Abstract
Sulfate-reducing bacteria (SRB) play a major role in the coupled biogeochemical cycling of sulfur and chalcophilic metal(loid)s. By implication, they can exert a strong influence on the speciation and mobility of multiple metal(loid) contaminants. In this study, we combined DsrAB gene sequencing and sulfur isotopic profiling to identify the phylogeny and distribution of SRB and to assess their metabolic activity in salt marsh sediments exposed to acid mine drainage (AMD) for over 100 years. Recovered dsrAB sequences from three sites sampled along an AMD flow path indicated the dominance of a single Desulfovibrio species. Other major sequence clades were related most closely to Desulfosarcina, Desulfococcus, Desulfobulbus, and Desulfosporosinus species. The presence of metal sulfides with low δ34S values relative to δ34S values of pore water sulfate showed that sediment SRB populations were actively reducing sulfate under ambient conditions (pH of ∼2), although possibly within less acidic microenvironments. Interestingly, δ34S values for pore water sulfate were lower than those for sulfate delivered during tidal inundation of marsh sediments. 16S rRNA gene sequence data from sediments and sulfur isotope data confirmed that sulfur-oxidizing bacteria drove the reoxidation of biogenic sulfide coupled to oxygen or nitrate reduction over a timescale of hours. Collectively, these findings imply a highly dynamic microbially mediated cycling of sulfate and sulfide, and thus the speciation and mobility of chalcophilic contaminant metal(loid)s, in AMD-impacted marsh sediments.
Salt marshes exhibit high primary production rates (1, 101) and form biogeochemical “transition zones” for nutrient production, transport, and cycling between terrestrial and coastal marine environments (41, 66, 100). These zones also serve to reduce the flux of potentially toxic metals in contaminated groundwater to estuaries (12, 99, 106). Both functions depend strongly on microbial activity, especially that of sulfate-reducing bacteria (SRB) (42, 62, 67). SRB recycle much of the sedimentary organic carbon pool in marsh sediments (42-44) and indirectly inhibit production of the greenhouse gas methane (37, 71). They can restrict the mobility of dissolved contaminant metals by inducing precipitation of poorly soluble metal sulfides, and studies have examined their use in constructed wetlands to bioremediate acid mine drainage (AMD) and other metalliferous waste streams (11, 35, 40, 46, 50, 76, 90, 94, 104). However, the high acidity and metal concentrations inherent to AMD can inhibit SRB growth (15, 88, 98), and preferential growth of iron- and sulfur-oxidizing bacteria over SRB has been observed in some treatment wetlands (39).
For natural salt marshes, 16S ribosomal nucleic acid- and phospholipid fatty acid (PLFA)-based analyses have shown that SRB commonly comprise a significant fraction of the microbial community (13, 24, 31, 34, 51, 58). Studies of salt marsh dissimilatory sulfite reductase genes (dsrAB), a highly conserved functional phylogenetic marker of prokaryotic sulfate reducers (49, 57, 102, 103, 107), have revealed both novel and deeply branching clades (3). Studies of mining-impacted sites at pH 2.0 to 7.8 (5, 7, 39, 70, 72, 77, 84), of soils and geothermal settings at a pH of ∼4 (55, 68), of metal-contaminated estuaries at pH 6.8 to 7.2 (65), and of hypersaline lakes at pH 7.5 (56) further outline the distribution and tolerance of specific groups and species of SRB under geochemically stringent conditions. Other findings point toward the existence of deltaproteobacteria in environments at a pH of ∼1 (10), although it is unknown if these include SRB. SRB diversity in salt marshes under long-term contamination by AMD has not been well investigated. Such studies may provide useful information for bioremediation projects in estuarine environments, as well as general insights into relationships between SRB physiology and the geochemistry of AMD.
We studied the diversity of SRB, based on phylogenetic analysis of recovered DsrAB gene sequences (∼1.9 kb), in natural salt marsh sediments of the San Francisco Bay impacted by AMD for over 100 years. Sulfur isotope ratio and concentration measurements of pore water sulfate and metal sulfide minerals provided information about the spatial and temporal extent of active bacterial sulfate reduction (BSR) in sediment cores taken from specific sites along an AMD flow path. Collectively, the results revealed a tidal marsh system characterized by rapidly cycling bacterial sulfate reduction and sulfide reoxidation associated with oscillating tidal inundation and groundwater infiltration.
MATERIALS AND METHODS
Site description, sampling, and preliminary geochemical analyses.
Stege Marsh, in the eastern central San Francisco Bay, consisted of tidal mudflats pre-1959 (see Fig. S1 in the supplemental material). From 1897 to 1970, the area received sulfuric acid from pyrite ore processing, mercury from explosive blasting cap manufacturing, and lead from paint production. Pyrite “cinders” containing high concentrations of As, Cd, Cu, Pb, Zn, and Hg have oxidized to several meters depth (95, 96). The cinders in the “AMD ponds” and tidal sloughs were deposited as meters-thick and centimeters-thick layers, respectively. The present study was conducted from late August 2002 to early September 2003, before and after dredging of the AMD pond. Three sites were hand cored at low tide along an AMD flow path (see Fig. S1B in the supplemental material) by using a stainless steel, hand-push coring device with sterilized 6-cm-diameter clear plastic inner sleeves. The first site (WSMA) was located within the western-most AMD pond prior to partial dredging of contaminated sediments by the site owner, the second within the upper tidal slough (WSMD), and the third within the lower tidal slough (WSMF), just above the intersection with a larger tidal channel (see Fig. S1B in the supplemental material). AMD pond sediments were resampled roughly 2 months after dredging (WSMO) and represented an “acid sulfate soil” setting. “Grab” samples were obtained from pyrite cinders exposed at the surface (see Fig. S1D in the supplemental material). All samples were transported to the lab at 4 to 10°C for processing within 4 to 5 h of recovery. Each core was bisected lengthwise, with one half archived at −80°C for mineralogical and water analyses and the other used for whole-community DNA extraction. Sediment total concentrations of As, Cd, Cu, Pb, Zn, and Hg from the AMD pond (prior to dredging) are shown in Table 1; most values exceeded regulatory standards by up to several orders of magnitude (32). Surface waters measured with a field pH meter and probe (Thermo Fisher Scientific, Waltham, MA) in both the western and eastern AMD ponds (see Fig. S1 in the supplemental material) exhibited pH values ranging from 1.8 to 3.1. pH values at depth within sediments were measured using a GeoProbe (Salina, KA) and coring instrumentation (95, 96). Total dissolved sulfide was measured with a portable spectrophotometer (Hach Co., Loveland, CO), using the method of Cline (21), in waters obtained from sediments via centrifugation and filtration through 0.2-μm-pore-diameter syringe filters (Terumo, Tokyo, Japan; Millipore Corp., Billerica, MA). Salinity values were calculated from field electrical conductivity and temperature data (74).
TABLE 1.
Metal concentrations in western Stege Marsh
| Sample location | Depth (cm) | Total concn (ppm or ppb)a |
pH | |||||
|---|---|---|---|---|---|---|---|---|
| As | Cd | Cu | Pb | Hg | Zn | |||
| AMD pond (predredge) | 0 | 1,020 | 2 | 193 | 37 | 1 | 517 | 3.1 |
| 20 | 746 | 3 | 745 | 289 | 28 | 945 | 4.8 | |
| 40 | 1,330 | 44 | 1,640 | 1,240 | 166 | 5,000 | 6.1 | |
| Upper tidal slough | 0 | 1,400 | 23 | 85 | 70 | 3 | 300 | 6.9 |
| 20 | 110 | 9 | 320 | 72 | 40 | 710 | 7.6 | |
| 40 | 4 | 3 | 15 | 7 | 0 | 28 | 7.8 | |
| Lower tidal slough | 0 | 23 | 7 | 93 | 80 | 1 | 260 | 7.4 |
| 20 | 190 | 10 | 500 | 140 | 22 | 1,300 | 8.2 | |
| 40 | 17 | 4 | 200 | 84 | 40 | 410 | 8.7 | |
| Main channel | 0 | 5 | 5 | 10 | 3 | 0 | 30 | 8.5 |
Typical upper range values, as determined by whole-sediment acid digestion and inductively coupled plasma optical emission spectroscopy (ICP-OES) or inductively coupled plasma mass spectrometry (ICP-MS) analysis (or cold-vapor atomic fluorescence spectrometry [for Hg]). Values for all sites are given as ppm, except for the “main channel” values, given in ppb.
