Abstract
A family 5 glycoside hydrolase from Clostridium phytofermentans was cloned and engineered through a cellulase cell surface display system in Escherichia coli. The presence of cell surface anchoring, a cellulose binding module, or a His tag greatly influenced the activities of wild-type and mutant enzymes on soluble and solid cellulosic substrates, suggesting the high complexity of cellulase engineering. The best mutant had 92%, 36%, and 46% longer half-lives at 60°C on carboxymethyl cellulose, regenerated amorphous cellulose, and Avicel, respectively.
The production of biofuels from nonfood cellulosic biomass would benefit the economy, the environment, and national energy security (17, 32). The largest technological and economical obstacle is the release of soluble fermentable sugars at prices competitive with those from sugarcane or corn kernels (17, 31). One of the approaches is discovering new cellulases from cellulolytic microorganisms, followed by cellulase engineering for enhanced performance on pretreated solid substrates. However, cellulase engineering remains challenging because enzymatic cellulose hydrolysis is complicated, involving heterogeneous substrates (33, 37), different action mode cellulase components (18), synergy and/or competition among cellulase components (36, 37), and declining substrate reactivity over the course of conversion (11, 26). Directed enzyme evolution, independent of knowledge of the protein structure and the enzyme-substrate interactions (6, 34), has been conducted to generate endoglucanase mutants, such as enhanced activities on soluble substrates (14, 16, 22), prolonged thermostability (20), changed optimum pH (24, 28), or improved expression levels (21). Here, we cloned and characterized a family 5 glycoside hydrolase (Cel5A) from a cellulolytic bacterium, Clostridium phytofermentans ISDg (ATCC 700394) (29, 30), and engineered it for enhanced thermostability.
Characterization of Cel5A.
The DNA fragment encoding the mature form of Cel5A (ABX41541) was cloned into vector pET-20b(+) to give plasmid pET20b-Cel5A (Fig. 1). Recombinant Cel5A by Escherichia coli BL21 Star (DE3) was highly soluble and purified by Bio-Rad Profinity IMAC Ni resins precharged with Ni2+ (Hercules, CA). The purified enzyme had an apparent molecular mass of about 45 kDa, and its optimal pH and temperature were 7.0 and 55°C on carboxymethyl cellulose (CMC), respectively. Recombinant Cel5A drastically decreased the viscosity of a 1% (wt/vol) CMC solution, suggesting that it is an endoglucanase (35). At pH 7.0, Cel5A had activities of 18.1 ± 0.8, 1.70 ± 0.04, and 0.24 ± 0.06 U/mg on CMC, regenerated amorphous cellulose (RAC) (33), and Avicel, respectively, but had no detectable activity on bacterial microcrystalline cellulose and xylan at 50°C.
FIG. 1.
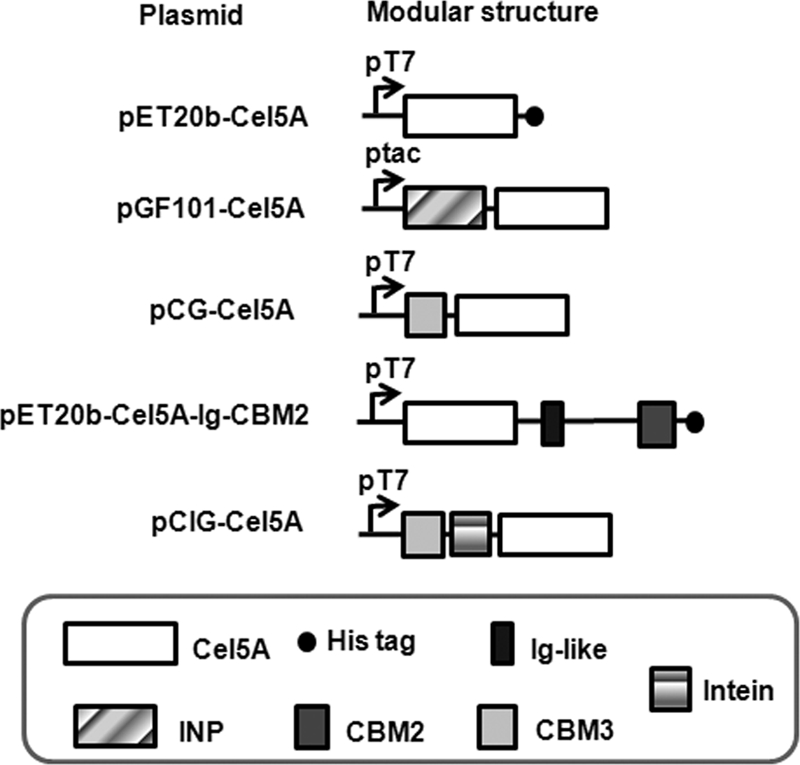
Schematic representation of the expression plasmids.
