Abstract
Low-G+C thermophilic obligate anaerobes in the class Clostridia are considered among the bacteria most resistant to genetic engineering due to the difficulty of introducing foreign DNA, thus limiting the ability to study and exploit their native hydrolytic and fermentative capabilities. Here, we report evidence of natural genetic competence in 13 Thermoanaerobacter and Thermoanaerobacterium strains previously believed to be difficult to transform or genetically recalcitrant. In Thermoanaerobacterium saccharolyticum JW/SL-YS485, natural competence-mediated DNA incorporation occurs during the exponential growth phase with both replicating plasmid and homologous recombination-based integration, and circular or linear DNA. In T. saccharolyticum, disruptions of genes similar to comEA, comEC, and a type IV pilus (T4P) gene operon result in strains unable to incorporate further DNA, suggesting that natural competence occurs via a conserved Gram-positive mechanism. The relative ease of employing natural competence for gene transfer should foster genetic engineering in these industrially relevant organisms, and understanding the mechanisms underlying natural competence may be useful in increasing the applicability of genetic tools to difficult-to-transform organisms.
The genera Thermoanaerobacter and Thermoanaerobacterium contain bacteria which are thermophilic, obligate anaerobes that specialize in polysaccharide and carbohydrate fermentation, producing primarily l-lactic acid, acetic acid, ethanol, CO2, and H2 (24, 27, 49). Taxonomically, they are distinguished from other anaerobic thermophilic clostridia by the ability to reduce thiosulfate to hydrogen sulfide or elemental sulfur (21). The majority of characterized Thermoanaerobacter and Thermoanaerobacterium strains have been isolated from hot springs and other thermal environments (20-22, 38, 47); however, they have also been isolated from canned foods (4, 10), soil (48), paper mills and breweries (41, 43), and deep subsurface environments (5, 13, 35), suggesting a somewhat ubiquitous environmental presence.
Representatives of the Thermoanaerobacter and Thermoanaerobacterium genera have been considered for biotechnological applications, such as conversion of lignocellulosic biomass to ethanol (8, 27) or other fuels and chemicals (3, 24). However, the branched fermentation pathways of these organisms generally require modification for industrial application. Several studies have investigated manipulating bioprocess and growth conditions to alter end product ratios and yields, but this has not resulted in reliable conditions to maximize the yield of a single end product (18, 25). Genetic engineering is likely necessary for commercial application of Thermanaerobacter or Thermoanaerobacterium species (26, 27, 44). As genetic systems for these bacteria have emerged (28, 45), increased product yields have been demonstrated by gene knockout of l-lactate dehydrogenase (9, 14), phosphotransacetylase and acetate kinase (40), and hydrogenase (39). Despite this recent progress, genetic transformation is still considered the greatest barrier for engineering these organisms (44).
In contrast, some of the bacteria most amenable to genetic manipulation are those exhibiting natural competence; for example, work with the naturally competent Streptococcus pneumoniae first established DNA as the molecule containing inheritable information (42). Naturally competent organisms are found in many bacterial phyla, although the overall number of bacteria known to be naturally competent is relatively small (16).
The molecular mechanisms of natural competence are often divided into two stages: early-stage genes that encode regulatory and signal cascades to control competence induction, and late-stage genes that encode the machinery of DNA uptake and integration (16). The Gram-positive late-stage consensus mechanism for DNA uptake and assimilation, elucidated primarily through work with Bacillus subtilis, occurs through several molecular machinery steps. First, DNA is believed to interact with a type IV pilus (T4P) or pseudopilus that brings it into close proximity of the cell membrane. The precise mechanism of this phenomenon is unclear; although components of the T4P in both Gram-positive and Gram-negative bacteria have been shown to bind DNA (7, 19), in specific studies, a full pilus structure has been either not observed or shown not to be essential during natural competence (6, 36). Two proteins, ComEA and ComEC, are then involved in creation and transport of single-stranded DNA across the membrane, where it is subsequently bound by CinA-localized RecA and either integrated into the genome or replicated at an independent origin, as for plasmid DNA (6).
Here, we report that several Thermoanaerobacter and Thermoanaerobacterium strains are naturally competent, characterize growth conditions conducive to natural competence, and identify genes in Thermoanaerobacterium saccharolyticum JW/SL-YS485 required for competence exhibition.