DNA extraction.
Subsamples of sediments (∼1 g [wet weight]) were taken at ∼1-cm intervals from the undisturbed center of one half of each core and homogenized manually with a sterile plastic spatula. Replicate ∼1-g subsamples were taken from homogenized sediments for genomic DNA extraction, using a PowerSoil DNA extraction kit (Mo Bio Laboratories, Inc., Carlsbad, CA), and aliquots of DNA were pooled from replicate extracts at roughly equivalent concentrations, as determined both visually by comparison to concentration standard markers in 1% agarose gels and by spectrophotometric analysis at 260 nm (Bio-Rad Laboratories, Inc., Hercules, CA), for PCR amplification (Thermo Hybaid Sprint; Thermo Fisher Scientific, Inc., Waltham, MA). We estimated that total DNA recoveries were 5 to 10 times lower from AMD pond sediments than from either of the tidal sloughs, on the basis of comparisons of sediment-extracted DNAs to the DNA concentration and size reference standards (Invitrogen, Carlsbad, CA).
PCR amplification.
Full-length (∼1.9 kb) dsrAB sequences were amplified from community DNA samples by using an equimolar mix of forward primers DSR1F, DSR1Fa, DSR1Fb, DSR1Fc, and DSR1Fd and reverse primers DSR4R, DSR4Ra, DSR4Rb, DSR4Rc, and DSR4Rd (107). Control amplifications were performed using DNA from Escherichia coli and PCR reagents only (negative control) or actively growing Desulfovibrio desulfuricans (positive control). PCR was performed using Platinum Taq Hi-Fidelity DNA polymerase (Applied Biosystems, Foster City, CA). Thermal cycling was performed using a hot start protocol of 95°C for 2 to 4 min followed by 30 cycles of denaturing at 94°C for 40 s, annealing at 50°C for 40 s, and extension at 68°C for 1.5 min. Final extension reactions were carried out for 7 min at 68°C. PCR products of ∼1.9 kb were excised from 1% agarose gels and purified using a MinElute gel elution kit (Qiagen Corp., Valencia, CA). Excised DsrAB genes were ligated into pGEM-T Easy vector (Promega, Stoughton, WI) for transformation of One-Shot chemically competent cells (Invitrogen Corp., Carlsbad, CA). Vector inserts were sequenced using modified T7 (5′-GTAATACGACTCACTATAGGGCGAATTGGG-3′) and standard M13R forward and reverse primers, respectively (MWG-Biotech, Inc., High Point, NC). Amplification of 16S rRNA genes was also performed, and the methodology and results can be found in the supplemental material. All PCR controls gave the expected results. All sequencing was performed at the DNA Sequencing Facility of the University of California, Berkeley, CA.
Cloning and phylogenetic analyses.
Clone sequences were screened by partial gene analysis, using BLAST (2) and the dsrAB pure culture database published by the University of Vienna's Department of Microbial Ecology (107 [http://www.microbial-ecology.net/]). DNA sequences of >1.7 kb were individually and manually aligned in ARB EDIT4 (69) against both pure culture and manually imported published dsrAB sequences from previous studies (3, 4, 20, 27, 30, 56, 65, 68, 81, 107). A composite consensus phylogenetic tree (Fig. 1) for all full taxonomically unique dsrAB sequences was synthesized from multiple trees constructed in the ARB software environment (69), using FITCH and PHYLIP protein maximum likelihood algorithms on inferred amino acid sequences of ≥582 positions. All percent sequence similarity values for full DsrAB genes were derived from consensus tree branch lengths. All clone sequences were tested for chimerism by comparative partial-sequence tree positioning (45). Fewer than 2% of all sequences were suspected to be chimeric, and these data were discarded. Additional checks for full-sequence protein similarity to known dsrAB-encoded amino acid sequences were performed using the Swiss Institute for Bioinformatics Swiss-Prot database, and all full sequences were manually inspected for highly conserved siroheme binding sites (103). Only one putatively nonfunctional dsrAB sequence was found in this manner, and this sequence was discarded. Clone sequences were classified into operational taxonomic units (OTUs) by using the public software package DOTUR (89), with a lower cutoff of 97% amino acid sequence similarity and a furthest-neighbor algorithm. Among 120 clones sequenced, 22 of 24 obtained full sequences were classified as unique OTUs. The remaining 2 full sequences and 106 partial gene sequences were grouped within these OTUs on the basis of full sequence analysis by DOTUR or because of partial sequence similarity by BLAST algorithms against the University of Vienna dsrAB database. OTUs were named after the first recovery of an included clone sequence and therefore may not be unique to the site (or enrichment culture) from which that sequence originated (e.g., WSMA38 was recovered from both the AMD pond and the upper tidal slough). 16S rRNA gene cloning and sequencing were also performed, and the results are discussed in the supplemental material. All full gene sequences were subjected to rarefaction and coverage analysis by use of DOTUR (see Fig. S2 in the supplemental material). Results from 16S rRNA gene phylogenetic analysis are presented as a consensus maximum likelihood tree (see Fig. S3 in the supplemental material).
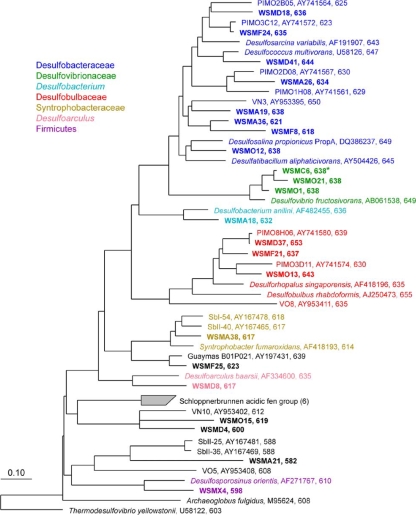
FIG. 1.
Consensus maximum likelihood tree of inferred dsrAB translated amino acid sequences. Numbers of amino acid sequences used for each alignment are shown after each clone/species name. The asterisk indicates the dominant sequence found in all dsrAB environmental clone libraries constructed in this study.
Culture.
Inocula for SRB culture were derived from pyrite cinders (WSMC) and AMD pond sediments resuspended in site water. SRB enrichment cultures were established from both types of inocula, using DSMZ410 (brackish Desulfovibrio) medium (www.dsmz.de) amended with lactate, pyruvate, fumarate, propionate, or hydrogen with acetate and CO2. SRB cultures were also grown in either ∼1.2 M NaCl (WSMX dsrAB sequences) or 132 μM Cu(II) (WSMU dsrAB sequences). For the latter, a copper toxicity assay medium (MTM) (88) was substituted for DSMZ410 medium and modified to include ∼1 mM FeSO4. All cultures were incubated in the dark at ∼25°C in 120-ml glass serum bottles (Bellco, Vineland, NJ) containing ∼50 ml of medium under either a 100% N2 or 80% H2-20% CO2 headspace and crimp sealed with aluminum caps over butyl rubber septa. Cells from positive growth enrichments were transferred to anaerobic 1.5% (wt/vol) agar roll tubes for attempted SRB species isolations. Selected colonies were picked individually with sterile glass pipettes for transfer to liquid culturing medium for further dilution to extinction and attempted isolation. Subsamples of the final positive growth dilution were stained with 4′,6-diamidino-2-phenylindole (DAPI) and evaluated by epifluorescence microscopy for their degree of morphological variability. Cultures that exhibited ≥90% morphological uniformity were further tested for species-level purity via PCR amplification and cloning of amplified DsrAB genes.
Dilution series culturing (22) was also conducted to estimate in situ SRB cell densities in pyrite cinders and acid pond sediments. Pyrite cinder- or sediment-derived inocula were injected through butyl rubber septa into anaerobic deep agar tubes of DSMZ410 medium in a dilution transfer series, similar to the approach used for attempted isolations from enrichment cultures, but following the “deep agar tube” enumeration method of Postgate (82). CFU within agar “plugs” were counted and multiplied by the dilution factor to obtain estimates of in situ SRB cell densities.
Isotope measurements.
