Abstract
Red-pigmented biofilms grow on rock and cobble surfaces present in anoxic hot springs located on Paoha Island in Mono Lake. The bacterial community was dominated (∼ 85% of 16S rRNA gene clones) by sequences from the photosynthetic Ectothiorhodospira genus. Scraped biofilm materials incubated under anoxic conditions rapidly oxidized As(III) to As(V) in the light via anoxygenic photosynthesis but could also readily reduce As(V) to As(III) in the dark at comparable rates. Back-labeling experiments with 73As(V) demonstrated that reduction to 73As(III) also occurred in the light, thereby illustrating the cooccurrence of these two anaerobic processes as an example of closely coupled arsenotrophy. Oxic biofilms also oxidized As(III) to As(V). Biofilms incubated with [14C]acetate oxidized the radiolabel to 14CO2 in the light but not the dark, indicating a capacity for photoheterotrophy but not chemoheterotrophy. Anoxic, dark-incubated samples demonstrated As(V) reduction linked to additions of hydrogen or sulfide but not acetate. Chemoautotrophy linked to As(V) as measured by dark fixation of [14C]bicarbonate into cell material was stimulated by either H2 or HS−. Functional genes for the arsenate respiratory reductase (arrA) and arsenic resistance (arsB) were detected in sequenced amplicons of extracted DNA, with about half of the arrA sequences closely related (∼98% translated amino acid identity) to those from the family Ectothiorhodospiraceae. Surprisingly, no authentic PCR products for arsenite oxidase (aoxB) were obtained, despite observing aerobic arsenite oxidation activity. Collectively, these results demonstrate close linkages of these arsenic redox processes occurring within these biofilms.
Oxyanions of the group 15 element arsenic, arsenate [As(V)] and arsenite [As(III)], have been known for millennia to be potent poisons. Despite its well-established toxicity to life, the phenomenon of arsenic resistance was discovered whereby some microorganisms maintain an otherwise “normal” existence in the presence of high concentrations of As(V) or As(III) (17, 29, 31). More recently it has become recognized that certain representatives from the bacterial and archaeal domains can actually exploit the electrochemical potential of the As(V)/As(III) redox couple (+130 mV) to gain energy for growth. This can be achieved either by employing As(III) as an autotrophic electron donor or by using As(V) as a respiratory electron acceptor (18, 21, 34). The latter phenomenon, although most commonly associated with chemoheterotrophy, can also employ inorganic substances like sulfide or H2. Indeed, As(V)-respiring anaerobes displaying a capacity for chemoautotrophy with these electron donors have been isolated and described (5, 7, 16). We recently reported that photoautotrophy is supported by As(III) in anoxic biofilms located in hot springs on Paoha Island in Mono Lake, CA (15). This process represented a novel means of As(III) oxidation achieved via anoxygenic photosynthesis occurring in certain photosynthetic bacteria (i.e., Ectothiorhodospira) and possibly within some cyanobacteria as well (e.g., “Oscillatoria”).
Whether or not a microbial habitat is overtly oxic or anoxic, or temporally shifts between these two states over a diel cycle, critical energy linkages between aerobes and anaerobes have long been known for the biogeochemical cycles of key elements, such as sulfur, iron, and nitrogen. Most prominently studied is the case of nitrogen, whereby an ecological coupling exists between the processes of nitrification and denitrification (9, 10, 28). The former process provides energy to aerobic nitrifiers, while the latter process consumes the nitrate produced by this reaction, thereby meeting the energy needs of the denitrifiers.
For arsenic, the detection of both As(III) oxidation and As(V) reduction in oxic and anoxic incubations of freshly collected periphyton suggested that an analogous coupled process may also occur for this element (12). Similarly, several uncontaminated soils in Japan displayed a capacity for either As(V) reduction or As(III) oxidation upon arsenic oxyanion amendment and whether they were incubated under oxic or anoxic conditions (39). A defined coculture consisting of an aerobic As(III) oxidizer (strain OL1) and an anaerobic As(V) respirer (strain Y5) was shown to function in this fashion under manipulated laboratory conditions of oxygen tension (26). We pursued the phenomenon of coupled arsenic metabolism further by using materials collected from the hot spring biofilms in Mono Lake, but we focused on examination of the cycling of arsenic under anoxic conditions.
In this paper we report results obtained by manipulated incubations of red-pigmented biofilms found in the hot springs of Paoha Island. Preliminary community characterizations of these biofilms show that they are dominated by Bacteria from the genus Ectothiorhodospira but also harbor an assemblage of Archaea related to the Halobacteriacaea. Incubation results have demonstrated the presence of the following arsenic metabolic activities: respiratory As(V) reduction, photosynthetic anaerobic As(III) oxidation, and aerobic As(III) oxidation, along with the ecophysiological conditions under which they occur. Surprisingly, we were unable to obtain authentic PCR products for arsenite oxidase genes (aoxB), despite observing aerobic As(III) oxidation activity. These biofilms serve as a model system for how anaerobic cycling of arsenic can be sustained with oxidation of As(III) by anoxygenic photosynthesis coupled to regeneration of this electron donor via dissimilatory As(V) reduction. The significance that such a light-driven anaerobic ecosystem may have played in the Archean Earth is discussed.
MATERIALS AND METHODS
Site description.
Paoha Island (37°59.633′N, 119°01.376′W) is located in Mono Lake, CA. The lake is geographically positioned on the eastern escarpment of the Sierra Nevada Mountains, bordering the western boundary of the Great Basin Desert. The island consists of uplifted lake bottom sediments that emerged from the lakebed about 500 years ago as a consequence of volcanic forces (33). A cinder cone is evident on its northern shore, while the hot springs we sampled are located on its southern shore and are also associated with previous episodes of volcanic activity. Volcanic rocks (e.g., pumice and breccia) are interspersed with the clay-like soils. The hot springs along with their active ebullition of reduced gases (e.g., methane, ethane, propane) in this region were described previously (19), and the fermentative alkalithermophile Anaerobranca californiensis was isolated from this location (4). The hot springs consisted of two primary types as indicated by temperature and color. Green-colored springs and small ponds were relatively rare, had high ambient temperatures (∼65°), and were dominated by cyanobacteria. The more abundant red springs and ponds had lower temperatures (∼45°C) and were dominated by purple photosynthetic bacteria; materials collected from these environments are the basis of this study. The spring waters were anoxic and contained abundant dissolved sulfide, ammonia, methane, and arsenite. Further details on the appearance and physical/chemical properties of these hot springs can be found elsewhere (15).