MATERIALS AND METHODS
Strains and plasmids.
Strains and plasmids used in this study are listed in Table 1. The replicating shuttle plasmid pMU131 contains a thermostable kanamycin resistance (Kanr) marker (28), the pUC origin of replication and ampicillin resistance marker, and a thermostable Gram-positive origin of replication isolated from a native plasmid of Thermoanaerobacterium saccharolyticum B6A-RI (N. Caiazza, A. Warner, and C. Herring, 19 March 2009, international patent application no. PCT/US2008/010545; N. Caiazza, C. Rice, A. Warner, D. Hogsett, and C. Herring, unpublished data; see Weimer et al. [47] for an earlier description of these native plasmids).
TABLE 1.
Plasmids and strains used in this study
| Plasmid/strain | Descriptiona | Source/reference |
|---|---|---|
| pMU131 | T. saccharolyticum-E. coli shuttle plasmid, Kanr Ampr | PCT/US2008/010545 |
| pMQ87 | Cloning plasmid for yeast homologous recombination, Genr, ura3 | Presque Isle culture collection |
| pSGD8 | l-ldh knockout plasmid with Kanr Ampr | 24 |
| pMU1966 | T. saccharolyticum T4P knockout vector, Eryr Genr, ura3 | This study |
| pMU1967 | T. saccharolyticum comEA knockout vector Eryr Genr, ura3 | This study |
| pMU1968 | T. saccharolyticum comEC knockout vector Eryr Genr, ura3 | This study |
| pMU1969 | T. saccharolyticum recA knockout vector Eryr Genr, ura3 | This study |
| TOP10 | E. coli cloning strain | Invitrogen |
| DSM 8691 | Thermoanaerobacterium saccharolyticum JW/SL-YS485 | DSMZ |
| ALK2 | T. saccharolyticum YS485 Δl-ldh Δpta Δack, Kanr Eryr | 27 |
| M1464 | T. saccharolyticum YS485 Δtfp, Eryr | This study |
| M1465 | T. saccharolyticum YS485 ΔrecA, Eryr | This study |
| M1466 | T. saccharolyticum YS485 ΔcomEC, Eryr | This study |
| M1467 | T. saccharolyticum YS485 ΔcomEA, Eryr | This study |
| ATCC 27405 | Clostridium thermocellum | Lynd lab |
| DSM 8903 | Caldicellulosiruptor saccharolyticus | DSMZ |
| ATCC 35047 | Thermoanaerobacter brockii | ATCC |
| DSM 2246 | Thermoanaerobacter ethanolicus JW200 | DSMZ |
| DSM 11426 | Thermoanaerobacter mathranii | DSMZ |
| ATCC 33223 | Thermoanaerobacter pseudethanolicus 39E | ATCC |
| DSM 10170 | Thermoanaerobacterium aotearoense | DSMZ |
| B6A | Thermoanaerobacterium saccharolyticum B6A | Paul Weimer |
| ATCC 49915 | Thermoanaerobacterium saccharolyticum B6A-RI | ATCC |
| ATCC 7956 | Thermoanaerobacterium thermosaccharolyticum | ATCC |
| ATCC 31960 | Thermoanaerobacterium thermosaccharolyticum HG-8 | ATCC |
| M0523 | Thermoanaerobacterium thermosaccharolyticum | Mascoma |
| M0524 | Thermoanaerobacterium thermosaccharolyticum | Mascoma |
| M0795 | Thermoanaerobacterium thermosaccharolyticum | Mascoma |
| DSM 7097 | Thermoanaerobacterium xylanolyticum | DSMZ |
| DSM 13642 | Thermoanaerobacterium zeae | DSMZ |
Kanr, kanamycin resistance; Ampr, ampicillin resistance; Eryr, erythromycin resistance; Genr, gentamicin resistance.
Media and growth conditions.
