in the Cambisol.
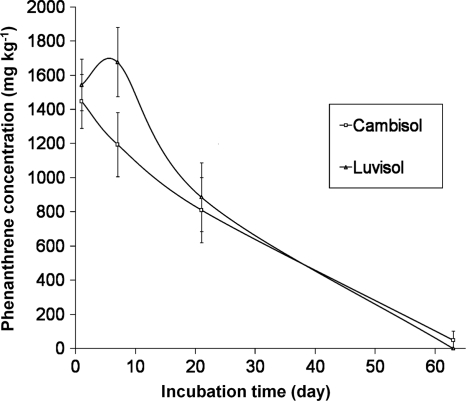
Concentrations of phenanthrene in both soils were followed at days 0, 7, 21, and 63 after spiking. No detectable phenanthrene was present in the Luvisol in the control set; small amounts (1.7 and 1.8 mg kg−1) were detected from two samples of the Cambisol controls. Seven days after spiking, the amount of phenanthrene extracted from the Cambisol, but not from the Luvisol, decreased. In the soil samples taken after 21 days, the amounts of phenanthrene extracted from both soils were similar, but only about 42% of the initial concentration in the soil was detectable. At day 63, no phenanthrene was extractable from the Luvisol while 49 ± 52 mg kg−1 was still detected in the Cambisol (Fig. 3).
FIG. 3.
Phenanthrene concentrations in the soil at different time points during incubation. The error bars indicate standard deviations of the four replicates of each treatment.
DISCUSSION
Genes coding for ring-hydroxylating dioxygenases are highly diverse, and thus, two important points were considered in order to develop primers to successfully amplify a wide range of RHDα genes from mixed template DNA. No mismatches in the last 7 bp at the 3′ end of each primer but two to six mismatches at the 5′ end were allowed. More importantly, to achieve amplification from mixed template DNA despite the mismatches at the 5′ end, a preamplification step with five cycles at low annealing temperature (46°C) was essential to generate templates that perfectly matched the primers. In contrast to other studies, primer validation was mainly based on sequencing cloned PAH-RHDα gene amplicons from two phenanthrene-contaminated soils. Sequence analysis confirmed not only the amplification of PAH-RHDα genes from Gram-positive and Gram-negative bacteria, but also high specificity of the primer system, as more than 90% of the sequences obtained for T63CP and T63LP were affiliated with PAH-RHDα genes. Only sequences obtained from the clone library of T21LP showed a rather high proportion of sequences (39%) that had no similarity to PAH-RHDα genes in GenBank. The amount of amplicon DNA obtained from T21LP was just visible on the agarose gel, indicating a low number of target sequences. Also, other authors reported increased proportions of nonspecific amplicons when PCR amplification was done from DNA with a low concentration of target genes (3). Without exposure to phenanthrene, no PAH-RHDα gene amplicons were detected by PCR and qPCR, and thus, the copy number was below the estimated detection limit of 104 copies g of soil−1. Other studies using different primer systems also reported the absence of PCR amplicons when TC DNA from noncontaminated soils or groundwater was used (3, 9, 14, 35). In contrast to the only previously published primer system (24) that also targeted RHDα genes of Gram-positive and Gram-negative bacteria, the amplified sequence that can be used for sequence analysis is much longer (approximately 280 bp compared to approximately 47 bp) and the primers specifically target PAH-RHDα genes.