Sediment splits from each depth and each core were dried overnight at ∼60°C to permit crushing to a fine powder. Splits were neither disaggregated nor agitated prior to drying so that exposure of reactive surface areas of acid-volatile sulfides (AVS) to oxidation to elemental sulfur (14) was minimized. Between 1 and 2 g of each powdered sample was processed by an AVS and chromium-reducible sulfide (CRS) extraction-distillation method (16). CRS extractions also recovered any small amounts of elemental sulfur produced during the initial drying of sediment splits. Sulfur recovered from each fraction (as Ag2S) was weighed and compared to initial powdered weights to estimate the concentrations of AVS and CRS recovered from each depth. A small amount of the sulfur isotope ratio standard NBS123 (http://www.nist.gov/) was subjected to this extraction process to test the efficiency of sulfur recovery with respect to mass conservation and/or isotopic fractionation. All acid extractions were performed at the Department of Geology of the University of California—Davis.
Pore waters were removed from each 4-cm interval of sediments (10-cm intervals in the AMD pond core due to the increased specific yield of pore water in the coarser cinders) from a replicate set of core splits by centrifugation and decanting (6,000 × g for 5 min, three times). Solutions extracted from each segment varied from 10 to 40 ml in volume (except for AMD pond sediments, which yielded 5- to 8-ml samples). Extracted solutions were centrifuged at 10,000 × g for 10 to 15 min per segment to collect remaining fine particles as pellets to be added to DNA extractions. Supernatant subaliquots of 1 ml were exposed dropwise to 4 M acetic acid until CO2 evolution ceased and then were mixed with 1 ml of 1 M BaCl2 to precipitate aqueous sulfate as barite for sulfur isotopic analysis (79). Barite samples were dried overnight at ∼60°C and crushed into fine powders. Sulfate in acid pond and tidal slough surface waters was processed in the same way. Remaining solutions for each depth were split for duplicate analyses of sulfate concentration by ion chromatography (Dionex Corp., Sunnyvale, CA). Sulfur isotopic analyses were conducted at UC Davis and UC Berkeley, using GV Micromass continuous-flow-isotope ratio mass spectrometers (CF-IRMS) preceded by elemental analyzers. International Atomic Energy Agency (IAEA) S-1, S-2, and S-3 standards (for δ34SV-CDT values [see Table S1 in the supplemental material]) were used for external and internal calibration. δ34S values are reported in parts per thousand, or per mille (‰), relative to Vienna-Cañon Diablo troilite (V-CDT) (28). Each standard's composition was determined as an unknown by three-point calibration to two other standards plus a well-characterized proxy (δ34SV-CDT = 5.6‰ ± 0.1‰). All uncertainty values represent 2 standard deviations from the mean.
Nucleotide sequence accession numbers.
The dsrAB sequences were deposited in GenBank (9) under accession numbers GU288599 to GU288620.
RESULTS
Typical pH values of surface waters across western Stege Marsh ranged from 2.1 to 5.6 in the AMD pond and from 5.6 to 7.2 and 7.6 to 8.6 in the upper and lower tidal sloughs, respectively. After dredging, the AMD pond area (acid sulfate soil) exhibited consistently lower pH values, ranging from 1.8 to 3.4. Total surface water AVS concentrations ranged from <3 μM in the AMD pond and acid sulfate soil to 4 to 5 μM in the upper and lower tidal sloughs. Pore waters obtained from depths of >4 cm yielded little dissolved sulfide (i.e., <3 μM).
The dsrAB sequences shown in Fig. 1 were derived from SRB families Desulfobacteraceae, Desulfovibrionaceae, Desulfobulbaceae, and Syntrophobacteraceae; the genera Desulfobacterium and Desulfoarculus; and the phylum Firmicutes. Sequences from some enrichment cultures were detected in the marsh sediment clone libraries. The phylogeny of all dsrAB clone sequences (and their closest relatives) is presented in Table 2. The site-specific distribution of unique dsrAB clone DNA sequences (OTUs) of >1.7 kb long (or ≥582 amino acids) and representative of the diversity in all sampling sites and positive growth enrichment cultures is presented in Table 3. Rarefaction and coverage analytical results are presented in Fig. S2 in the supplemental material and are discussed in the supplemental material.
TABLE 2.
Phylogeny of Stege Marsh dsrAB sequences
| Clade | Cluster | OTUa | Most related dsrAB sequence(s) (GenBank accession no.) | % Similarity |
|---|---|---|---|---|
| 1 | Desulfobacteraceae | D18 | PIMO2B05 (AY741564), Desulfosarcina variabilis (AF191907) | 71 |
| F24 | PIMO3C12 (AY741572), Desulfosarcina variabilis (AF191907) | 78 | ||
| D41 | Desulfococcus multivorans (U58126) | 83 | ||
| A26 | PIMO2D08 (AY741561) | 81 | ||
| A19 | VN3 (AY953395) | 70 | ||
| O12 | Desulfosalina propionicus PropA (DQ386237) | 85 | ||
| A36 | VN3 (AY953395) | 67 | ||
| F8 | VN3 (AY953395) | 55 | ||
| 2 | Desulfovibrionaceae | O1 | Desulfovibrio fructosivorans (AB061538) | 89 |
| O21 | Desulfovibrio fructosivorans (AB061538) | 83 | ||
| C6 | Desulfovibrio fructosivorans (AB061538) | 87 | ||
| 3 | Desulfobacterium | A18 | Desulfobacterium anilini (AF482455) | 81 |
| 4 | Desulfobulbaceae | D37 | PIMO8H06 (AY741580) | 99 |
| F21 | PIMO8H06 (AY741580) | 81 | ||
| O13 | PIMO3D11 (AY741574) | 81 | ||
| 5 | Syntrophobacteraceae/Desulfoarculus | A38 | Sbll-40 (AY167465) | 82 |
| D8 | Desulfoarculus baarsii (AF334600) | 70 | ||
| F25 | Guaymas B01P021 (AY197431) | 90 | ||
| 6 | Deeper branches | O15 | VN10 (AY953402) | 62 |
| D4 | VN10 (AY953402) | 68 | ||
| A21 | Sbll-36 (AY167469) | 45 | ||
| X4 | Desulfosporosinus orientis (AF271767) | 71 |
A, AMD pond; C, pyrite cinder enrichment culture (DSM410 amended with ∼1 mM fumarate); D, upper tidal slough; F, lower tidal slough; O, acid sulfate soils (dredged AMD pond sediments); U, 132 μM Cu(II) enrichment culture (modified MTM [88]); X, ∼1.2 M NaCl in DSM410 medium amended with ∼10 mM lactate.
TABLE 3.
Distribution of dsrAB sequences from western Stege Marsh
| OTU | No. of occurrencesa |
Total no. of occurrences | Group | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A | C | D | F | O | U | X | |||
| WSMA21 | 1 | 1 | I | ||||||
| WSMA18 | 6 | 1 | 2 | 1 | 10 | I | |||
| WSMA36 | 2 | 1 | 1 | 4 | I | ||||
| WSMA19 | 1 | 1 | 2 | ||||||
| WSMA38 | 1 | 1 | 2 | ||||||
| WSMD37 | 1 | 1 | 1 | 3 | |||||
| WSMF24 | 1 | 1 | 1 | 1 | 4 | ||||
| WSMC6 | 16 | 15 | 14 | 6 | 7 | 58 | |||
| WSMD18 | 2 | 2 | II | ||||||
| WSMD41 | 2 | 2 | II | ||||||
| WSMD4 | 1 | 1 | II | ||||||
| WSMO13 | 1 | 1 | 2 | II | |||||
| WSMA26 | 2 | 8 | 1 | 11 | II | ||||
| WSMF8 | 2 | 2 | III | ||||||
| WSMF21 | 1 | 1 | III | ||||||
| WSMF25 | 1 | 1 | III | ||||||
| WSMD8 | 1 | 2 | 3 | III | |||||
| WSMO1 | 1 | 1 | IV | ||||||
| WSMO12 | 1 | 1 | IV | ||||||
| WSMO15 | 1 | 1 | IV | ||||||
| WSMO21 | 1 | 1 | IV | ||||||
| WSMX4 | 4 | 3 | 7 | ||||||
| Total | 31 | 15 | 35 | 16 | 15 | 4 | 4 | 120 | |
A, AMD pond; C, pyrite cinder enrichment culture (DSM410 amended with ∼1 mM fumarate); D, upper tidal slough; F, lower tidal slough; O, acid sulfate soils (dredged AMD pond sediments); U, 132 μM Cu(II) enrichment culture (modified MTM [88]); X, ∼1.2 M NaCl in DSM410 medium amended with ∼10 mM lactate. A total of 120 full and partial DsrAB genes were sequenced.