All culturing of thermophilic bacteria was performed in modified DSMZ medium 122, containing per liter 5.0 g cellobiose, 1.3 g (NH4)2SO4, 2.6 g MgCl2 6 · H2O, 1.43 g KH2PO4, 1.8 g K2HPO4, 0.13 gCaCl2 2 · H2O, 6.0 g Na-β-glycerophosphate, 0.00013 g FeSO4 7 · H2O, 4.5 g yeast extract, 0.002 g resazurin, 0.5 g l-cysteine-HCl, and 10 g agarose for solid media. The pH was adjusted to 6.7 with 10 N NaOH or 72% (wt/vol) H2SO4 if necessary. Chemicals were obtained from Sigma-Aldrich, and yeast extract was obtained from BD Difco. Cultures were grown at 55°C, unless otherwise noted, in an anaerobic chamber (COY Labs, Grass Lake, MI). For selection of erythromycin-resistant colonies of T. saccharolyticum, a medium pH of 6.1 and an incubation temperature of 50°C were used. Escherichia coli was grown in LB medium with kanamycin at 50 μg/ml or gentamicin at 25 μg/ml for plasmid selection and maintenance. Saccharomyces cerevisiae was grown on solid CM minus uracil media for plasmid selection (37). Stock cultures of thermophilic strains were prepared from cultures grown to exponential or early stationary phase by the addition of 5% dimethyl sulfoxide (DMSO) and frozen at −80°C.
Natural genetic competence.
Natural competence transformations were conducted in an anaerobic chamber by inoculation of 10 ml of medium with 1 to 3 μl of a frozen stock culture. After mixing, 1-ml aliquots were transferred to tubes containing 250 ng DNA suspended in 10 mM Tris buffer, pH 8.0, at a concentration of approximately 50 ng/μl. pMU131 plasmid DNA prepared in E. coli TOP10 (Invitrogen, Madison, WI) was used for natural competence tests of different species. Different types of DNA used to transform T. saccharolyticum were prepared as described in Results. The tubes were then incubated at 55°C for 16 to 18 h to an optical density at 600 nm (OD600) of 0.6 to 1.0. Dilutions of the transformation culture were mixed with liquid agar at 55°C containing the appropriate antibiotic concentration, poured into petri dishes, allowed to solidify at room temperature, and incubated at 55°C in a moisture-retaining container until colony formation. Negative controls were performed by the exclusion of DNA. Putative transformants were tested for the presence of the kanamycin marker via PCR with primers X00860 and X00861, and 16S sequencing was performed to confirm culture identity with primers X00050 and X00051 (Table 2).
TABLE 2.
Primers used in this study
| Primer no. | 5′-3′ sequence |
|---|---|
| X00004 | GGGTTTATCGACCTTGGTTCGTGACATTGTGGGC |
| X00021 | TGCTGCTTCTGTTCTTGACC |
| X00050 | AGAGTTTGATCCTGGCTCAG |
| X00051 | ACGGCTACCTTGTTACGACTT |
| X00861 | ACCACCTATGATGTGGAACGGGAA |
| X00862 | TTTCTCCCAATCAGGCTTGATCCC |
| X00957 | GGGCATTTAACGACGAAACTGGCT |
| X00958 | ACATCTGTGGTATGGCGGGTAAGT |
| X01177 | GCTCATGAACCCAAAGTTGCAAAGC |
| X01178 | CCCTCCTGCATTGCCTACAAAGTA |
| X08154 | TGCTGTCAAGAGCTGTGTCCTCAT |
| X08155 | AACTTCACTTCGCCAGCAGTTGTC |
| X08160 | TTGATGGCACTTTGCTCCCTGTTG |
| X08161 | CAGCCACACTAAATCCTGGGACAA |
| X08268 | CAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGGAGTCTTTCGCAATAAGAGGCAAC |
| X08151 | GGTTTATCGACCTGCAACCCAGTCAATAATGAAGCTACTATCAA |
| X08269 | TTGATAGTAGCTTCATTATTGACTGGGTTGCAGGTCGATAAACC |
| X08270 | AGAGCCGCTGGATTTATCGTTGGATTAGTAACGTGTAACTTTCC |
| X08152 | GGAAAGTTACACGTTACTAATCCAACGATAAATCCAGCGGCTCT |
| X08271 | GTGAGCGGATAACAATTTCACACAGGAAACAGCTATGACCATGCCGATACCGAATCAACCTGGA |