Sample collection and processing.
The red-colored biofilms of several of the ∼45°C springs were sampled in August 2008, October 2008, and April 2009. Biofilm-covered rocks and clay cobbles were collected from the ponds, placed in self-sealing plastic bags, and transported back to the shore for processing. The biofilm was removed from the rock surfaces with a sterile toothbrush and rinsed with a sterile mineral salts medium (see below). The resulting slurry was collected in 150-ml serum bottles as thick suspensions, which were bubbled with N2 and crimp sealed. Slurries were stored in the dark at 5°C prior to use in experiments and remained viable for several months, yielding reproducible experimental results over time with regard to arsenic metabolism. For DNA analyses, biofilm was scraped from the rocks with a sterile spatula, transferred into a sterile microcentrifuge tube, and immediately placed on dry ice and stored at −80°C until extraction.
Incubation experiments.
The composition of the artificial hot spring medium, designed to mimic the pH and salt composition of the Paoha Island hot spring water, was as follows (with concentrations in g liter−1 shown in parentheses): NaCl (20), (NH4)2SO4 (0.1), KH2PO4 (0.08), K2HPO4 (0.15), MgSO4·7H2O (0.025), H3BO3 (0.62), Na2CO3 (5.3), NaHCO3 (2.1), Na2WO4 (0.00001), and trace elements solution described by Widdel et al. (38) (5 ml liter−1). The pH was adjusted to 9.3 with 6 N HCl, bubbled for 30 min with O2-free N2, and 9 ml of medium was dispensed into anoxic Balch tubes (∼25 ml), which were then crimp sealed and sterilized by autoclaving (121°C at 250 kPa for 60 min). All amendments of electron donors and acceptors were made by syringe injection from sterile, anoxic stock solutions. Tubes for aerobic incubation experiments were prepared without N2 and capped with a permeable foam stopper to allow exchange with the atmosphere. Each tube was inoculated with 1 ml of the red biofilm slurry collected from the hot spring (see above). Tubes were incubated at 45°C, with or without shaking, and illuminated with a standard 25-W tungsten light bulb (illumination intensity, ∼10 to 20 μE·m−2·s−1). Dark-incubated controls and samples from arsenate reduction experiments were completely wrapped in aluminum foil. For electron donor experiments the red biofilm material was centrifuged, resuspended, and washed three times with a mineral salts medium (see above) before final suspension in the same medium. A temperature range experiment for light- and dark-incubated tubes containing biofilm was conducted as outlined above in incubators set over a range of temperatures spanning from 4 to 70°C. The photosynthetic bacterium Ectothiorhodospira strain PHS-1 was also examined for its temperature range for growth under light with As(III) conditions by cultivation procedures outlined previously (15).
DNA extraction, PCR, and 16S rRNA gene clone library construction and analysis.
Microbial genomic DNA was extracted from two Paoha red mat samples (referred to as SDT and Mordor, collected in August 2008) using the FastDNA spin kit for soil (MP Biomedicals, Santa Ana, CA) according to the manufacturer's instructions. The extracted DNA concentration was determined by absorption at 260 nm by using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Eighty-five to 129 μg DNA (g of mat)−1 (wet weight) was obtained. All PCR mixtures contained 50 ng of template DNA, 1× PCR buffer (10 mM Tris-Cl, 50 mM KCl, 2 mM MgCl2, pH 9.0), 0.1 mM each deoxynucleoside triphosphate, 0.2 μM each primer, and 1 U of Taq DNA polymerase in a 50-μl final volume. Amplifications were performed in a PTC-200 PCR machine (MJ Research) as follows: initial denaturation for 3 min at 94°C, 35 cycles with denaturation (30 s at 94°C), annealing (30 s at 55°C), and extension (30 s at 72°C), and a final extension step at 72°C for 5 min. Bacterial and archaeal assemblage compositions were analyzed by 16S rRNA gene clone libraries. 16S rRNA genes were amplified with a bacteria-specific 27F and 1492R primer set (3a) and archaea-specific A21F and A958R primers (3). All PCRs were performed in triplicate, and PCR products were combined prior to construction of clone libraries. For each of the two libraries (Bacteria and Archaea), 96 randomly selected clones were grouped via random fragment length polymorphism (RFLP) analysis by using 4-base restriction enzymes (MspI and HhaI) as previously described (14). Representative clones were sequenced and classified by BLASTn analysis relative to the NCBI nonredundant nucleotide database as previously described (14). Singlet clones (i.e., RFLP groups represented by only a single clone) were generally not included for sequence analysis.
Detection, cloning, and sequence analysis of arsenic functional genes.
Arsenate respiratory reductase genes (arrA) were PCR amplified as described above with primers HAArrA-D1F and HAArrA-G2R (13) at an empirically determined annealing temperature of 53.5°C; a single 550-bp PCR product was obtained. Arsenite transporter genes (arsB) were PCR amplified as above with primers darsB1F and darsB1R (1) at an empirically determined annealing temperature of 59°C; a PCR product of the predicted 750-bp size was obtained. For the arsenite oxidase gene (aoxB), multiple bands were present, including the predicted band sizes of 1,100 bp (aoxBM1-2F and aoxBM3-2R primer pair [24]; 55°C annealing temperature) or 670 bp (aoxB forward and aoxB reverse primer pair [C. Saltikov, personal communication]; 58°C annealing temperature). A PCR product of the expected size was excised for clone library construction.
Clone libraries were constructed as described above for 16S rRNA genes. Clones were grouped by RFLP analysis as described for 16S rRNA genes, and representative clones for each group were sequenced. Only groups containing two or more clones were sequenced. Sequence similarities were determined by BLASTx analysis against the NCBI nonredundant protein database (11). Several putative arrA clones and all putative aoxB clones failed to show significant similarity to anything in the GenBank database: these sequences were excluded from further analyses.