Upon spiking Luvisol or Cambisol with phenanthrene, the abundance of PAH-RHDα gene-carrying populations must have increased, as PCR products were obtained only from these soil samples. Furthermore, the increased abundance of soil bacteria carrying RHDα genes indicated their involvement in the degradation of phenanthrene. Also, in other studies, the relative abundance of PAH-RHDα genes was correlated with the level of PAH contamination (4, 9, 14, 35). Microbial communities inhabiting the two soils studied showed strikingly different abundances and diversities of PAH-RHDα genes after exposure to the same concentrations of phenanthrene under identical incubation conditions. The diversity of PAH-RHDα genes in Luvisol was rather low, and all sequences from the T21LP clone library of PAH-RHDα gene amplicons had the highest nucleotide identity with the phnAc gene of the Burkholderia strain Eh-1, a strain that was originally isolated from a phenanthrene enrichment of a contaminated soil from a former coal gasification plant in Wisconsin (33). These amplicons were obtained even though sequences belonging to this phnAc gene cluster have 2 to 4 mismatches with the forward primer (PAH-RHDα-396f) and 4 to 5 with the reverse primer (PAH-RHDα-696r) (see Fig. S1 in the supplemental material). While the clone library of T63LP was still dominated by phnAc, a completely different situation was encountered for the PAH-RHDα gene library obtained for the Cambisol. This clone library had much higher diversity (Fig. 1 and Table 2; see Table S4 in the supplemental material), and the PAH-RHDα genes detected were derived from high-G+C Gram-positive bacteria (Mycobacterium and Rhodococcus), Alphaproteobacteria (Agrobacterium and Novosphingobium), and Betaproteobacteria (Burkholderia and Acidovorax). Although several of the sequences showed high sequence similarities to known PAH-RHDα gene sequences, many sequences had only rather low similarities (BlastN and BlastX) to sequences in GenBank, indicating novel genes not yet described. Exposure to phenanthrene in the Luvisol resulted in more than 1 order of magnitude higher PAH-RHDα gene copy numbers in the Luvisol than in the Cambisol detected at day 63. Whether the initial abundance of PAH-RHDα genes in the Luvisol was already higher at the beginning of the experiment or whether the differences were due to the more rapidly growing phnAc-carrying population remains unclear, as in both soils, the abundance of PAH-RHDα genes was below the detection limit. Adaptation of soil microbial communities to PAHs is assumed to occur through induction of enzymes involved in the biodegradation of PAHs or an increase in the number of bacteria with degradative capacities (23). The high abundance of a fast-growing population carrying the phnAc gene in the Luvisol might have prevented sufficient growth of slower-growing populations carrying PAH-RHDα genes and their detection in the Luvisol library. In contrast to the Luvisol, the response of the bacterial communities in the Cambisol to phenanthrene was delayed, as amplicons were obtained only at day 63 but a much higher diversity of PAH-RHDα genes was observed. A high proportion of the genes (36.4%) that were detected in response to phenanthrene were affiliated with PAH-RHDα genes from mycobacteria. Nine of the sequences displayed high similarity to genes identified on the recently completed genome sequence of Mycobacterium gilvum PYR-GCK (7, 19). Recently, Fredslund et al. (11) demonstrated surface sliding motility of M. gilvum and discussed the implications for PAH degradation in contaminated soil. Furthermore, PAH-degrading mycobacteria were shown to be enriched on PAH-containing soil components (32).
The differences in the responses of the bacterial communities of both soils to phenanthrene contamination might also be related to the different soil properties (Table 1). The composition and heterogeneity of soil organic matter can greatly influence the mineralization of PAHs (5, 23, 26). However, the phenanthrene sorption experiments performed in this study (see Fig. S4 in the supplemental material) showed no significant difference in phenanthrene sorption between the two soils. This implies that the difference in clay content had no effect on phenanthrene sorption and that the organic matter content was probably the only factor determining phenanthrene sorption. The two soils have similar C/N ratios and land use histories and therefore probably similar organic matter compositions. On the other hand, the differences in soil properties are assumed to be an important factor shaping soil microbial communities. Soil type-dependent differences in the bacterial community composition are well documented (28).
In conclusion, the bacterial community compositions present in the two soils, as well as soil texture-related factors (e.g., porosity or the matric potential), might have contributed to the different responses. Furthermore, the batch experiment also showed that many degradative genes will remain undiscovered in soil metagenomics unless enrichments are performed to increase the abundance of rare populations carrying these genes. Thus, it is important to keep in mind that PCR-based detection with a novel primer system will also provide insights into the degradative potentials of bacterial populations that occur with 104 or more gene copies per gram of soil. Thus, soil enrichments could be an important means to uncover the biodegradative potentials of uncommon populations.
Supplementary Material
Acknowledgments
This work was supported by DFG SPP1315 (SM59/8-1, SP255/19-1, and KO1035/33-1).
We thank I.-M. Jungkurth for carefully checking our manuscript.