Improvement of the thermostability of Cel5A by directed evolution.
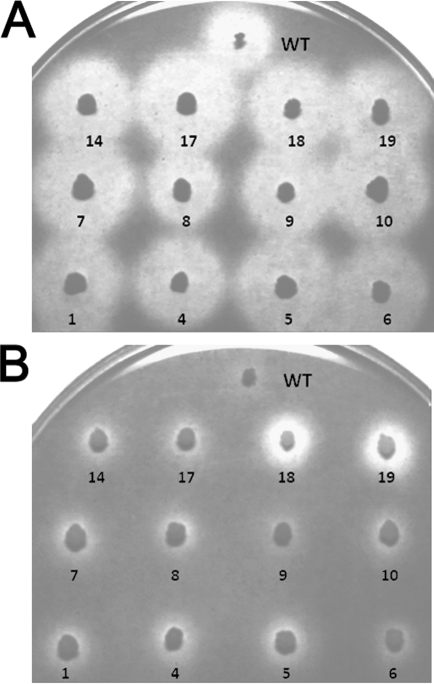
For potential cellulase recycling (38), Cel5A was engineered for better thermostability. A random cel5A mutant library generated by error-prone PCR (4) was expressed by plasmid pGF101-Cel5A (Fig. 1) so that Cel5A was displayed on the cell surface of E. coli by fusing with an outer membrane ice nucleation protein (INP) (12). After overnight incubation at 37°C, the colonies on the agar plates without isopropyl-β-d-thiogalactopyranoside (IPTG) were duplicated onto fresh LB plates containing 50 μM IPTG. The duplicate plates were incubated at room temperature for about 40 h and then heated at 70°C for 20 min. The plates were subsequently overlaid with soft agar containing 0.5% CMC and incubated at 37°C overnight. After Congo red staining and washing, the size of the yellow halo zone around a colony reflected endoglucanase activity on CMC (35). Approximately 20 colonies with halo zones larger than those of the wild type after the heat treatment were identified from ca. 20,000 colonies. The selected colonies were retrieved from the original plates, and their thermostabilities were further confirmed (Fig. 2). After DNA sequencing, five mutants with two or three amino acid substitutions (see Table S1 in the supplemental material) were selected for further characterization.
FIG. 2.
Congo red staining of E. coli JM109 colonies surface displaying engineered C. phytofermentans endoglucanase mutants on LB-Amp-IPTG agar plates overlaid with a CMC soft agar layer. Colonies selected from a library of putatively positive mutants without heat treatment (A) and with heat treatment (70°C, 20 min) (B).
CBM addition to Cel5A mutants.