Radioassays.
Radioassay incubations were prepared in artificial hot spring media as described above with the exception of those used to measure H14CO3− fixation, which were prepared with a lower content of NaHCO3 (0.5 mM) plus Na2CO3 (1.0 mM) to permit better cellular incorporation of 14C. Killed control slurries for each experiment were prepared by autoclaving (121°C, 250 kPa, 1 h) prior to radioisotope amendment. For 73As(V) reduction, slurries of biofilm material were prepared with 2 mM As(III) plus 10 μM As(V) (as unlabeled arsenic) and subsequently injected with 1.11 × 106 Βq HNa273AsO4 (100 μl; specific activity, 2.29 × 106 Bq/mmol; Brookhaven National Laboratory, Upton, NY). These slurries were incubated at 43°C for 8 h and were subsampled by using an N2-flushed syringe each hour (0.3 ml). The radiolabeled arsenic species in each subsample were separated on ion exchange columns and quantified by gamma spectrometry to measure the reduction of 73As(V) to 73As(III) as described by Oremland et al. (20). Experiments designed to assess the linkage of arsenic biotransformations to chemoheterotrophic or chemoautotrophic processes in the biofilm community were conducted with either [2-14C]acetate (1.85 × 106 Bq; specific activity, 2.04 × 107 Bq/mmol; American Radiolabeled Chemicals, Inc., St. Louis, MO) or NaH14CO3 (3.7 × 104 Bq; specific activity, 1.85 × 107 Bq/mmol; American Radiolabeled Chemicals, Inc., St. Louis, MO). To investigate heterotrophic oxidation of organic electron donors during As(V) reduction, we monitored 14CO2 production from [14C]acetate in both light- and dark-incubated slurries (43°C for 7 days) using radio-gas chromatographic methods (12). Assimilation of [2-14C]acetate (i.e., methyl group labeled) and chemoautotrophic 14CO2 fixation during As(V) reduction were measured in suspensions of biofilm cells that had been washed, centrifuged, and decanted three times with anoxic media. Cell suspensions (5-ml total volume) were prepared under either a N2 or H2 headspace, received either no As(V) amendment or 5 mM As(V), and were incubated at 43°C in the dark. All tubes that received [2-14C]acetate were also amended with unlabeled sodium acetate (1.5 mM), whereas some replicate tubes that were injected with NaH14CO3 received 3 mM sulfide as a potential electron donor. After 2 to 7 days the experiments were terminated and the cells from the radiolabeled samples were collected on nylon filters (pore size, 0.45 μm). The filters were acid fumed in a desiccator (8 h), and residual radioactivity was counted with a liquid scintillation spectrometer (20).
Analytical determinations.
The arsenic speciation and concentration and acetate concentration were determined by high-performance liquid chromatography (HPLC) (5). Sulfide was determined spectrophotometrically (2).
Nucleotide sequence accession numbers.
All sequences have been deposited in the NCBI database (accession numbers GU974346 to GU974368). 16S rRNA gene sequences were deposited in GenBank with the accession numbers GU974346 to GU974357; ArrA genes were deposited in GenBank with the accession numbers GU974358 to GU974368.
RESULTS
Microbial composition of biofilms.
Only two types of Bacteria were prevalent in the biofilms. Based on a BLASTn search of the GenBank database, the dominant clone group was most closely related to the 16S rRNA gene of Ectothiorhodospira sp. (Table 1). Kulp et al. (15) found that an isolate from the genus Ectothiorhodospira, strain PHS-1, was capable of growth via anoxygenic photosynthetic oxidation of arsenite from these same biofilms. The only other sequence, from a single clone, found in this library was related to the Firmicute Anaerobranca gottshalkii (Table 1). The Archaea were slightly more diverse in the biofilm: four different archaeal groups were observed. All archaeal sequences were affiliated with the Euryarchaeota phylum, and most sequences were related to the family Halobacteriaceae. All of the archaeal sequences are related to organisms or genes obtained from hypersaline environments.
TABLE 1.
Identification of representative RFLPs of bacterial and archaeal 16S rRNA gene clone libraries from the Paoha red mat
| Clone library | % of RFLP type | Nearest neighbor of representative clone | GenBank accession no. | Identity (%) |
|---|---|---|---|---|
| Bacteria | 93.0 | Ectothiorhodospira sp. AM4 16S rRNA gene | EU252492 | 985/999 (98) |
| 1.0 | Anaerobranca gottschalkii 16S rRNA gene | AF203703 | 958/1,008 (95) | |
| Archaea | 39.6 | Haloterrigena turkmenica DSM 5511 16S rRNA gene | AB004878 | 875/916 (95) |
| 35.2 | Uncultured archaeon clone SA10 16S rRNA gene | EU722677 | 886/910 (97) | |
| 6.6 | Uncultured haloarchaeon clone TX4CA_15 16S rRNA gene | EF690570 | 863/917 (94) | |
| 13.2 | Uncultured archaeon partial 16S rRNA gene, clone CG154 | FM210828 | 580/587 (98) |
Biofilm incubation experiments.
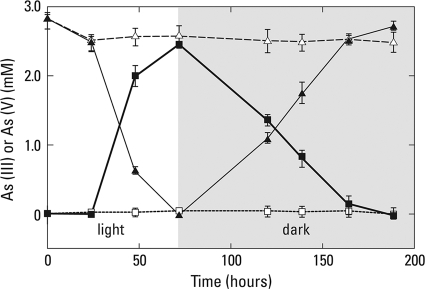
Light-incubated anoxic slurries completely oxidized ∼3.0 mM As(III) to As(V) over an ∼72-h period (Fig. 1). When the samples were shifted to a dark regimen the opposite reaction occurred, namely, the immediate reduction of the accumulated As(V) completely back to As(III). The rate of As(V) reduction observed over the subsequent 100 h was roughly comparable (about 25% slower) to the initial rate of anaerobic As(III) oxidation. When a light regimen was reimposed upon these samples after 200 h of incubation, an immediate but even lower rate of As(III) oxidation was observed and again was followed by an episode of As(V) reduction during dark incubation at a further-diminished rate (data not shown). Killed controls incubated under the same conditions demonstrated neither As(III) oxidation nor As(V) reduction.