Enumeration of cells in deep agar tube dilutions of inocula derived from cinder and AMD pond sediments yielded >9.3 × 102 and >2.3 × 102 SRB cells ml−1 sediment slurry, respectively. These estimates were ≥7 orders of magnitude lower than those obtained for tidal slough dilution series. Fumarate-amended WSMC and lactate-amended WSMX and WSMU enrichment cultures exhibited SRB growth after several days to roughly 2 weeks. In cases of increased salinity and dissolved copper concentration, SRB growth was observed in media containing up to 1.2 M NaCl and 132 μM Cu(II), respectively. A control culture of Desulfovibrio desulfuricans G20 did not grow in copper concentrations of >16 μM. WSMX and WSMU enrichment cultures contained both vibrio- and rod-shaped cell morphologies, with an apparent selection for rod-shaped cells as the cultures aged from several days to 2 weeks. Subsequent attempts to isolate SRB species from either enrichment medium resulted in mixed cultures or cocultures only. However, aged enrichment cultures seemed to be dominated, on the basis of partial gene sequencing, by a single dsrAB OTU (WSMX4) most closely related to Desulfosporosinus orientis (Fig. 1; Table 2).
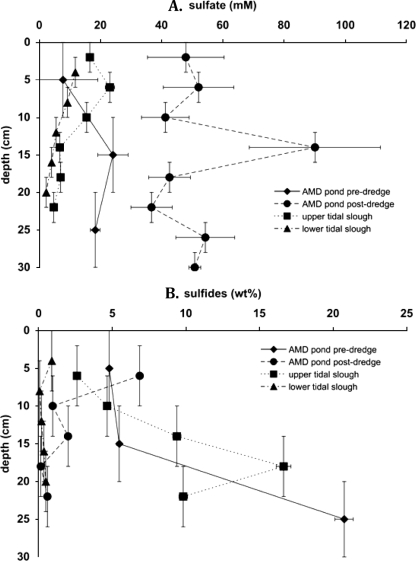
Pore water sulfate and CRS concentrations as a function of sediment depth are shown in Fig. 2. Sulfate concentrations in the AMD pond area increased and remained nearly constant with depth before and after dredging, respectively. Tidal slough sulfate concentrations decreased nearly linearly with depth.
FIG. 2.
Concentration profiles of pore water sulfate (A) and extractable sulfide minerals (B) with depth for Stege Marsh sampling sites. x axis uncertainties represent 2 standard deviations from the mean and estimated weighing precisions for sulfate and sulfides, respectively. y axis uncertainties reflect scales of homogenization of samples obtained from sediment coring.
Interlaboratory comparisons of IAEA silver sulfide standards showed good agreement (see Table S1 in the supplemental material). A total of >93% ± 1% of an NBS123 test sample was recovered by acid extraction and gave a δ34SV-CDT composition of ∼15‰. The difference from the published value of 14.3‰ (23) was a result of calibration to the IAEA standards (28). Results from sample analyses showed large variability and suggested complex sulfur isotope systematics. The δ34SV-CDT composition of AMD pond predredging surface water sulfate obtained at low tide was 1.4‰ ± 0.1‰, roughly the same as that of the original pyrite ore from which cinder deposits were derived (δ34SV-CDT = 1.14‰ ± 0.1‰). The δ34SV-CDT of sulfate from lower slough surface water was 19.9‰ ± 0.1‰, which is slightly lower than the generally accepted value for seawater (∼21‰) or the value for seawater sulfate (22.6‰) calibrated to the IAEA standards used in this study (25). All sediment and pore water sulfur isotope data here are reported as δ34SV-CDT, with subscripts indicative of the sample type (e.g., SO42−, AVS, CRS, etc.).
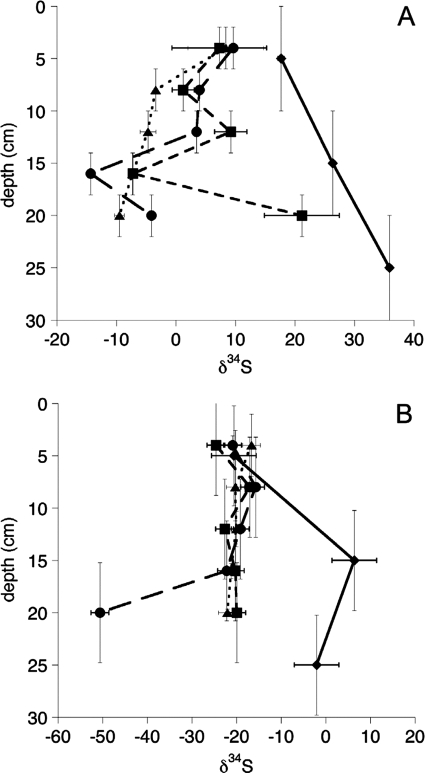
Sulfur isotope compositions for all other aqueous sulfates and for CRS are plotted versus depth in Fig. 3. For all but the AMD pond predredging samples, we observed highly variable δ34S values with depth. Only for samples from depths of >5 cm for the AMD pond predredging samples and of ∼20 cm for upper slough pore waters were δ34SSO4 values obtained that approached that of seawater. Otherwise, negative excursions in δ34SSO4 (as low as −14.3‰) from that of seawater were observed in the AMD pond postdredging samples and in the upper and lower tidal sloughs. With the exception of depths below 10 to 15 cm in the AMD pond predredging and upper slough samples, δ34SCRS compositions clustered around −21‰. Exceptions occurred for the AMD pond and acid sulfate soils at >10- and ∼20-cm depths, where δ34SCRS became significantly heavier (up to 10‰) and lighter (down to −51‰), respectively.
FIG. 3.
Sulfur isotope ratio profiles of pore water sulfate (A) and chromium-reducible sulfides (B). ⧫, AMD pond predredging; •, AMD pond postdredging; ▪, upper tidal slough; ▴, lower tidal slough. x axis uncertainties represent 2 standard errors of the mean (2 or 3 replicates), with the exception of the AMD pond predredging and upper tidal slough CRS values, where the uncertainties represent the largest value for 2 standard errors of the mean for IAEA standards obtained during analysis of these samples. y axis uncertainties reflect scales of homogenization of samples obtained from sediment coring. For reference, the δ34S value of AMD sulfate is 1.14‰ ± 0.1‰, and the δ34S value of seawater is ≈21‰.
DISCUSSION
Within the marsh, pH values varied significantly over relatively short lateral distances along the AMD flow path (see Fig. S1 in the supplemental material). This pH gradient resulted from both tidal and groundwater dilution (or buffering by dissolved bicarbonate) of the AMD stream. Where AMD pond sediments were exposed by dredging, pH values ranged from <2 to ∼4, independent of the location within this area. Tidal flooding, however, could have created less acidic and/or oxidizing microenvironments that offered more hospitable conditions for SRB growth (8). The result was most likely a complex spatial and temporal environment with respect to acidity and metal concentrations, two primary determinants of SRB species distribution and metabolic activity (60, 63, 88). The growth of SRB detected in the AMD pond (before and after dredging) may have been restricted to these microenvironments, as observed or inferred by previous studies of oxidizing marine sediments (48), acidic mine tailings (33, 83), and AMD-treated permeable reactive barriers (8), until conditions suitable for broader colonization could be reestablished.
The four sampling sites (AMD pond, acid sulfate soils, and upper and lower tidal sloughs) contained different but overlapping populations of close SRB relatives (Table 3). Although the detection of some OTUs at multiple sites and times could simply have reflected recent changes in geochemical conditions, we saw a clear separation of two OTU groups, associated with the upper (group II) and lower (group III) tidal sloughs. Although we cannot rule out undersampling of dsrAB diversity due to PCR amplification bias or undersequencing, our results showed a wider apparent distribution of Desulfobacteraceae in both the uncontaminated and heavily contaminated sediments, while Desulfobulbaceae relatives were less commonly observed.
The clear difference between groups I and IV (present in pre- and postdredged AMD pond sediments) indicated selection for different SRB types based on the pH, metal(loid) concentrations, and salinity associated with each site. The detection of a Desulfosalina relative (O12) supported some degree of selection for halotolerance. Other OTUs (O15, D4, and A21) related to acidic fen soil SRB clones may represent an acid-tolerant group with few or no cultured representatives.