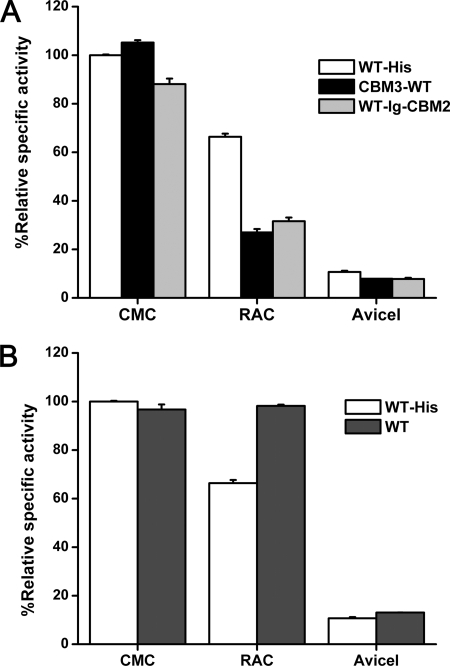
The five selected mutants that had potentially improved thermostability were evaluated as fusion proteins, each with a family 3 cellulose binding module (CBM3) from the Clostridium thermocellum scaffoldin (10, 19) added to the N terminus (Fig. 1) because (i) CBM3 is well-known to enhance hydrolysis of endoglucanase on insoluble cellulosic substrates (2, 7) and (ii) CBM3 helps heterologous protein expression in E. coli (10, 19). We first added a CBM3 to the N terminus (Fig. 1) of each selected mutant and found that only two (m1 and m18) were more thermostable than wild-type Cel5A. Substrate prebinding experiments suggested that cellulase thermostability was greatly affected by whether the enzyme was immobilized or free with the substrates (see Table S2 in the supplemental material). We also evaluated wild-type Cel5A with a His tag at the C terminus (WT-His) or a CBM3 at the N terminus (CBM3-WT) (Fig. 3 A). The specific activities of WT-His Cel5A on RAC and Avicel were 66% and 11% of that on CMC, respectively, due to a decreasing cellulose accessibility to cellulase in the order of CMC, RAC, and Avicel (11, 37). Adding an N-terminal CBM3 to Cel5A did not influence its activity on soluble substrate CMC but decreased its activity on insoluble substrates RAC and Avicel (Fig. 3A). This surprising result does not agree with previous reports that addition of CBM3 enhanced endoglucanase activities on solid cellulosic substrates (2) and that removal of CBM3 greatly decreased the enzyme activity on solid substrates (7). We speculated that this negative CBM addition effect could be attributed to a short linker length (8 amino acids) between CBM3 and Cel5A. Therefore, we constructed plasmid pET20b-Cel5A-Ig-CBM2 (Fig. 1) for expressing a Cel5A fusion protein (WT-Ig-CBM2) that has a C-terminal sequence (containing a 41-amino-acid linker, an Ig-like module, and a CBM2) from another C. phytofermentans family 5 glycoside hydrolase, Cel5B (GenBank accession number YP_001560295). WT-Ig-CBM2 did not show any significant improvement of activities on insoluble substrates compared to CBM3-WT (Fig. 3A), implying that the addition of a CBM might not always enhance endoglucanase activity on solid substrates. We further investigated the effects of the His tag on Cel5A activities on various substrates (Fig. 3B). Wild-type Cel5A without the His tag was expressed and purified from plasmid pCIG-Cel5A (Fig. 1). Removing the His tag increased Cel5A-specific activities on RAC and Avicel by 48% and 22%, respectively, but had no significant effect on its activity on CMC (Fig. 3B), suggesting that His tag effects on Cel5A activities were substrate dependent.
FIG. 3.
Effects of the CBM tags (A) and His tag (B) on the molar specific activities of wild-type Cel5A on CMC, RAC, and Avicel.
Because of the negative effects of CBMs and His tags on Cel5A activities, we expressed the tag-free wild-type Cel5A based on plasmid pCIG-Cel5A (Fig. 1) to produce mutants m1 (N144I/N291K) and m18 (E158V/V245G) and their combination mutant m1-m18 (N144I/N291K/E158V) by site-directed mutagenesis. Table 1 shows that m1, m18, and m1-m18 had activities similar to those of wild-type Cel5A on both soluble and insoluble substrates. On CMC, m1-m18 presented a nearly doubled half-life (14.8 min) compared to the wild type (7.7 min); on RAC and Avicel, m1-m18 exhibited 36% and 46% increased half-lives on RAC and Avicel, respectively.
TABLE 1.