| X08272 | CAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGCAATTCTTGGCTCACATGGGCCTT |
| X08157 | GGTTTATCGACCTGCATTTCTCCCACCGTCAATCCCAAGA |
| X08273 | TCTTGGGATTGACGGTGGGAGAAATGCAGGTCGATAAACC |
| X08274 | ACTACTTCTCCATCTGGCTGTCCATTAGTAACGTGTAACTTTCC |
| X08158 | GGAAAGTTACACGTTACTAATGGACAGCCAGATGGAGAAGTAGT |
| X08275 | GTGAGCGGATAACAATTTCACACAGGAAACAGCTATGACCCCGAAACTGTCGTGCAATCATGGA |
| X08721 | TTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGATGAAACTGCTGTTGTTGGCGACC |
| X08722 | GGTTTATCGACCTGCATAAACCCGACAATGATGCCGGTTG |
| X08723 | CAACCGGCATCATTGTCGGGTTTATGCAGGTCGATAAACC |
| X08724 | CAGGACTCTGCGATTGATTATCGGTTAGTAACGTGTAACTTTCC |
| X08725 | GGAAAGTTACACGTTACTAACCGATAATCAATCGCAGAGTCCTG |
| X08726 | CGGATAACAATTTCACACAGGAAACAGCTATGACCTTCCAGCTCCAATTGCACCAGATG |
| X08727 | ATATGGCCTCTTAAATGGCGGTGC |
| X08728 | TGCCAGAGCCACCAGCAATTTCAA |
| X08729 | TTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGCGCGGCCAGCAATCTTGGTAAATA |
| X08730 | GGTTTATCGACCTGCAAAATCCATTCCCAACAAGCGGAGC |
| X08731 | GCTCCGCTTGTTGGGAATGGATTTTGCAGGTCGATAAACC |
| X08732 | TGCCCAAGCCTTATGTCGCCATATTTAGTAACGTGTAACTTTCC |
| X08733 | GGAAAGTTACACGTTACTAAATATGGCGACATAAGGCTTGGGCA |
| X08734 | GTGAGCGGATAACAATTTCACACAGGAAACAGCTACGGGCATAATTTGTGAGCCATCCA |
| X08735 | TTTCCGGGAGAGACAGAGGATGAA |
| X08736 | TACTGCAGTTTACTGGGTCTTGTGGG |
Transformation frequency during batch growth.
Exponentially growing T. saccharolyticum cells were diluted to an OD600 of 0.03 in fresh medium, and each hour 1-ml subcultures were mixed with 250 ng pMU131 DNA and grown under the same conditions as the main culture. After an hour of incubation, 2 units of DNase (New England Biolabs, Ipswich, MA) was added to the subcultures to hydrolyze free DNA, and the mixture was incubated for an additional hour to allow expression of the kanamycin resistance marker. Subcultures were then diluted and plated in nonselective and kanamycin-containing solid media to determine the transformation frequency.
Plasmid and knockout strain construction.
Plasmids were constructed by S. cerevisiae-based in vivo recombination cloning (37) using the S. cerevisiae-E. coli shuttle plasmid pMQ87. Knockout plasmids were isolated by miniprep (Qiagen, Germantown, MD) in E. coli TOP10 cells prior to transformation in T. saccharolyticum. Primers used to construct knockout plasmids are shown on plasmid maps in Fig. S1 in the supplemental material and the primer sequences in Table 2.
Nucleotide sequence accession number.
The sequences reported in this paper have been deposited in the GenBank database (accession no. GU479453 [T4P region], GU479454 [comEA region], GU479455 [comEC region], and GU479456 [cinA recA region]).
RESULTS
Determination of natural competence.
To transform T. saccharolyticum JW/SL-YS485, we previously used a hybrid chemical-electrotransformation protocol (28) that includes incubation with isonicotinic acid hydrazide to weaken cell walls, cell harvesting, washing, electropulsing, and an outgrowth period in fresh media prior to plating with a selective antibiotic. This protocol first came into question when a no-pulse control experiment yielded more transformants than one which included an electrical pulse. It was subsequently determined that the only essential step of the protocol was the cell outgrowth period, leading us to conclude that T. saccharolyticum JW/SL-YS485 is naturally competent.
Transformation with different DNA types.