FIG. 1.
Time course of anaerobic As(III) oxidation and As(V) reduction in Paoha Island biofilm slurry samples incubated under alternating regimens of light and dark. Dark symbols represent live samples, and open symbols are heat-killed controls. Symbols: ▴, As(III); ▪, As(V). Symbols represent the means of three slurries, and bars indicate ±1 standard deviation.
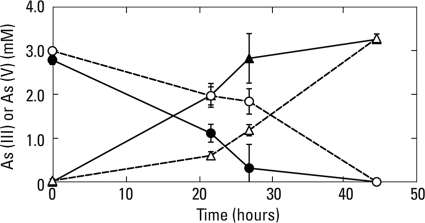
The above results suggested that both As(III) oxidation as well as As(V) reduction by biofilm materials were potentially capable of a cooccurrence when incubated in the light under imposed anoxia. To further reinforce this point, we conducted a radiolabel experiment employing 73As(V) added to samples that were incubated in the light with 2 mM As(III) and 10 μM As(V). After 2 h of incubation, 96% (±3% standard deviation; n = 3) of the added 73As(V) had been reduced to 73As(III), while no such activity was noted in killed controls, where all the counts remained as 73As(V). In another experiment, biofilm material incubated in the light or dark under oxic conditions demonstrated oxidation of As(III) to As(V) (Fig. 2) at rates equivalent to those observed under comparable anoxic conditions (Fig. 1). Constant shaking of the samples to promote O2 exchange did not alter these rates (data not shown).
FIG. 2.
Time course of aerobic As(III) oxidation for biofilm slurries incubated in the light (open symbols) or the dark (closed symbols). Symbols: ○, As(III); Δ, As(V). Symbols represent the means of three slurries, and bars indicate ±1 standard deviation.
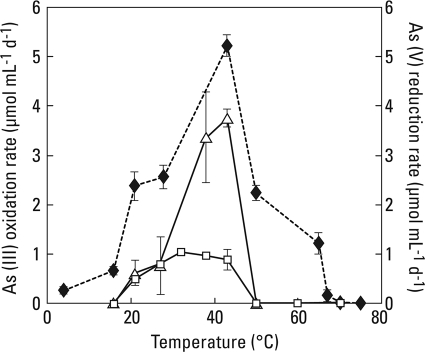
The temperature ranges and optima for anaerobic As(V) reduction in the dark and anaerobic As(III) oxidation in the light were considerably different (Fig. 3). Dark As(V) reduction occurred over a very broad temperature range (4 to 70°C) with an optimum evident at 43°C, while light As(III) oxidation occurred over a narrow range (20 to 43°C) but with a comparable optimum. By comparison, Ectothiorhodospira strain PHS-1, which was isolated from this same red biofilm (15), had a temperature range comparable to that of the light-incubated biofilm material.
FIG. 3.
Temperature range experiment for anaerobic biofilm materials and strain PHS-1. Symbols: ⧫, As(V) reduction rates for dark-incubated biofilm; Δ, As(III) oxidation rates for light-incubated slurries; □, As(III) oxidation rates for light-incubated strain PHS-1. All symbols represent the mean rates calculated for three incubated samples, with error bars indicating ±1 standard deviation.
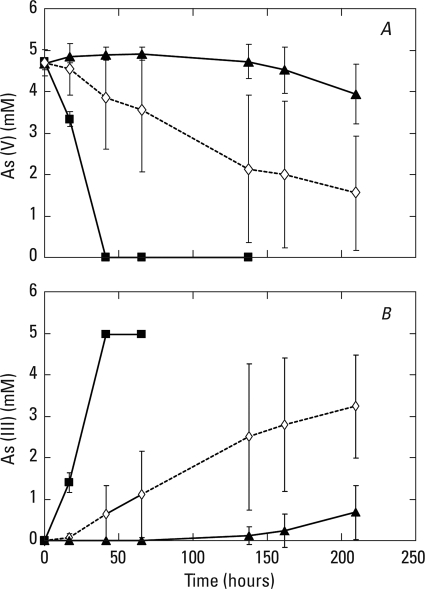
Washed biofilm anaerobic incubations with added electron donors.
Arsenate was completely reduced to As(III) in dark-incubated washed samples amended with hydrogen, while ∼50% of the added As(V) was reduced to As(III) over the same time period in samples amended with 2 mM sulfide (Fig. 4). The large errors associated with the sulfide-amended samples were likely due to the variable kinetics of formation and destruction of various thioarsenic intermediates which are soluble at this high pH (7, 20). No As(V) reduction occurred in these experiments without any added electron donor. In a separate experiment, neither unamended controls nor the addition of 1.5 mM acetate to samples demonstrated any detectable As(V) reduction (data not shown). Light-incubated live slurries oxidized [2-14C]acetate to 14CO2 after a 40 h of incubation period. The percentage of [2-14C]acetate oxidized for the light-incubated samples (n = 3; ±1 standard deviation) for the indicated conditions was as follows: no additions, 41.7 (±5.3); with 1 mM As(III), 39.7 (±2.7); with 1 mM As(V), 36.0 (±2.7). In contrast, there was no detectable [2-14C]acetate oxidation in dark-incubated live samples maintained under the same conditions (data not shown) or in any of the heat-killed controls incubated either in the light or dark with As(V) or As(III) amendments (data not shown).
FIG. 4.
Rates of As(V) reduction (A) to As(III) (B) in washed biofilm materials incubated under anaerobic conditions in the dark. Symbols: ▴, samples without added electron donor; ⋄, samples amended with 2 mM sulfide; ▪, samples incubated under an atmosphere of 100% H2. Symbols represent the means of three slurries, and bars indicate ±1 standard deviation.