Reducing-end release and half-life times of wild-type Cel5A and mutants m1, m18, and m1-m18 at 60°Ca
| Wild type or mutant (amino acid substitutions) | CMC |
RAC |
Avicel |
|||
|---|---|---|---|---|---|---|
| RE releaseb (μmol/mg protein) | t1/2 (min) | RE releaseb (μmol/mg protein) | t1/2 (min) | RE releaseb (μmol/mg protein) | t1/2 (min) | |
| WT | 95 ± 2 | 7.7 ± 1.1 | 95.9 ± 0.5 | 3.6 ± 0.1 | 12.8 ± 0.1 | 4.8 ± 0.3 |
| m1 (N144I/N291K) | 97 ± 4 | 9.1 ± 0.7 | 84.7 ± 0.1 | 4.6 ± 0.2 | 13.2 ± 0.3 | 6.6 ± 0.4 |
| m18 (E158V/V425G) | 101 ± 2 | 7.9 ± 0.7 | 95.1 ± 4.6 | 3.2 ± 0.2 | 13.8 ± 0.4 | 4.5 ± 0.3 |
| m1-m18 (N144I/N291K/E158V) | 95 ± 2 | 14.8 ± 1.7 | 84.7 ± 0.7 | 4.9 ± 0.3 | 12.7 ± 1.6 | 7.0 ± 0.7 |
RE, reducing end; t1/2, half-life. Results are means ± standard deviations.
In a 30-min reaction.
Complexity of cellulase engineering.
Removal of the CBMs from natural cellulases drastically decreased their activities on solid substrates (1, 3, 5, 8, 13, 15, 23), and addition of the C. thermocellum CBM to C. thermocellum endoglucanase CelD significantly improved its activity on solid cellulosic materials (2). However, our research shows that addition of the C. thermocellum CBM3 or the Ig-CBM2 fragment of C. phytofermentans Cel5B to C. phytofermentans Cel5A did not enhance its activities on solid substrates (Fig. 3A). One possible reason is that Cel5A might not interact efficiently with the CBM for generating a proper conformation for enhanced activity on insoluble substrates (25, 27). Some of the putative thermostable mutants that were displayed on the cell surface were found to be nonthermostable when purified in a free form (Fig. 2; see also Table S2 in the supplemental material), suggesting that the cell surface display technology may be more suitable for screening enzyme mutants for whole-cell catalysis.
Previous directed evolution studies successfully enhanced endoglucanase catalytic efficiency (14, 16, 22) based on assays on soluble cellulose derivatives. Because there is no clear relationship between the endoglucanase activities on soluble substrates and those on solid cellulose substrates (9, 34), the development of high-throughput cellulase assays on solid cellulosic substrates is urgently needed.
Supplementary Material
Acknowledgments
This work was supported mainly by the DOE BioEnergy Science Center (BESC) and partially by the USDA Bioprocessing and Biodesign Center and DuPont Young Professor Award to Y.-H. P. Zhang. Z. Zhang was partially supported by the China Scholar Council.
Footnotes
Published ahead of print on 28 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Boraston, A. B., E. Kwan, P. Chiu, R. A. J. Warren, and D. G. Kilburn. 2003. Recognition and hydrolysis of noncrystalline cellulose. J. Biol. Chem. 278:6120-6127. [DOI] [PubMed] [Google Scholar]
- 2.Carrard, G., A. Koivula, H. Soderlund, and P. Beguin. 2000. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. U. S. A. 97:10342-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrard, G., and M. Linder. 1999. Widely different off rates of two closely related cellulose-binding domains from Trichoderma reesei. Eur. J. Biochem. 262:637-643. [DOI] [PubMed] [Google Scholar]
- 4.Cirino, P. C., K. M. Mayer, and D. Umeno. 2003. Generating mutant libraries using error-prone PCR. Methods Mol. Biol. 231:3-9. [DOI] [PubMed] [Google Scholar]
- 5.Coutinho, J. B., N. R. Gilkes, D. G. Kilburn, R. A. J. Warren, and J. R. C. Miller. 1993. The nature of the cellulose-binding domain affects the activities of a bacterial endoglucanase on different forms of cellulose. FEMS Microbiol. Lett. 113:211-218. [Google Scholar]
- 6.Farinas, E. T., T. Bulter, and F. H. Arnold. 2001. Directed enzyme evolution. Curr. Opin. Biotechnol. 12:545-551. [DOI] [PubMed] [Google Scholar]
- 7.Gilad, R., L. Rabinovich, S. Yaron, E. A. Bayer, R. Lamed, H. J. Gilbert, and Y. Shoham. 2003. CelI, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall, J., G. W. Black, T. Ferenci, S. J. Millward-Sadler, B. R. S. Ali, G. P. Hazlewood, and H. J. Gilber. 1995. The non-catalytic cellulose-binding domain of a novel cellulase from Pseudomonas fluorescens subsp. cellulosa is important for the efficient hydrolysis of Avicel. Biochem. J. 309:749-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Himmel, M. E., W. S. Adney, J. O. Baker, R. A. Nieves, and S. R. Thomas. 1996. Cellulases: structure, function and applications, p. 143-161. In C. E. Wyman (ed.), Handbook on bioethanol. Taylor & Francis, Washington, DC.