In contrast to the differentiated groups, other OTUs were detected under highly variable environmental conditions (A19, A38, C6, D37, and F24). Thus, they appeared to be versatile SRB which were also resilient to AMD contamination. Furthermore, several clones (groups II and III) represented relatives of substrate-versatile species such as Desulfococcus multivorans (105) and Desulfoarculus baarsii (29). These phylotypes occurred on multiple branches of the dsr tree (Fig. 1), but none belonged to the Desulfovibrionaceae or deeper branches. OTU F24 was most closely related to Desulfosarcina, a substrate-versatile, completely oxidizing SRB often identified in natural and contaminated coastal and marine settings (20, 38, 59, 75, 80, 85, 86). Desulfosarcina was consistently found, for example, in Plum Island dsr libraries and may be common to salt marsh ecosystems in general (3, 58). Desulfosarcina spp. are also capable of oxidizing aromatic hydrocarbons (18) that contaminate many urban wetlands (54).
Desulfovibrio species are common SRB in coastal marine sediments (87) and were the dominant group of bacteria detected in this study. OTU C6, the most common dsr sequence recovered from multiple sites, may represent a highly adaptive Desulfovibrio species. The closest relative, Desulfovibrio fructosivorans, is known for its unique ability among SRB to oxidize fructose, a root exudate of the dominant salt marsh grass Spartina alterniflora (78).
OTU X4, which is most closely related to Desulfosporosinus orientis, dominated high-salinity enrichment cultures. The most-concentrated Cu(II)-containing cultures were also dominated by the rod-shaped morphotype and endospores associated with Desulfosporosinus spp. In contrast, growth of a Desulfovibrio desulfuricans G20 control culture was significantly inhibited by a Cu(II) concentration of ∼16 μM (88; this study). Copper tolerance may have been achieved in part by sorption of Cu2+ to some cells (19, 73). Although it was undetected in environmental samples, X4 represents a species cultivated from marsh sediments that exhibits some degree of tolerance to the copper and salinity concentrations associated with AMD contamination and periodic desiccation. Furthermore, sulfate reduction under acidic conditions (pH 3.8 to 4.2) has been demonstrated for a Desulfosporosinus orientis relative (55).
In contrast to previous findings (49), the 16S rRNA and dsrAB gene phylogenies of salt marsh whole-community DNA extracts were inconsistent. Fewer 16S rRNA gene sequences with known or likely sulfate-reducing closest relatives were obtained than were dsrAB sequences from the same sediment samples. For example, no Desulfobulbus, Syntrophobacter, Desulfoarculus, or Desulfosporosinus 16S rRNA gene sequences were detected in samples from which related dsrAB sequences were successfully recovered. This result supports the use of dsrAB genes as a measure of SRB diversity in environmental studies.
Sulfur isotope measurements allowed indirect assessment of SRB metabolic activity. Negative δ34SCRS values supported a primary role for SRB in the production of authigenic metal sulfides. The clustering of δ34SCRS values around −21‰ suggests the dominance of a particular metabolic guild of SRB species (26), consistent with our detection of a single dominant dsrAB phylotype (WSMC6, a close relative of Desulfovibrio fructosivorans). An excursion to lower values in the AMD pond postdredging (acid sulfate soil) likely reflects a contribution to sulfur redox cycling from sulfur-disproportionating bacteria (17). Indeed, vertical stratification of potential oxidants insufficiently represents a system in which brackish tidewaters and fresher groundwaters exchange diurnally, especially within marsh sloughs (97) or where bioturbation can increase sediment structural complexity (61). Higher δ34SCRS values approaching or exceeding 0‰ in the AMD pond (predredging) clearly reflected increased contributions from abiotically formed pyrite associated with cinder deposits, consistent with the observed increase in the concentration (wt%) of CRS in this site at certain depths.
Interestingly, most δ34SSO4 values suggested that the tidal slough sediments exhibited open-system behavior with respect to sulfate, consistent with an influx of sulfate-depleted groundwater during periods of tidal retreat. The data suggest that large quantities of biogenic (isotopically lighter) bisulfide (HS−) must have been reoxidized chemically or microbially to produce δ34SSO4 values lower than that of seawater. This observation is consistent with reports that ≥95% of biogenic sulfide is recycled in salt marsh sediments, with roughly half coming from dissolved S(II) (92). Bacteria such as Thiobacillus denitrificans are well known to couple the reoxidation of solid-phase sulfide (FeS) to nitrate reduction (6), thereby extending S(II) oxidation from the oxic to the suboxic zone in flooded sediments.
We noted that 16S rRNA gene sequences amplified from Stege Marsh sediments (see Fig. S3 in the supplemental material) revealed close relatives of the epsilon- and gammaproteobacterial subgroups of Thiomicrospira (note that the epsilonproteobacterium Thiomicrospira denitrificans was reclassified as Sulfurimonas denitrificans [93]). This chemolithoautotrophic genus can also couple the oxidization of aqueous sulfide to the reduction of nitrate (36), producing intermediate redox-state sulfur species and sulfate (53, 64), similar to Thiobacillus denitrificans, and Sulfurimonas denitrificans was the closest relative to several of our 16S rRNA gene clones. Nitrate concentrations in Stege Marsh varied from 11 to 44 mg/liter in groundwater inputs (47) to below the detection limits in slough sediment pore waters (∼0.5 mg/liter). These observations collectively suggested that sulfide oxidation coupled to nitrate reduction by anaerobic sulfide-oxidizing bacteria (such as Sulfurimonas) could explain observed trends in sediment and surface water sulfate and nitrate concentrations. Recent studies highlight the importance of anaerobic sulfide-oxidizing bacteria in the oxidative (re)cycling of reduced sulfur species (91).
The 16S rRNA gene sequence data also revealed a contingent of environmental clones most closely related to Halothiobacillus (52) (see Fig. S3 in the supplemental material), a halotolerant obligate aerobe capable of oxidizing sulfide. We noted that pore water salinity during high tides typically approached 42 to 47 mM NaCl, and it increased in some places to roughly 1.2 M during the lowest tides. These increased salinity levels are interpreted to have selected for close relatives of Halothiobacillus. Therefore, sulfate concentration profiles in the tidal slough sediments reflected a combination of microbial seawater sulfate consumption (via BSR) and biogenic bisulfide reoxidation (both aerobic and anaerobic), with periodic tidal retreat and groundwater influx.
In contrast, sulfate concentrations in the AMD pond remained consistently higher due to the input of oxidizing cinders, even during the lowest tides. This observation was most evident in the acid sulfate soil-like AMD pond sediments after dredging, where cinders and authigenic sulfides were most exposed to oxidative weathering. Values of δ34SSO4 and δ34SCRS for AMD pond sediments reflected more closed-system characteristics, and both seawater sulfate and biogenic chromium-reducible sulfides evidently contained some isotopic “contamination,” from AMD-derived sulfuric acid and cinders, respectively.
In conclusion, phylogenetic analyses of recovered dsrAB sequences revealed the association of distinct SRB groups within the marsh community. Some of these SRB groups appeared uniquely associated with an environment, while others spanned geochemical gradients. Both clone libraries and sulfur isotopic data supported the existence of a metabolically active SRB community throughout the marsh, including within the most acidic sediments. More active and resilient SRB species play a greater role in influencing the linked biogeochemical cycles of sulfur and chalcophilic metal(loid)s in Stege Marsh and other brackish wetlands. The efficiency of metal sulfide precipitation is tempered by microbial sulfide reoxidation occurring on diurnal timescales and mediated by the activities of both aerobic and anaerobic sulfide-oxidizing bacteria.
Supplementary Material
Acknowledgments
We thank Karl Hans and Anna Moore (UC Berkeley Environmental Health and Safety) for permission to work in the RFS Stege Marsh during active remediation operations, Stephen Andrews and Bill Berry (UC Berkeley Department of Earth and Planetary Science) for the use of field sampling equipment, Miza Moreau for fieldwork assistance, and Alexander Loy (University of Vienna) for advice on dsrAB sequence data analysis. We thank three anonymous reviewers for their excellent and helpful comments.
This research was funded by the U.S. Department of Energy's Basic Energy Sciences program under grant DE-AC02-05CH11231.
Footnotes
Published ahead of print on 14 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alongi, D. M. 1998. Coastal ecosystem processes. CRC Marine Science Series, vol. 3. CRC Press, Boca Raton, FL.