Assimilation levels of both [14C]acetate and [14C]bicarbonate were stimulated during As(V) reduction in dark-incubated samples (Table 2). For [14C]bicarbonate, this was particularly evident when As(V) reduction was driven by the inclusion of either sulfide or H2 as chemoautotrophic electron donors. [14C]acetate assimilation was also strongly enhanced in the presence of both H2 and As(V) (the sulfide condition was not tested). In a separate experiment, assimilation of 12.7 ± 3.6% [14C]acetate also occurred in light-incubated slurries but was not stimulated by the presence of arsenic oxyanions (data not shown).
TABLE 2.
[14C]acetate and [14C]bicarbonate assimilation in biofilm cells after incubation for 48 h in the darka
| Addition(s) | % 14C assimilatedb |
|
|---|---|---|
| Acetate | Bicarbonate | |
| None | 0.54 (0.01) | 0.04 (0.00) |
| 5 mM As(V) | 0.74 (0.09) | 0.12 (0.03) |
| H2 | 0.56 (0.12) | 0.17 (0.06) |
| 5 mM As(V) + H2 | 2.56 (0.30) | 0.32 (0.07) |
| 5 mM + H2, killed | 0.35 (0.10) | 0.06 (0.02) |
| 3 mM sulfide | NDc | 0.07 (0.02) |
| 5 mM As(V) + 3 mM sulfide | ND | 0.41 (0.09) |
| As(V) + sulfide, killed | ND | 0.04 (0.02) |
Assimilation of [14C]acetate in the light was 12.7% ± 3.6%.
Values in parentheses represent standard deviations of triplicate samples.
ND, not determined.
Arsenic functional genes (arrA, aoxB, and arsB).
We assessed the diversity of known arsenic cycling and resistance genes for dissimilatory arsenate respiratory reductase (arrA), the arsenite oxidase large subunit (aoxB), and a gene encoding a membrane subunit of arsenic oxyanion translocation pump (arsB). In the case of aoxB, we obtained a PCR product of the correct size with two separate sets of PCR primers, but the sequence analysis indicated that the product was not an aoxB. It is not clear if this result was due to the inefficacy of the primers for this particular application or to the alternate explanation, that the aerobic As(III) oxidation activity observed was due to an aoxB-independent mechanism.
Eight groups of arrA clones were observed based on RFLP analysis, and five were confirmed as arrA-like genes by sequence analysis (Table 3). The dominant group, representing 48% of clones, was nearly identical (98% translated amino acid similarity) to an Ectothiorhodospira sp. PHS-1 DMSO reductase family oxidoreductase, a protein family that includes respiratory arsenate reductases. This finding is consistent with the results from the 16S rRNA gene clone library and supports our observation that Ectothiorhodospira sp. dominates this mat. The other four groups are closely related (>85% translated amino acid identity) to arrA genes previously obtained from Mono Lake samples and other soda lake systems.
TABLE 3.
Identification of representative arsenate respiratory reductase genes from the Paoha red mat based on BLASTx analysis and the GenBank nonredundant protein database
| Representative clone | Nearest neighbor | GenBank accession no. | No. of clones | Identity (%) | Reference |
|---|---|---|---|---|---|
| SDT-arrA-13 | Ectothiorhodospira sp. PHS-1 DMSO reductase family oxidoreductase | ACF76560 | 23 | 86/87 (98) | 15 |
| SDT-arrA-16 | Arsenate respiratory reductase (uncultured bacterium) | ABJ53058 | 4 | 126/129 (97) | 13 |
| SDT-arrA-24 | Molybdopterin dinucleotide binding region (bacterium S5) | EFC66482 | 3 | 116/135 (85) | 32 |
| SDT-arrA-4 | Arsenate respiratory reductase | ABU48654 | 4 | 121/123 (98) | 14 |
| SDT-arrA-8 | Arsenate respiratory reductase (uncultured bacterium) | ABJ53060 | 2 | 113/129 (87) | 13 |
Only a single clone type was detected from the mat sample for the arsenic oxyanion translocation pump gene (arsB). The sequence was matched to an arsenical pump membrane protein of Halorhodospira halophila SL1 and Thioalkalivibrio sp. HL-EbGR7 with 80% (607 of 750 bp) and 78% nucleic acid similarity (592 of 758 bp) based on BLASTn analysis. Both of these genera are members of the Ectothiorhodospiracaea and are closely related to the genus Ectothiorhodospira. This observation provides further corroboration that members of this group are the dominant arsenic cycling organisms in the mats.
DISCUSSION
In the Paoha Island biofilms, immediate anoxygenic photosynthetic As(III) oxidation occurs in the light followed by an immediate shift to As(V) reduction in the dark (Fig. 1). The fact that the cyclic nature of the phenomenon persisted with time even as the rates of both processes diminished indicates the overall robustness of the primary pattern. The dampening of the rates with the passage of incubation time is explained by the senescence of the populations and toxin accumulation in what is essentially a batch culture system. Back-labeling experiments with 73As(V) also illustrated that both anaerobic arsenotrophic processes [photosynthetic As(III) oxidation and As(V) reduction] could occur simultaneously in the light. Hence, the observed oxidation of As(III) to As(V) carried out by anoxygenic photosynthesis represented a net rate and illustrated the close ecophysiological linkage of the two processes. Curiously, a capacity for aerobic As(III) oxidation was also observed in the samples (Fig. 2). Although the chemistry of the spring waters is strongly reducing, with millimolar ambient concentrations of strong reductants like sulfide and ammonia (15), the springs and ponds are quite shallow. Hence, portions of the biofilms (or the planktonic microbes) can become directly exposed to the atmosphere, so our detection of oxygen-dependent As(III) oxidation within this community is not surprising.