- 10.Hong, J., X. Ye, Y. Wang, and Y.-H. P. Zhang. 2008. Bioseparation of recombinant cellulose binding module-protein by affinity adsorption on an ultra-high-capacity cellulosic adsorbent. Anal. Chem. Acta 621:193-199. [DOI] [PubMed] [Google Scholar]
- 11.Hong, J., X. Ye, and Y.-H. P. Zhang. 2007. Quantitative determination of cellulose accessibility to cellulase based on adsorption of a nonhydrolytic fusion protein containing CBM and GFP with its applications. Langmuir 23:12535-12540. [DOI] [PubMed] [Google Scholar]
- 12.Jung, H.-C., S. Ko, S.-J. Ju, E.-J. Kim, M.-K. Kim, and J.-G. Pan. 2003. Bacterial cell surface display of lipase and its randomly mutated library facilitates high-throughput screening of mutants showing higher specific activities. J. Mol. Catal. B Enzym. 26:177-184. [Google Scholar]
- 13.Jung, H., D. B. Wilson, and L. P. Walker. 2002. Binding mechanisms for Thermobifida fusca Cel5A, Cel6B, and Cel48A cellulose-binding modules on bacterial microcrystalline cellulose. Biotechnol. Bioeng. 80:380-392. [DOI] [PubMed] [Google Scholar]
- 14.Kim, Y. S., H. C. Jung, and J. G. Pan. 2000. Bacterial cell surface display of an enzyme library for selective screening of improved cellulase variants. Appl. Environ. Microbiol. 66:788-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehtio, J., J. Sugiyama, M. Gustavsson, L. Fransson, M. Linder, and T. T. Teeri. 2003. The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc. Natl. Acad. Sci. U. S. A. 100:484-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, L., X. Meng, P. Liu, Y. Hong, G. Wu, X. Huang, C. Li, J. Dong, L. Xiao, and Z. Liu. 2009. Improved catalytic efficiency of endo-β-1,4-glucanase from Bacillus subtilis BME-15 by directed evolution. Appl. Microbiol. Biotechnol. 82:671-679. [DOI] [PubMed] [Google Scholar]
- 17.Lynd, L. R., M. S. Laser, D. Bransby, B. E. Dale, B. Davison, R. Hamilton, M. Himmel, M. Keller, J. D. McMillan, J. Sheehan, and C. E. Wyman. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169-172. [DOI] [PubMed] [Google Scholar]
- 18.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murashima, K., A. Kosugi, and R. H. Doi. 2003. Solubilization of cellulosomal cellulases by fusion with cellulose-binding domain of noncellulosomal cellulase EngD from Clostridium cellulovorans. Proteins 50:620-628. [DOI] [PubMed] [Google Scholar]
- 20.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Thermostabilization of cellulosomal endoglucanase EngB from Clostridium cellulovorans by in vitro DNA recombination with non-cellulosomal endoglucanase EngD. Mol. Microbiol. 45:617-626. [DOI] [PubMed] [Google Scholar]
- 21.Nakazawa, H., K. Okada, T. Onodera, W. Ogasawara, H. Okada, and Y. Morikawa. 2009. Directed evolution of endoglucanase III (Cel12A) from Trichoderma reesei. Appl. Microbiol. Biotechnol. 83:649-657. [DOI] [PubMed] [Google Scholar]
- 22.Peralta-Yahya, P., B. T. Carter, H. Lin, H. Tao, and V. W. Cornish. 2008. High-throughput selection for cellulase catalysts using chemical complementation. J. Am. Chem. Soc. 130:17446-17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole, D. M., A. J. Durrant, G. P. Hazlewood, and H. J. Gilbert. 1991. Characterization of hybrid proteins consisting of the catalytic domains of Clostridium and Ruminococcus endoglucanases, fused to Pseudomonas non-catalytic cellulose-binding domains. Biochem. J. 279:787-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin, Y., X. Wei, X. Song, and Y. Qu. 2008. Engineering endoglucanase II from Trichoderma reesei to improve the catalytic efficiency at a higher pH optimum. J. Biotechnol. 135:190-195. [DOI] [PubMed] [Google Scholar]