T. saccharolyticum was transformable by replicating plasmid and homologous recombination-based chromosomal integration vectors (Table 3), and like other naturally competent organisms, can be transformed with genomic DNA containing a selectable genotype (17, 23). pSGD8 (9; see also Fig. S1 in the supplemental material), a nonreplicating knockout vector containing 1.2 kb of upstream homology and 0.4 kb of downstream homology to the l-ldh locus, transformed T. saccharolyticum as circular DNA and after an AclI/EcoRI digestion that created a linear fragment. The linear digested plasmid was confirmed by agarose gel analysis to contain a fragment with the kanamycin resistance marker and flanking homology regions. Evidence of genome integration after transformation was determined by PCR (Fig. 1).
TABLE 3.
Transformation efficiency of T. saccharolyticum JW/SL-YS485 with different DNA typesa
| DNA type | Transformed cells per μg DNA | Transformed cells per μg Kan gene DNA |
|---|---|---|
| pMU131 | 2.5E+05 | 1.2E+06 |
| ALK2 gDNA | 2.0E+02 | 4.6E+05b |
| pSGD8 | 5.1E+04 | 2.2E+05 |
| pSGD8 AclI/EcoRIc | 5.7E+03 | 2.4E+04 |
Transformation efficiency as a function of total DNA and of DNA encoding the kanamycin resistance marker.
Estimate based on a genome size of 3.0 Mb.
Plasmid digested to produce a linear DNA fragment containing the kanamycin resistance gene and flanking regions with homology to the l-ldh locus.
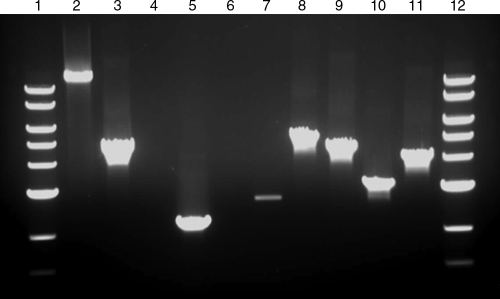
FIG. 1.
PCRs used to confirm kanamycin marker integration in the T. saccharolyticum genome. Primers were external to homologous recombination regions for the l-ldh locus of pSGD8 and the pta ack Kanr locus of ALK2. Lane 1, NEB 10-kb ladder; lane 2, wild-type with l-ldh external primers X01177 and X01178; lanes 3 to 5, Kanr colonies transformed with the knockout plasmid pSGD8 with l-ldh external primers; lanes 6 to 8, Kanr colonies transformed with restriction-digested pSGD8 linear vector with l-ldh external primers. Predicted sizes are 3,863 bp for wild type and 5,160 bp for kanamycin resistance marker integration. Lane 9, wild type with pta ack external primers X00004 and X00021; lanes 10 to 12, Kanr colonies transformed with ALK2 genomic DNA with pta ack external primers. Predicted sizes are 3,209 bp for wild type and 4,245 bp for kanamycin resistance marker integration.
Natural competence occurs during exponential growth.
No obvious induction event was required to bring T. saccharolyticum cells into the competent state beyond growth in a typical laboratory medium. Figure 2 shows the transformation frequency of T. saccharolyticum with the replicating plasmid pMU131 (see Materials and Methods) throughout batch growth. The transformation frequency was highest during early exponential growth and declined until reaching the stationary phase, in which the transformation frequency was below the limit of detection (8.0E−9 transformants per CFU). In this experiment DNA was incubated with cells for 1 h before DNase treatment to discern the effect of growth phase on transformation frequency, whereas for all other experiments described here cells were incubated with DNA for 16 to 18 h prior to plating on selective media (see Materials and Methods). In our hands, transformation efficiencies were highest when DNA was added at low initial cell densities (1E3 to 1E5 cells/ml), and cells were plated on selective medium prior to the onset of the stationary phase. Efficiencies were lowest when DNA was added at higher cell densities (1E8 cells/ml), and cells were plated after entering the stationary phase.
FIG. 2.
Transformation efficiency of T. saccharolyticum JW/SL-YS485 during batch growth. ⋄, optical density; ▪, transformation frequency. Exponentially growing cells were transferred into fresh media at an initial OD of 0.03. To evaluate transformation efficiency, 1 ml of culture was transferred into a new tube containing 250 ng pMU131, incubated for 1 h before addition of DNase, and incubated for an additional hour to allow expression of the kanamycin resistance marker. Cells were then serially diluted on selective and nonselective media to determine transformation efficiency. Transformation efficiency data points are plotted at the time of DNase addition.
Natural competence in related bacteria.