The temperature range experiments indicated that different microbial populations were responsible for observed light-driven As(III) oxidation versus the anaerobic (dark) As(V) reduction (Fig. 3). The light-incubated biofilms had a narrow range of activity that abruptly ended at 50°C, and these temperature responses were very similar to that displayed by the pure culture of Ectothiorhodospira strain PHS-1 which was originally isolated from these materials (15). This finding is consistent with our observation that the bacterial 16S rRNA genes in this mat are dominated by Ectothiorhodospira closely related to strain PHS-1. In contrast, dark As(V) reduction occurred over a much broader temperature range, which implied the involvement of physiologically different anaerobes. Since arsenate reduction is not a monophyletic trait, it is unclear based on 16S rRNA gene analysis which organisms are carrying out this process in the Paoha biofilms. That all three experimental incubations displayed optimal activities proximate to the in situ temperatures of the hot springs themselves (∼45°C) illustrates their adaptation to the prevailing environmental conditions.
We conducted a variety of incubation experiments designed to explore what electron donors could be available to drive the observed As(V) reduction. Clearly, addition of either sulfide or hydrogen to washed biofilm cells resulted in immediate and pronounced rates of As(V) reduction over unamended samples (Fig. 4). The facts that these biofilms are constantly bathed in spring water that contains ∼5 mM sulfide (15) and that the springs themselves are permeated by active ebullition of natural gas seeps that contain traces of H2 (19) indicate they are best adapted to harness these readily available inorganic materials to drive the observed As(V) reduction.
Acetate is a key terminal substrate in important anaerobic processes like methanogenesis and sulfate reduction. Nevertheless, we did not observe any stimulation of As(V) reduction with acetate amendment (see text), implying that chemoheterotrophy did not drive As(V) reduction. The role of acetate was better explained by the radiolabel experimental results (see Results).
No oxidation of [14C]acetate occurred in the dark, nor was any 14CH4 detected, observations that further support its lack of linkage to chemoheterotrophy and As(V) reduction. However, we did observe considerable oxidation activity in the light. There was no significant noticeable effect of As(V) or As(III) addition to samples. These results suggest that acetate itself was employed as an electron donor to sustain anoxygenic photosynthesis and that photoheterotrophy was responsible for its oxidation to 14CO2. Indeed, the fact that we also observed the largest amount of [14C]acetate incorporation into light-incubated materials, which had 5-fold-higher activity than comparable dark-incubated ones (Table 2), further substantiates this interpretation. Members of the genus Ectothiorhodospira, like E. shaposhnikovii, can grow in the light using acetate in lieu of reduced sulfur compounds to sustain anoxygenic photosynthesis (23), and we have observed such anaerobic growth with Ectothiorhodospira strain PHS-1 (S. E. Hoeft, unpublished data).
We previously demonstrated that these biofilms incorporate [14C]bicarbonate during light-exposed incubation and that this process was stimulated by the presence of As(III) or sulfide as evidence for their robust anoxygenic photosynthesis (15). We now observed incorporation of [14C]bicarbonate into dark-incubated samples and that this activity was stimulated by the presence of As(V), with the highest counts in samples that also received the inorganic electron donors sulfide or H2 (Table 2). This further underscores the fact that dissimilatory As(V) reduction was carried out by chemoautotrophic bacteria. Additions of H2 or sulfide to anoxic sediment slurries from salt-saturated, alkaline Searles Lake markedly stimulated dissimilatory As(V) reduction (22). Hydrogen or sulfide can serve as electron donors for the chemoautotrophic growth of diverse arsenate-respiring prokaryotes, including strain Y5 (16), enrichment cultures from Mono Lake (7), the Deltaproteobacterium strain MLMS-1 (5), “Halarsenatibacter silvermanii” (36), and Pyrobaculum arsenaticum of the Crenarchaea (8). The dark incorporation of [14C]acetate was stimulated by the presence of H2 plus As(V) and was similar to observations made with many hydrogenotrophic anaerobes that can readily use acetate as a source of reduced carbon when provided with molecular hydrogen. Thus, this appears to extend to the arsenate-respiring bacteria.
16S rRNA gene clone libraries indicate that the dominant Bacteria in this sample are closely related to Ectothiorhodospira strain PHS-1, an anoxygenic photosynthetic arsenite-oxidizing organism. That this dominant gene sequence belongs to an organism playing a similar role in this environment is supported by our analysis of arrA and arsB gene clone libraries. The largest single group of arrA clones (48%) was closely related to an arrA gene from Ectothiorhodospira strain PHS-1 (Table 3). We recently proposed that the respiratory arsenate reductase of strain PHS-1, and presumably the very similar sequence found in the biofilms, functions in vivo as a de facto arsenite oxidase (15), in part based on its similarity to the gene from Alkalilimnicola ehrlichii (6), which was recently demonstrated to work in this manner by Richey et al. (27). The other arrA amplicons were arrA genes more closely related to other arsenate-reducing bacteria and not closely related to Ectothiorhodospira. Therefore, these clones were likely associated with microorganisms that carried out the respiratory As(V) reduction detected in our dark incubations (Fig. 1, 3, and 4). It is not at all clear from our data set, however, which biofilm microbes were associated with these arrA amplicons. All arsenite efflux gene (arsB) clone sequences were identical and were closely related to those from organisms in the family Ectothiorhodospiracaea. Arsenate-respiring bacteria, like Shewanella strain ANA-3, have been shown to have both the As(V) respiratory and the full arsC complement of As(V) resistance genes (30); thus, this gene may be associated either with arsenic-resistant microbes not involved in redox cycling of arsenic or (more likely) the Ectothiorhodospira spp. that dominate this mat. The Archaea found in the 16S rRNA gene clone libraries are all related to extremely halophilic Archaea. There is no indication that any of these Archaea is involved in arsenic cycling in this system, and they are likely present due to the high salt content of the Paoha hot spring environment.
An unexpected result was our inability to detect arsenite oxidase genes related to the aoxB amplicon despite our clear detection of this metabolic activity in oxic biofilm samples (Fig. 2). This could have been merely due to the fact that primers were not quite suited for this particular environment. However, an alternative explanation is that the observed aerobic As(III) oxidation was carried out by a novel mechanism. If this were the case, the isolation of new types of aerobic As(III)-oxidizing microbes may be possible from this system, and the attendant follow-up biochemical investigations may provide insight into their means of As(III) oxidation.