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bahr, M., B. C. Crump, V. Klepac-Ceraj, A. Teske, M. L. Sogin, and J. E. Hobbie. 2005. Molecular characterization of sulfate-reducing bacteria in a New England salt marsh. Environ. Microbiol. 7:1175-1185. [DOI] [PubMed] [Google Scholar]
- 4.Baker, B. J., D. P. Moser, B. J. MacGregor, S. Fishbain, M. Wagner, N. K. Fry, B. Johnson, N. Speolstra, S. Loos, K. Takai, B. Sherwood Lollar, J. Fredrickson, D. Balkwill, T. C. Onstott, C. F. Wimpee, and D. A. Stahl. 2003. Related assemblages of sulphate-reducing bacteria associated with ultradeep gold mines of South Africa and deep basalt aquifers of Washington State. Environ. Microbiol. 5:267-277. [DOI] [PubMed] [Google Scholar]
- 5.Batten, K. M., and K. M. Scow. 2003. Sediment microbial community composition and methylmercury pollution at four mercury mine-impacted sites. Microb. Ecol. 46:429-441. [DOI] [PubMed] [Google Scholar]
- 6.Beller, H. R., P. S. G. Chain, T. E. Letain, A. Chakicherla, F. W. Larimer, P. M. Richardson, M. A. Coleman, A. P. Wood, and D. P. Kelly. 2006. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J. Bacteriol. 188:1473-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-David, E. A., P. J. Holden, D. J. M. Stone, B. D. Harch, and L. J. Foster. 2004. The use of phospholipid fatty acid analysis to measure impact of acid rock drainage on microbial communities in sediments. Microb. Ecol. 48:300-315. [DOI] [PubMed] [Google Scholar]
- 8.Benner, S. G., W. D. Gould, and D. W. Blowes. 2000. Microbial populations associated with the generation and treatment of acid mine drainage. Chem. Geol. 169:435-448. [Google Scholar]
- 9.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2008. GenBank. Nucleic Acids Res. 36:D25-D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bond, P. L., G. K. Druschel, and J. F. Banfield. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boothman, C., S. Hockin, D. E. Holmes, G. M. Gadd, and J. R. Lloyd. 2006. Molecular analysis of a sulphate-reducing consortium used to treat metal-containing effluents. Biometals 19:601-609. [DOI] [PubMed] [Google Scholar]
- 12.Bricker, S. B. 1993. The history of Cu, Pb, and Zn inputs to Narragansett Bay, Rhode Island as recorded by salt marsh sediments. Estuaries 16:589-607. [Google Scholar]
- 13.Burke, D. J., E. P. Hamerlynck, and D. Hahn. 2002. Interactions among plant species and microorganisms in salt marsh sediments. Appl. Environ. Microbiol. 68:1157-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton, E. D., R. T. Bush, L. A. Sullivan, R. K. Hocking, D. R. G. Mitchell, S. G. Johnston, R. W. Fitzpatrick, M. Raven, S. McClure, and L. Y. Jang. 2009. Iron-monosulfide oxidation in natural sediments: resolving microbially mediated S transformations using XANES, electron microscopy, and selective extractions. Environ. Sci. Technol. 43:3128-3134. [DOI] [PubMed] [Google Scholar]
- 15.Cabrera, G., R. Pérez, J. M. Gomez, A. Ábalos, and D. Cantero. 2006. Toxic effects of dissolved heavy metals on Desulfovibrio vulgaris and Desulfovibrio sp. strains. J. Hazard. Mater. A 135:40-46. [DOI] [PubMed] [Google Scholar]
- 16.Canfield, D. E., R. Raiswell, J. T. Westrich, C. M. Reaves, and R. A. Berner. 1986. The use of chromium reduction in the analysis of reduced inorganic sulfur in sediments and shales. Chem. Geol. 54:149-155. [Google Scholar]
- 17.Canfield, D. E. 2001. Biogeochemistry of sulfur isotopes. Rev. Mineral. 43:607-636. [Google Scholar]
- 18.Chakraborty, R., and J. D. Coates. 2004. Anaerobic degradation of monoaromatic hydrocarbons. Appl. Microbiol. Biotechnol. 64:437-446. [DOI] [PubMed] [Google Scholar]
- 19.Chang, J.-S., R. Law, and C.-C. Chang. 1997. Biosorption of lead, copper and cadmium by biomass of Pseudomonas aeruginosa PU21. Water Res. 31:1651-1658. [Google Scholar]
- 20.Chin, K. J., M. L. Sharma, L. A. Russell, K. R. O'Neill, and D. R. Lovley. 2007. Quantifying expression of a dissimilatory (bi)sulfite reductase gene in petroleum-contaminated marine harbor sediments. Microb. Ecol. 55:489-499. [DOI] [PubMed] [Google Scholar]
- 21.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 22.Crawford, R. L., and P. Olson. 1978. Microbial catabolism of vanillate: decarboxylation to guaiacol. Appl. Environ. Microbiol. 36:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowe, D. E., and R. G. Vaughan. 1996. Characterization and use of isotopically homogenous standards for in situ laser microprobe analysis of 34S/32S ratios. Am. Mineral. 81:187-193. [Google Scholar]
- 24.Crump, B. C., C. S. Hopkinson, M. L. Sogin, and J. E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis, A. S., D. A. Clague, R. A. Zierenberg, C. G. Wheat, and B. L. Cousens. 2003. Sulfide formation related to changes in the hydrothermal system on Loihi Seamount, Hawai'i, following the seismic event in 1996. Can. Mineral. 41:457-472. [Google Scholar]
- 26.Detmers, J., V. Brüchert, K. S. Habicht, and J. Kuever. 2001. Diversity of sulfur isotope fractionations by sulfate-reducing prokaryotes. Appl. Environ. Microbiol. 67:888-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding, T., S. Valkiers, H. Kipphardt, P. De Bièvre, P. D. P. Taylor, R. Gonfiantini, and R. Krouse. 2001. Calibrated sulfur isotope abundance ratios of three IAEA sulfur isotope reference materials and V-CDT with a reassessment of the atomic weight of sulfur. Geochim. Cosmochim. Acta 65:2433-2437. [Google Scholar]
- 29.Drzyzga, O., J. Küver, and K.-H. Blotevogel. 1993. Complete oxidation of benzoate and 4-hydroxybenzoate by a new sulfate-reducing bacterium resembling Desulfoarculus. Arch. Microbiol. 159:109-113. [DOI] [PubMed] [Google Scholar]
- 30.Dubilier, N., C. Mülders, T. Ferdelman, D. de Beer, A. Pernthaler, M. Klein, M. Wagner, C. Erséus, F. Thiermann, J. Krieger, O. Giere, and R. Amann. 2001. Endosymbiotic sulphate-reducing and sulphate-oxidizing bacteria in an oligochaete worm. Nature 411:298-302. [DOI] [PubMed] [Google Scholar]
- 31.Edgcomb, V. P., J. H. McDonald, R. Devereux, and D. W. Smith. 1999. Estimation of bacterial cell numbers in humic acid-rich salt marsh sediments with probes directed to 16S ribosomal DNA. Appl. Environ. Microbiol. 63:1516-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Environmental Protection Agency. 2003. National primary drinking water standards. EPA 816-F-03-016. Office of Water (4606M), EPA, Washington, DC. www.epa.gov/safewater.