To test whether the natural competence phenomenon was unique to T. saccharolyticum YS485 among related bacteria, 16 other strains were tested for the ability to be transformed with the replicating plasmid pMU131. No optimization of the transformation protocol was made beyond determination of the minimum concentration of kanamycin required to eliminate spontaneous colony formation. As shown in Table 4, a total of 13 strains exhibited natural competence, three of which were Thermoanaerobacterium thermosaccharolyticum strains isolated by Mascoma Corporation. Transformation frequencies ranged from 1.0E−3 to 1.9E−6 transformants per CFU. For each transformation, three colonies were checked for the presence of the kanamycin marker by PCR (Fig. 3), and a 16S sequence was amplified using universal primers, sequenced, and compared to that of the original starting culture. In no case was there evidence of spontaneously kanamycin-resistant colony formation or a transformable contaminant within the tested culture. Thermoanaerobacterium zeae, Thermoanaerobacter mathranii, Caldicellulosiruptor saccharolyticus, and Clostridium thermocellum were not transformed with this protocol. However, the ability of these strains to become naturally competent cannot be excluded based on this result, as several factors could result in a lack of transformants, such as the pMU131 resistance marker or replication origin not functioning in the host organism, an unmet condition for competence induction, or a native mechanism for limiting foreign DNA, such as a restriction or CRISPR system (29, 46).
TABLE 4.
Transformation frequencies of Thermoanaerobacter and Thermoanaerobacterium bacteriaa
| Strain | Transformants per CFU | Kan (μg/ml) |
|---|---|---|
| Thermoanaerobacterium saccharolyticum JW/SL-YS485 DSM 8691 | 1.4E−04 | 200 |
| Thermoanaerobacter ethanolicus JW200 DSM 2246 | 1.0E−03 | 1,000 |
| Thermoanaerobacterium thermosaccharolyticum M0523 | 2.8E−04 | 200 |
| Thermoanaerobacterium thermosaccharolyticum M0524 | 4.2E−05 | 200 |
| Thermoanaerobacterium aotearoense DSM 10170 | 1.5E−04 | 1,000 |
| Thermoanaerobacterium thermosaccharolyticum HG-8 ATCC 31960 | 1.2E−04 | 200 |
| Thermoanaerobacterium saccharolyticum B6A | 2.1E−04 | 200 |
| Thermoanaerobacterium saccharolyticum B6A-RI ATCC 49915 | 1.7E−04 | 200 |
| Thermoanaerobacterium thermosaccharolyticum M0795 | 7.1E−05 | 200 |
| Thermoanaerobacterium xylanolyticum DSM 7097 | 1.6E−05 | 200 |
| Thermoanaerobacterium thermosaccharolyticum ATCC 7956 | 1.2E−05 | 200 |
| Thermoanaerobacter pseudethanolicus 39E ATCC 33223 | 6.3E−05 | 400 |
| Thermoanaerobacter brockii ATCC 35047 | 1.9E−06 | 1,000 |
Bacteria were transformed with the replicating plasmid pMU131 as described in Materials and Methods.
FIG. 3.
PCRs with primers X00861 and X00862 of Kanr colonies designed to amplify a 603-bp region in the kanamycin resistance marker. First and last lanes on each row were loaded with NEB 1-kb DNA ladder. Internal gel lanes are grouped four per strain: the first three per group are colonies transformed with pMU131, and the fourth is a reaction with cells from the same strain that was not transformed. Strain order is as follows: (first row, left to right) Thermoanaerobacter brockii ATCC 35047, Thermoanaerobacter ethanolicus JW200 DSM 2246, Thermoanaerobacter pseudethanolicus 39E ATCC 33223, Thermoanaerobacterium aotearoense DSM 10170, Thermoanaerobacterium saccharolyticum B6A, Thermoanaerobacterium saccharolyticum B6A-RI ATCC 49915, Thermoanaerobacterium saccharolyticum JW/SL-YS485 DSM 8691, (second row, left to right) Thermoanaerobacterium thermosaccharolyticum ATCC 7956, Thermoanaerobacterium thermosaccharolyticum HG-8 ATCC 31960, Thermoanaerobacterium thermosaccharolyticum M0523, Thermoanaerobacterium thermosaccharolyticum M0524, Thermoanaerobacterium thermosaccharolyticum M0795, Thermoanaerobacterium xylanolyticum DSM 7097.