We previously hypothesized that As(III) oxidation achieved by anoxygenic photosynthesis extended back to the Archean eon (∼3.8 billion years ago), and the consequent accumulation of As(V) would have opened immediate niches for anaerobes capable of As(V) respiration (15). By extension, dissimilatory arsenate reduction could also be viewed as an “ancient” phenomenon in the realm of Earth's geobiological evolution, which over time gave rise to the broad phylogenetic diversity of this trait among the prokaryotes (21, 35). Contemporary biological communities exposed to arsenic should therefore exhibit close cyclical linkages between As(III) oxidation and As(V) reduction. These cyclical reactions are summarized in Fig. 5. While this cyclic phenomenon has been demonstrated for shifting oxic/anoxic experimental biomes (12, 26), it has not as yet been shown for permanently anaerobic ecosystems. The early Earth was anoxic, and hence emerging anaerobic microbial ecosystems exposed to sunlight may have been specifically structured to the diel rhythms of light and dark. Bacterial anoxygenic photosynthesis is thought to be an ancient process, perhaps extending back in time to the Archean eon (37). The Paoha Island biofilms are dominated by members of the Ectothiorhodospira (Table 1). They offer us a glimpse into the Earth's distant past of the potential influence of anoxygenic photosynthesis upon primordial arsenic cycling.
FIG. 5.
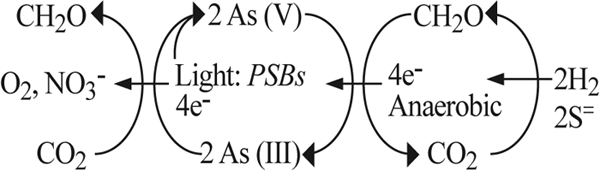
Schematic representation of microbial arsenic cycling that occurs in contemporary ecosystems like the Paoha Island red biofilms. Under reducing conditions, chemoautotrophic dissimilatory As(V) reduction is fueled by inorganic electron donors like hydrogen or sulfide, which results in the fixation of inorganic carbon into cell material. Chemoheterotrophic metabolism can also fuel As(V) reduction (although not evident in these biofilms), resulting in the oxidation of the organic matter to CO2. On the oxidative side, As(III) generated by the above reactions, or from waters originally emanating from the hot springs themselves, can be biologically oxidized to As(V) in the presence of strong oxidants like O2 (or NO3−). Alternatively, in the presence of light photosynthetic bacteria (PSBs) use As(III) as an electron donor to drive anoxygenic photosynthesis, resulting in the formation of As(V) and the fixation of inorganic carbon into cell material.
Acknowledgments
This work was funded by a grant from NASA Exobiology Program and by the USGS.
We are grateful to L.G. Miller, S. Baesman, C. Culbertson, and J. Switzer Blum for assistance with field sample collection and processing. We thank F. Wolfe-Simon and J. Coates for their constructive comments on an earlier version of the manuscript, and we are grateful to four anonymous reviewers for their help in improving this paper.
Footnotes
Published ahead of print on 28 May 2010.
REFERENCES
- 1.Achour, A. R., P. Bauda, and P. Billard. 2007. Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res. Microbiol. 158:128-137. [DOI] [PubMed] [Google Scholar]
- 2.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-459. [Google Scholar]
- 3.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-201. In E. Stackebrandt and M. Goodfellow (ed.), Sequencing and hybridization techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 4.Gorlenko, V., A. Tsapin, Z. Namsarev, T. Teal, T. Tourova, D. Engler, R. Mielke, and K. Nealson. 2004. Anaerobranca californiensis sp. nov., an anaerobic, alkalithermophilic, fermentative bacterium isolated from a hot spring on Mono Lake. Int. J. Syst. Evol. Micobiol. 54:739-743. [DOI] [PubMed] [Google Scholar]
- 5.Hoeft, S. E., T. R. Kulp, J. F. Stolz, J. T. Hollibaugh, and R. S. Oremland. 2004. Dissimilatory arsenate reduction with sulfide as the electron donor: experiments with Mono Lake water and isolation of strain MLMS-1, a chemoautotrophic arsenate-respirer. Appl. Environ. Microbiol. 70:2741-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoeft, S. E., J. Switzer Blum, J. F. Stolz, F. R. Tabita, B. Witte, G. M. King, J. M. Santini, and R. S. Oremland. 2007. Alkalilimnicola ehrlichii, sp. nov., a novel, arsenite oxidizing haloalkaliphilic γ-proteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int. J. Syst. Evol. Microbiol. 57:504-512. [DOI] [PubMed] [Google Scholar]
- 7.Hollibaugh, J. T., C. Budinoff, R. A. Hollibaugh, B. Ransom, and N. Bano. 2006. Sulfide oxidation coupled to arsenate reduction by a diverse microbial community in a soda lake. Appl. Environ. Microbiol. 72:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huber, R., M. M. Sacher, A. Vollmann, H. Huber, and D. Rose. 2000. Respiration of arsenate and selenate by hyperthermophilic archaea. Syst. Appl. Microbiol. 23:305-314. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins, M. C., and M. Kemp. 1984. The coupling of nitrification and denitrification in two estuarine sediments. Limnol. Oceanogr. 29:609-619. [Google Scholar]