- 33.Fortin, D., and T. J. Beveridge. 1997. Microbial sulfate reduction within sulfidic mine tailings: formation of diagenetic Fe sulfides. Geomicrobiol. J. 14:1-21. [Google Scholar]
- 34.Frischer, M. E., J. M. Danforth, M. A. Newton Healy, and F. M. Saunders. 2000. Whole-cell versus total RNA extraction for analysis of microbial community structure with 16S rRNA-targeted oligonucleotide probes in salt marsh sediments. Appl. Environ. Microbiol. 66:3037-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gammons, C. H., and A. K. Frandsen. 2001. Fate and transport of metals in H2S-rich waters at a treatment wetland. Geochem. Trans. 2:1. [Google Scholar]
- 36.Gevertz, D., A. J. Telang, G. Voordouw, and G. E. Jenneman. 2000. Isolation and characterization of strains CVO and FWKO B, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl. Environ. Microbiol. 66:2491-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giani, L., K. Dittrich, A. Martsfield-Hartmann, and G. Peters. 1996. Methanogenesis in saltmarsh soils of the North Sea coast of Germany. Eur. J. Soil Sci. 47:175-182. [Google Scholar]
- 38.Gillan, D. C., B. Danis, P. Pernet, G. Joly, and P. Dubois. 2005. Structure of sediment-associated microbial communities along a heavy-metal contamination gradient in the marine environment. Appl. Environ. Microbiol. 71:679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hallberg, K. B., and D. B. Johnson. 2005. Microbiology of a wetland ecosystem constructed to remediate mine drainage from a heavy metal mine. Sci. Total Environ. 338:53-66. [DOI] [PubMed] [Google Scholar]
- 40.Harris, M. A., and S. Ragusa. 2000. Bacterial mitigation of pollutants in acid drainage using decomposable plant material and sludge. Environ. Geol. 40:195-215. [Google Scholar]
- 41.Hines, M. E. 2006. Microbially mediated redox cycling at the oxic-anoxic boundary in sediments: comparison of animal and plants habitats. Water Air Soil Pollut. 6:523-536. [Google Scholar]
- 42.Howarth, R. W. 1993. Microbial processes in salt-marsh sediments, p. 239-259. In T. E. Ford (ed.), Aquatic microbiology: an ecological approach. Blackwell, Oxford, United Kingdom.
- 43.Howarth, R. W., and J. M. Teal. 1979. Sulfate reduction in a New England salt marsh. Limnol. Oceanogr. 24:999-1013. [Google Scholar]
- 44.Howarth, R. W., and A. Giblin. 1983. Sulfate reduction in the salt marshes at Sapelo Island, Georgia. Limnol. Oceanogr. 28:70-82. [Google Scholar]
- 45.Hugenholtz, P., and T. Huber. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst. Evol. Microbiol. 53:289-293. [DOI] [PubMed] [Google Scholar]
- 46.Johnson, D. B., and K. B. Hallberg. 2005. Biogeochemistry of the compost bioreactor components of a composite acid mine drainage passive remediation system. Sci. Total Environ. 338:81-93. [DOI] [PubMed] [Google Scholar]
- 47.Jones & Associates, Inc., Environmental Consultants. 1990. Removal site evaluation of mercury in soil and groundwater at former mercury fulminate facility (report submitted to the University of California, Richmond Field Station, 21 May 1990). University of California—Berkeley Environmental Health and Safety Office, Berkeley, CA.
- 48.Jorgensen, B. B. 1977. Bacterial sulfate reduction within reduced microniches of oxidized marine sediments. Mar. Biol. 41:7-17. [Google Scholar]
- 49.Joulian, C., N. B. Ramsing, and K. Ingvorsen. 2001. Congruent phylogenies of most common small-subunit rRNA and dissimilatory sulfite reductase gene sequences retrieved from estuarine sediments. Appl. Environ. Microbiol. 67:3314-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaksonen, A. H., J. J. Plumb, W. J. Robertson, P. D. Franzmann, J. A. E. Gibson, and J. A. Puhakka. 2004. Culturable diversity and community fatty acid profiling of sulfate-reducing fluidized bed reactors treating acidic metal-containing wastewater. Geomicrobiol. J. 21:469-480. [Google Scholar]
- 51.Keith-Roach, M. J., N. D. Bryan, R. D. Bardgett, and F. R. Livens. 2002. Seasonal changes in the microbial community of a saltmarsh, measured by phospholipid fatty acid analysis. Biogeochemistry 60:77-96. [Google Scholar]
- 52.Kelly, D. P., and A. P. Wood. 2000. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50:511-516. [DOI] [PubMed] [Google Scholar]
- 53.Kelly, D. P., and A. P. Wood. 2001. The chemolithotrophic prokaryotes. Springer-Verlag, New York, NY.
- 54.Kimbrough, K. L., and R. M. Dickhut. 2006. Assessment of polycyclic aromatic hydrocarbon input to urban wetlands in relation to adjacent land use. Mar. Pollut. Bull. 52:1355-1363. [DOI] [PubMed] [Google Scholar]
- 55.Kimura, S., K. B. Hallberg, and D. B. Johnson. 2006. Sulfidogenesis in low pH (3.8-4.2) media by a mixed population of acidophilic bacteria. Biodegradation 17:57-65. [DOI] [PubMed] [Google Scholar]
- 56.Kjeldsen, K. U., A. Loy, T. F. Jakobsen, T. R. Thomsen, M. Wagner, and K. Ingvorsen. 2007. Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah). FEMS Microbiol. Ecol. 60:287-298. [DOI] [PubMed] [Google Scholar]
- 57.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klepac-Ceraj, V., M. Bahr, B. C. Crump, A. P. Teske, J. E. Hobbie, and M. F. Polz. 2004. High overall diversity and dominance of microdiverse relationships in salt marsh sulphate-reducing bacteria. Environ. Microbiol. 6:686-698. [DOI] [PubMed] [Google Scholar]
- 59.Knittel, K., A. Boetius, A. Lemke, H. Eilers, K. Lochte, O. Pfannkuche, and P. Linke. 2003. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia Margin, Oregon). Geomicrobiol. J. 20:269-294. [Google Scholar]
- 60.Kolmert, A., and D. B. Johnson. 2001. Remediation of acidic waste waters using immobilized, acidophilic sulfate-reducing bacteria. J. Chem. Technol. Biotechnol. 76:836-843. [Google Scholar]
- 61.Koretsky, C. M., P. Van Cappellen, T. J. Dichristina, J. E. Kostka, K. L. Lowe, C. M. Moore, A. N. Roychoudhury, and E. Viollier. 2005. Salt marsh pore water geochemistry does not correlate with microbial community structure. Estuar. Coast. Shelf Sci. 62:233-251. [Google Scholar]
- 62.Kostka, J. E., A. Roychoudhury, and P. Van Cappellen. 2002. Rates and controls of anaerobic microbial respiration across spatial and temporal gradients in saltmarsh sediments. Biogeochemistry 60:49-76. [Google Scholar]
- 63.Küsel, K., U. Roth, T. Trinkwalter, and S. Peiffer. 2001. Effect of pH on the anaerobic microbial cycling of sulfur in mining-impacted freshwater lake sediments. Environ. Exp. Bot. 46:213-223. [Google Scholar]
- 64.Lane, D. J., D. A. Stahl, G. J. Olsen, D. J. Heller, and N. R. Pace. 1985. Phylogenetic analysis of the genera Thiobacillus and Thiomicrospira by 5S rRNA sequences. J. Bacteriol. 163:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leloup, J., L. Quillet, T. Berthe, and F. Petit. 2006. Diversity of the dsrAB (dissimilatory sulfite reductase) gene sequences retrieved from two contrasting mudflats of the Seine estuary, France. FEMS Microbiol. Ecol. 55:230-238. [DOI] [PubMed] [Google Scholar]
- 66.Levin, L. A., D. F. Boesch, A. Covich, C. Dahm, C. Erséus, K. C. Ewel, R. T. Kneib, A. Moldenke, M. A. Palmer, P. Snelgrove, D. Strayer, and J. Marcin Weslawski. 2001. The function of marine critical transition zones and the importance of sediment biodiversity. Ecosystems 4:430-451. [Google Scholar]
- 67.Lowe, K. L., T. J. DiChristina, A. N. Roychoudhury, and P. Van Cappellen. 2000. Microbiological and geochemical characterization of microbial Fe(III) reduction in salt marsh sediments. Geomicrobiol. J. 17:163-178. [Google Scholar]
- 68.Loy, A., K. Küsel, A. Lehner, H. J. Drake, and M. Wagner. 2004. Microarray and functional gene analysis of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal co-occurrence of recognized and novel lineages. Appl. Environ. Microbiol. 70:6998-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Macalady, J. L., E. E. Mack, D. C. Nelson, and K. M. Scow. 2000. Sediment microbial community structure and mercury methylation in mercury-polluted Clear Lake, California. Appl. Environ. Microbiol. 66:1479-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Magenheimer, J. F., T. R. Moore, G. L. Chmura, and R. J. Daoust. 1996. Methane and carbon dioxide flux from a macrotidal salt marsh, Bay of Fundy, New Brunswick. Estuaries 19:139-145. [Google Scholar]
- 72.Morales, T. A., M. Dopson, R. Athar, and R. B. Herbert, Jr. 2005. Analysis of bacterial diversity in acidic pond water and compost after treatment of artificial acid mine drainage for metal removal. Biotechnol. Bioeng. 90:543-551. [DOI] [PubMed] [Google Scholar]
- 73.Mullen, D., D. C. Wolf, F. G. Ferris, T. J. Beveridge, C. A. Fleming, and G. W. Bailey. 1989. Bacterial sorption of heavy metals. Appl. Environ. Microbiol. 55:3143-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muller, T. J. 1999. Determination of salinity, p. 41-74. In K. Grasshoff, K. Kremling, and M. Ehrhardt (ed.), Methods of seawater analysis, 3rd ed. Wiley-VCH, New York, NY.