Footnotes
Published ahead of print on 21 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baran, S., and P. Oleszczuk. 2002. Chromatographic determination of polycyclic aromatic hydrocarbons (PAH) in sewage sludge, soil, and sewage sludge-amended soils. Pol. J. Environ. Studies 11:609-615. [Google Scholar]
- 3.Bordenave, S., M. Goni-Urriza, C. Vilette, S. Blanchard, P. Caumette, and R. Duran. 2008. Diversity of ring-hydroxylating dioxygenases in pristine and oil contaminated microbial mats at genomic and transcriptomic levels. Environ. Microbiol. 10:3201-3211. [DOI] [PubMed] [Google Scholar]
- 4.Cébron, A., M. P. Norini, T. Beguiristain, and C. Leyval. 2008. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 73:148-159. [DOI] [PubMed] [Google Scholar]
- 5.Celis, R., H. De Jonge, L. W. De Jonge, M. Real, M. C. Hermosin, and J. Cornejo. 2006. The role of mineral and organic components in phenanthrene and dibenzofuran sorption by soil. Eur. J. Soil Sci. 57:308-319. [Google Scholar]
- 6.Chain, P. S., V. J. Denef, K. T. Konstantinidis, L. M. Vergez, L. Agullo, V. L. Reyes, L. Hauser, M. Cordova, L. Gomez, M. Gonzalez, M. Land, V. Lao, F. Larimer, J. J. LiPuma, E. Mahenthiralingam, S. A. Malfatti, C. J. Marx, J. J. Parnell, A. Ramette, P. Richardson, M. Seeger, D. Smith, T. Spilker, W. J. Sul, T. V. Tsoi, L. E. Ulrich, I. B. Zhulin, and J. M. Tiedje. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. U. S. A. 103:15280-15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean-Ross, D., and C. E. Cerniglia. 1996. Degradation of pyrene by Mycobacterium flavescens. Appl. Microbiol. Biotechnol. 46:307-312. [DOI] [PubMed] [Google Scholar]
- 8.Debruyn, J. M., and G. S. Sayler. 2009. Microbial community structure and biodegradation activity of particle-associated bacteria in a coal tar contaminated creek. Environ. Sci. Technol. 43:3047-3053. [DOI] [PubMed] [Google Scholar]
- 9.Flocco, C. G., N. C. Gomes, C. W. Mac, and K. Smalla. 2009. Occurrence and diversity of naphthalene dioxygenase genes in soil microbial communities from the maritime Antarctic. Environ. Microbiol. 11:700-714. [DOI] [PubMed] [Google Scholar]
- 10.Food and Agriculture Organization of the United Nations. 2006. World soil resources reports. FAO, Rome, Italy.
- 11.Fredslund, L., K. Sniegowski, L. Y. Wick, C. S. Jacobsen, R. De Mot, and D. Springael. 2008. Surface motility of polycyclic aromatic hydrocarbon (PAH)-degrading mycobacteria. Res. Microbiol. 159:255-262. [DOI] [PubMed] [Google Scholar]
- 12.Gerzabek, M. H., R. S. Antil, I. Kogel-Knabner, H. Knicker, H. Kirchmann, and G. Haberhauer. 2006. How are soil use and management reflected by soil organic matter characteristics: a spectroscopic approach. Eur. J. Soil Sci. 57:485-494. [Google Scholar]
- 13.Gomes, N. C., L. R. Borges, R. Paranhos, F. N. Pinto, E. Krögerrecklenfort, L. C. Mendonca-Hagler, and K. Smalla. 2007. Diversity of ndo genes in mangrove sediments exposed to different sources of polycyclic aromatic hydrocarbon pollution. Appl. Environ. Microbiol. 73:7392-7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes, N. C., I. A. Kosheleva, W. R. Abraham, and K. Smalla. 2005. Effects of the inoculant strain Pseudomonas putida KT2442 (pNF142) and of naphthalene contamination on the soil bacterial community. FEMS Microbiol. Ecol. 54:21-33. [DOI] [PubMed] [Google Scholar]
- 15.Habe, H., and T. Omori. 2003. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 67:225-243. [DOI] [PubMed] [Google Scholar]
- 16.Johnsen, A. R., L. Y. Wick, and H. Harms. 2005. Principles of microbial PAH-degradation in soil. Environ. Pollut. 133:71-84. [DOI] [PubMed] [Google Scholar]
- 17.Kanaly, R. A., and S. Harayama. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, S. J., O. Kweon, R. C. Jones, R. D. Edmondson, and C. E. Cerniglia. 2008. Genomic analysis of polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Biodegradation 19:859-881. [DOI] [PubMed] [Google Scholar]