- 25.Sonan, G. K., V. Receveur-Brechot, C. Duez, N. Aghajari, M. Czjzek, R. Haser, and C. Gerday. 2007. The linker region plays a key role in the adaptation to cold of the cellulase from an Antarctic bacterium. Biochem. J. 407:293-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.South, C. R., D. A. L. Hogsett, and L. R. Lynd. 1995. Modeling simultaneous saccharification and fermentation of lignocellulose to ethanol in batch and continuous reactors. Enzyme Microb. Technol. 17:797-803. [Google Scholar]
- 27.Violot, S., N. Aghajari, M. Czjzek, G. Feller, G. K. Sonan, P. Gouet, C. Gerday, R. Haser, and V. Receveur-Bréchot. 2005. Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering. J. Mol. Biol. 348:1211-1224. [DOI] [PubMed] [Google Scholar]
- 28.Wang, T., X. Liu, Q. Yu, X. Zhang, Y. Qu, P. Gao, and T. Wang. 2005. Directed evolution for engineering pH profile of endoglucanase III from Trichoderma reesei. Biomol. Eng. 22:89-94. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, X.-Z., N. Sathitsuksanoh, and Y.-H. P. Zhang. 2010. Glycoside hydrolase family 9 processive endoglucanase from Clostridium phytofermentans: heterologous expression, characterization, and synergy with family 48 cellobiohydrolase. Bioresour. Technol. 101:5534-5538. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, X.-Z., Z.-M. Zhang, Z. Zhu, N. Sathitsuksanoh, Y. Yang, and Y.-H. P. Zhang. 2010. The non-cellulosomal family 48 cellobiohydrolase from Clostridium phytofermentans ISDg: heterologous expression, characterization, and processivity. Appl. Microbiol. Biotechnol. 86:525-533. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Y.-H. P. 2008. Reviving the carbohydrate economy via multi-product biorefineries. J. Ind. Microbiol. Biotechnol. 35:367-375. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y.-H. P. 2009. A sweet out-of-the-box solution to the hydrogen economy: is the sugar-powered car science fiction? Energy Environ. Sci. 2:272-282. [Google Scholar]
- 33.Zhang, Y.-H. P., J.-B. Cui, L. R. Lynd, and L. R. Kuang. 2006. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidences from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 7:644-648. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y.-H. P., M. Himmel, and J. R. Mielenz. 2006. Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 24:452-481. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Y.-H. P., J. Hong, and X. Ye. 2009. Cellulase assays. Methods Mol. Biol. 581:213-231. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Y.-H. P., and L. R. Lynd. 2006. A functionally-based model for hydrolysis of cellulose by fungal cellulase. Biotechnol. Bioeng. 94:888-898. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, Y.-H. P., and L. R. Lynd. 2004. Toward an aggregated understanding of enzymatic hydrolysis of cellulose: noncomplexed cellulase systems. Biotechnol. Bioeng. 88:797-824. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, Z., N. Sathitsuksanoh, and Y.-H. P. Zhang. 2009. Direct quantitative determination of adsorbed cellulase on lignocellulosic biomass with its application to study cellulase desorption for potential recycling. Analyst 134:2267-2272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.