- 75.Mussmann, M., K. Ishii, R. Rabus, and R. Amann. 2005. Diversity and vertical distribution of cultured and uncultured Deltaproteobacteria in an intertidal mud flat of the Wadden Sea. Environ. Microbiol. 7:405-418. [DOI] [PubMed] [Google Scholar]
- 76.Neculita, C. M., G. J. Zagury, and B. Bussiére. 2007. Passive treatment of acid mine drainage in bioreactors using sulfate-reducing bacteria: critical review and research needs. J. Environ. Qual. 36:1-16. [DOI] [PubMed] [Google Scholar]
- 77.Nicomrat, D., W. A. Dick, and O. H. Tuovinen. 2006. Assessment of the microbial community in a constructed wetland that receives acid coal mine drainage. Microb. Ecol. 51:83-89. [DOI] [PubMed] [Google Scholar]
- 78.Pakulski, J. D. 1986. Release of reducing sugars and dissolved organic carbon from Spartina alterniflora Loisel in a Georgia salt marsh. Estuar. Coast. Shelf Sci. 22:385-394. [Google Scholar]
- 79.Paytan, A., M. Kastner, E. E. Martin, J. D. Macdougall, and T. Herbert. 1993. Marine barite as a monitor of seawater strontium isotope composition. Nature 366:445-449. [Google Scholar]
- 80.Pérez-Jiménez, J. R., L. Y. Young, and L. J. Kerkhof. 2001. Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsrAB) genes. FEMS Microbiol. Ecol. 35:145-150. [DOI] [PubMed] [Google Scholar]
- 81.Pérez-Jiménez, J. R., and L. J. Kerkhof. 2005. Phylogeography of sulfate-reducing bacteria among disturbed sediments, disclosed by analysis of the dissimilatory sulfite reductase genes (dsrAB). Appl. Environ. Microbiol. 71:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Postgate, J. R. 1984. The sulphate-reducing bacteria, 2nd ed., p. 35. Cambridge University Press, Cambridge, United Kingdom.
- 83.Praharaj, T., and D. Fortin. 2004. Indicators of microbial sulfate reduction in acidic sulfide-rich mine tailings. Geomicrobiol. J. 21:457-467. [Google Scholar]
- 84.Rampinelli, L. R., R. D. Azevedo, M. C. Teixeira, R. Guerra-Sá, and V. A. Leão. 2008. A sulfate-reducing bacterium with unusual growing capacity in moderately acidic conditions. Biodegradation 19:613-619. doi: 10.1007/s10532-007-9166-y. [DOI] [PubMed] [Google Scholar]
- 85.Ravenschlag, K., K. Sahm, C. Knoblauch, B. B. Jorgensen, and R. Amann. 2000. Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine Arctic sediments. Appl. Environ. Microbiol. 66:3592-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rooney-Varga, J., R. Devereux, R. S. Evans, and M. E. Hines. 1997. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl. Environ. Microbiol. 63:3895-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sahm, K., B. J. MacGregor, B. B. Jorgensen, and D. A. Stahl. 1999. Sulphate reduction and vertical distribution of sulphate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ. Microbiol. 1:65-74. [DOI] [PubMed] [Google Scholar]
- 88.Sani, R. K., B. M. Peyton, and L. T. Brown. 2001. Copper-induced inhibition of growth of Desulfovibrio desulfuricans G20: assessment of its toxicity and correlation with those of zinc and lead. Appl. Environ. Microbiol. 67:4765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sierra-Alvarez, R., J. Hollingsworth, and M. S. Zhou. 2007. Removal of copper in an integrated sulfate-reducing bioreactor-crystallization reactor system. Environ. Sci. Technol. 41:1426-1431. [DOI] [PubMed] [Google Scholar]
- 91.Sievert, S. M., K. M. Scott, M. G. Klotz, P. S. G. Chain, L. J. Hauser, J. Hemp, M. Hugler, M. Land, A. Lapidus, F. W. Larimer, S. Lucas, S. A. Malfatti, F. Meyer, I. T. Paulsen, Q. Ren, J. Simon, and the USF Genomics Class. 2008. Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 74:1145-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Swider, K. T., and J. E. Mackin. 1989. Transformations of sulfur compounds in marsh-flat sediments. Geochim. Cosmochim. Acta 53:2311-2323. [Google Scholar]
- 93.Takai, K., M. Suzuki, S. Nakagawa, M. Miyazaki, Y. Suzuki, F. Inagaki, and K. Horikoshi. 2006. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 56:1725-1733. [DOI] [PubMed] [Google Scholar]
- 94.Tsukamoto, T. K., H. A. Killion, and G. C. Miller. 2004. Column experiments for microbiological treatment of acid mine drainage: low temperature, low-pH and matrix investigations. Water Res. 38:1405-1418. [DOI] [PubMed] [Google Scholar]
- 95.URS Corporation. 2000. Field sampling and analysis results, University of California, Berkeley, Richmond Field Station/Stege Marsh, Richmond, California. University of California—Berkeley Environmental Health and Safety Office, Berkeley, CA.
- 96.URS Corporation. 2002. Conceptual remedial action plan, marsh portion of subunit 2B, Richmond Field Station/Stege Marsh, Richmond, California. University of California—Berkeley Environmental Health and Safety Office, Berkeley, CA.
- 97.Ursino, N., S. Silvestri, and M. Marani. 2004. Subsurface flow and vegetation patterns in tidal environments. Water Resour. Res. 40:W05115. doi: 10.1029/2003WR002702. [DOI] [Google Scholar]
- 98.Utgikar, V. P., B. Y. Chen, N. Chaudhary, H. H. Tabak, J. R. Haines, and R. Govind. 2001. Acute toxicity of heavy metals to acetate-utilizing mixed cultures of sulfate-reducing bacteria: EC100 and EC50. Environ. Toxicol. Chem. 20:2662-2669. [PubMed] [Google Scholar]
- 99.Valette-Silver, N. J. 1993. The use of sediment cores to reconstruct historical trends in contamination of estuarine and coastal sediments. Estuaries 16:577-588. [Google Scholar]
- 100.Valiela, I., and J. M. Teal. 1979. The nitrogen budget of a salt marsh ecosystem. Nature 280:652-656. [Google Scholar]
- 101.Vernberg, F. J. 1993. Salt marsh processes: a review. Environ. Toxicol. Chem. 12:2167-2195. [Google Scholar]
- 102.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductase supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wagner, M., A. Loy, M. Klein, N. Lee, N. B. Ramsing, D. A. Stahl, and M. W. Friedrich. 2005. Functional marker genes for identification of sulfate-reducing prokaryotes. Methods Enzymol. 397:469-489. [DOI] [PubMed] [Google Scholar]
- 104.Webb, J. S., S. McGinness, and H. M. Lappin-Scott. 1998. Metal removal by sulphate-reducing bacteria from natural and constructed wetlands. J. Appl. Microbiol. 84:240-248. [DOI] [PubMed] [Google Scholar]
- 105.Widdel, F. 1980. Anaerober Abbau von Fettsäuren und Benzoesäure durch neu isolierte Arten sulfatreduzierender Bakterien. Thesis. University of Göttingen, Göttingen, Germany.
- 106.Windhom, H. L., S. J. Schropp, F. D. Calder, J. D. Ryan, R. G. Smith, Jr., L. C. Burney, F. G. Lewis, and C. H. Rawlinson. 1989. Natural trace metal concentrations in estuarine and coastal marine sediments of the southeastern United States. Environ. Sci. Technol. 23:314-320. [Google Scholar]
- 107.Zverlov, V., M. Klein, S. Lücker, M. W. Friedrich, J. Kellermann, D. A. Stahl, A. Loy, and M. Wagner. 2005. Lateral gene transfer of dissimilatory (bi)sulfite reductase revisited. J. Bacteriol. 187:2203-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.