- 19.Kim, Y. H., K. H. Engesser, and C. E. Cerniglia. 2005. Numerical and genetic analysis of polycyclic aromatic hydrocarbon-degrading mycobacteria. Microb. Ecol. 50:110-119. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., M. Nei, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd-Jones, G., A. D. Laurie, D. W. F. Hunter, and R. Fraser. 1999. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMS Microbiol. Ecol. 29:69-79. [Google Scholar]
- 22.Lozada, M., J. P. Riva Mercadal, L. D. Guerrero, W. D. Di Marzio, M. A. Ferrero, and H. M. Dionisi. 2008. Novel aromatic ring-hydroxylating dioxygenase genes from coastal marine sediments of Patagonia. BMC Microbiol. 8:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macleod, C. J. A., and K. T. Semple. 2002. The adaptation of two similar soils to pyrene catabolism. Environ. Pollut. 119:357-364. [DOI] [PubMed] [Google Scholar]
- 24.Ni Chadhain, S. M., R. S. Norman, K. V. Pesce, J. J. Kukor, and G. J. Zylstra. 2006. Microbial dioxygenase gene population shifts during polycyclic aromatic hydrocarbon biodegradation. Appl. Environ. Microbiol. 72:4078-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Organization for Economic Cooperation and Development. 2000. Test no. 106: adsorption-desorption using a batch equilibrium method. OECD guidelines for testing chemicals. OECD Publications, Paris, France.
- 26.Reid, B. J., C. J. MacLeod, P. H. Lee, A. W. Morriss, J. D. Stokes, and K. T. Semple. 2001. A simple 14C-respirometric method for assessing microbial catabolic potential and contaminant bioavailability. FEMS Microbiol. Lett. 196:141-146. [DOI] [PubMed] [Google Scholar]
- 27.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smalla, K., M. Oros-Sichler, A. Milling, H. Heuer, S. Baumgarte, R. Becker, G. Neuber, S. Kropf, A. Ulrich, and C. C. Tebbe. 2007. Bacterial diversity of soils assessed by DGGE, T-RFLP and SSCP fingerprints of PCR-amplified 16S rRNA gene fragments: do the different methods provide similar results? J. Microbiol. Methods 69:470-479. [DOI] [PubMed] [Google Scholar]
- 29.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 30.Takai, K., and K. Horikoshi. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66:5066-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiel, V., S. C. Neulinger, T. Staufenberger, R. Schmaljohann, and J. F. Imhoff. 2007. Spatial distribution of sponge-associated bacteria in the Mediterranean sponge Tethya aurantium. FEMS Microbiol. Ecol. 59:47-63. [DOI] [PubMed] [Google Scholar]
- 32.Uyttebroek, M., P. Breugelmans, M. Janssen, P. Wattiau, B. Joffe, U. Karlson, J. J. Ortega-Calvo, L. Bastiaens, A. Ryngaert, M. Hausner, and D. Springael. 2006. Distribution of the Mycobacterium community and polycyclic aromatic hydrocarbons (PAHs) among different size fractions of a long-term PAH-contaminated soil. Environ. Microbiol. 8:836-847. [DOI] [PubMed] [Google Scholar]
- 33.Vacca, D. J., W. F. Bleam, and W. J. Hickey. 2005. Isolation of soil bacteria adapted to degrade humic acid-sorbed phenanthrene. Appl. Environ. Microbiol. 71:3797-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, M. S., C. Bakermans, and E. L. Madsen. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yagi, J. M., and E. L. Madsen. 2009. Diversity, abundance, and consistency of microbial oxygenase expression and biodegradation in a shallow contaminated aquifer. Appl. Environ. Microbiol. 75:6478-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yagi, J. M., D. Sims, T. Brettin, D. Bruce, and E. L. Madsen. 2009. The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar-contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ. Microbiol. 11:2253-2270. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, H. W., C. L. Guo, Y. S. Wong, and N. F. Tam. 2006. Genetic diversity of dioxygenase genes in polycyclic aromatic hydrocarbon-degrading bacteria isolated from mangrove sediments. FEMS Microbiol. Lett. 262:148-157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.