- 10.Jensen, K., N. P. Sloth, N. Risgaard-Petersen, S. Rysgaard, and N. P. Revsbech. 1994. Estimation of nitrification and denitrification from microprofiles of oxygen and nitrate in model sediments systems. Appl. Environ. Microbiol. 60:2094-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rRNDB: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulp, T. R., S. E. Hoeft, and R. S. Oremland. 2004. Redox transformations of arsenic oxyanions in periphyton communities. Appl. Environ. Microbiol. 70:6428-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulp, T. R., S. E. Hoeft, L. G. Miller, C. Saltikov, J. Nilsen, S. Han, B. Lanoil, and R. S. Oremland. 2006. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl. Environ. Microbiol. 72:6514-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulp, T. R., S. Han, C. W. Saltikov, B. D. Lanoil, K. Zargar, and R. S. Oremland. 2007. Effects of imposed salinity gradients on dissimilatory arsenate reduction and other microbial processes in sediments from two California soda lakes. Appl. Environ. Microbiol. 73:5130-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulp, T. R., S. E. Hoeft, M. Asao, M. T. Madigan, J. T. Hollibaugh, J. C. Fisher, J. F. Stolz, C. W. Culbertson, and R. S. Oremland. 2008. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science 321:967-970. [DOI] [PubMed] [Google Scholar]
- 16.Liu, A., E. Garcia-Dominguez, E. D. Rhine, and L. Y. Young. 2004. A novel arsenate respiring isolate that can utilize aromatic substrates. FEMS Microbiol. Ecol. 48:323-332. [DOI] [PubMed] [Google Scholar]
- 17.Mukhopadhyay, R., B. P. Rosen, L. T. Phung, and S. Silver. 2002. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol. Rev. 26:311-325. [DOI] [PubMed] [Google Scholar]
- 18.Newman, D. K., D. Ahmann, and F. M. M. Morel. 1997. A brief review of microbial arsenate respiration. Geomicrobiol. J. 15:255-268. [Google Scholar]
- 19.Oremland, R. S., L. G. Miller, and M. Whiticar. 1987. Sources and flux of natural gases from Mono Lake, California. Geochim. Cosmochim. Acta 51:2915-2929. [Google Scholar]
- 20.Oremland, R. S., P. R. Dowdle, S. Hoeft, J. O. Sharp, J. K. Schaefer, L. G. Miller, J. S. Blum, R. L. Smith, N. S. Bloom, and D. Wallschlaeger. 2000. Bacterial dissimilatory reduction of arsenate and sulfate in meromictic Mono Lake, California. Geochim. Cosmochim. Acta 64:3073-3084. [Google Scholar]
- 21.Oremland, R. S., and J. F. Stolz. 2003. The ecology of arsenic. Science 299:939-944. [DOI] [PubMed] [Google Scholar]
- 22.Oremland, R. S., T. R. Kulp, J. Switzer Blum, S. E. Hoeft, S. Baesman, L. G. Miller, and J. F. Stolz. 2005. A microbial arsenic cycle in a salt-saturated, extreme environment. Science 308:1305-1308. [DOI] [PubMed] [Google Scholar]
- 23.Pfennig, N., and H. G. Trüper. 1976. The phototrophic bacteria, p. 24-75. In R. E. Buchanan and N. E. Gibbons (ed.), Bergey's manual of determinative bacteriology, 8th ed. Williams & Wilkins Co., Baltimore, MD.
- 24.Quéméneur, A. Heinrich-Salmeron, D. Muller, D. Lièvremont, M. Jauzein, P. N. Bertin, F. Garrido, and C. Joulian. 2008. Diversity surveys and evolutionary relationships of aoxB genes in aerobic arsenite-oxidizing bacteria. Appl. Environ. Microbiol. 74:4567-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reference deleted.
- 26.Rhine, E. D., E. Garcia-Dominguez, C. D. Phelps, and L. Y. Young. 2005. Environmental microbes can speciate and cycle arsenic. Environ. Sci. Technol. 39:9569-9573. [DOI] [PubMed] [Google Scholar]
- 27.Richey, C., P. Chovanec, S. Hoeft, R. S. Oremland, P. Basu, and J. F. Stolz. 2009. Respiratory arsenate reductase as a bidirectional enzyme. Biochem. Biophys. Res. Commun. 382:298-302. [DOI] [PubMed] [Google Scholar]
- 28.Risgaard-Petersen, N. 2003. Coupled nitrification-denitrification in autotrophic and heterotrophic sediments: on the influence of benthic microalgae. Limnol. Oceanogr. 48:93-105. [Google Scholar]
- 29.Rosen, B. 2002. Biochemistry of arsenic detoxification. FEBS Lett. 529:86-92. [DOI] [PubMed] [Google Scholar]
- 30.Saltikov, C. W., R. Wildman, Jr., and D. K. Newman. 2005. Expression dynamics of arsenic respiration and detoxification in Shewanella sp. strain ANA-3. J. Bacteriol. 187:7390-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silver, S., and L. T. Phung. 2005. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, B., E. Chyun, P. Jaffe, and B. B. Ward. 2009. Molecular methods to detect and monitor dissimilatory arsenate-respiring bacteria (DARB) in sediments. FEMS Microb. Ecol. 68:108-117. [DOI] [PubMed] [Google Scholar]
- 33.Stine, S. 1984. Late Holocene lake level fluctuations and island volcanism at Mono Lake, California, p. 21-49. In Geologic guide to Aspen Valley, Mono Lake, Mono Craters, and Inyo Craters. Genny Smith Books, Palo Alto, CA.
- 34.Stolz, J. F., and R. S. Oremland. 1999. Bacterial respiration of selenium and arsenic. FEMS Microbiol. Rev. 23:615-627. [DOI] [PubMed] [Google Scholar]
- 35.Stolz, J. F., P. Basu, and R. S. Oremland. 2010. Microbial arsenic metabolism: new twists on an old poison. Microbe 5:53-59. [Google Scholar]
- 36.Switzer Blum, J., S. Han, Brian Lanoil, C. Saltikov, B. Witte, F. R. Tabita, S. Langley, T. J. Beveridge, John F. Stolz, L. Jahnke, and R. S. Oremland. 2009. Halarsenatibacter silvermanii strain SLAS-1T, gen. nov., sp. nov., ecophysiology of an extremely halophilic, facultative chemoautotrophic arsenate-respirer of the Halanaerobiales isolated from Searles Lake, California. Appl. Environ. Microbiol. 75:1950-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tice, M. M., and D. R. Lowe. 2004. Photosynthetic microbial mats in the 3,416-Myr-old ocean. Nature 431:549-552. [DOI] [PubMed] [Google Scholar]
- 38.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
- 39.Yamamura, S., M. Watanabe, N. Yamamoto, K. Sei, and M. Ile. 2009. Potential for microbially mediated redox transformations and mobilization of arsenic in uncontaminated soils. Chemosphere 77:169-174. [DOI] [PubMed] [Google Scholar]