Gram-positive competence homologues are required for natural competence.
To begin elucidation of the natural competence mechanism in T. saccharolyticum, gene knockouts were made in loci with high similarity to genes involved in natural competence in other Gram-positive bacteria. Knockouts of a putative T4P locus (of which only one was identified on the genome), comEA, comEC, and a cinA recA locus were made using an erythromycin resistance marker. Deletions with chromosomal integration of both flanking regions were confirmed by PCR with primers external to the areas of homologous recombination (Fig. 4). The subsequent knockout strains were assayed for transformability with the replicating plasmid pMU131. As shown in Table 5, the ΔT4P, ΔcomEA, and ΔcomEC strains had transformation frequencies below the limit of detection, while the ΔcinA ΔrecA strain had a 250-fold reduction in transformation efficiency compared to the wild type.
FIG. 4.
PCRs used to confirm erythromycin marker integration in the T. saccharolyticum genome. Lane 1, NEB 10-kb ladder; lane 2, wild type with T4P external primers X08727, X08728, predicted size 12,981 bp; lane 3, M1464 with T4P external primers, predicted size 4,669 bp; lane 4, wild type with cinA recA downstream external primer X08736 and erythromycin internal primer X00957, no predicted band; lane 5, M1465 with primers X08736 and X00957, predicted size 2,191 bp; lane 6, wild type with cinA recA upstream external primer X08735 and erythromycin internal primer X00958, no predicted band; lane 7, M1465 with primers X08735 and X00958, predicted size 2,692 bp (internal and external primers were used to verify M1465, as the erythromycin gene replaced a similarly sized fragment of the cinA recA locus); lane 8, wild-type with comEC external primers X08160 and X08161, predicted size 5,217 bp; lane 9, M1466 with comEC external primers, predicted size 4,519 bp; lane 10, wild type with comEA external primers X08154 and X08155, predicted size 3,165 bp; lane 9, M1467 with comEA external primers, predicted size 4,187 bp; lane 12, NEB 10-kb ladder.
TABLE 5.
Transformation frequencies of T. saccharolyticum JW/SL-YS485 and mutants
| Strain | Description or genotype | Transformants per CFUa |
|---|---|---|
| JW/SL-YS485 | Wild type | 1.4E−04 |
| M1464 | ΔT4P (or1949 to or1961) | ND |
| M1465 | ΔcinA ΔrecA (or1848 and or1849) | 5.7E−07 |
| M1466 | ΔcomEC (or2075) | ND |
| M1467 | ΔcomEA (or2100) | ND |
ND, not detected, below detection limit of 5.4E−09.
DISCUSSION
We were unable to identify previous reports of natural competence in members of the class Clostridia, although the 80 or so prokaryotic species known to be naturally competent are widely distributed phylogenetically (16). With 13 of the 15 tested Thermoanaerobacter and Thermoanaerobacterium strains demonstrating natural competence in this study, the phenomenon is apparently widespread among these organisms.
Most studied naturally competent bacterial species induce competence in response to external factors, such as pheromone density (quorum sensing) or stringent nutritional conditions; prominent examples include that of Streptococcus pneumoniae and Bacillus subtilis (11, 15). T. saccharolyticum falls within the smaller subset of studied bacteria, including Acinetobacter calcoaceticus, Neisseria gonorrhoeae, Deinococcus radiodurans, and the cyanobacteria Synechococcus and Chlorobium, that are naturally competent during the exponential growth phase without the requirement of special stimuli (16, 33). As seen in Fig. 2, the transformation frequency for T. saccharolyticum is highest during early growth in fresh medium and decreases toward zero as the stationary phase is reached. Further study of the physiology and regulation of natural competence in T. saccharolyticum will be required for a better understanding of how and why this organism enters the competent state.
With the protocol reported here, the transformation efficiency of T. saccharolyticum JW/SL-YS485 with pMU131 was observed to be 1.4E−4 transformants per CFU. Thermoanaerobacter ethanolicus JW200 had the highest transformation frequency at 1.0E−3 transformants per CFU, while Thermoanaerobacter brockii had the lowest at 1.9E−6 transformants per CFU. DNA concentration, divalent cation concentration, pH, temperature, carbon source, exposure time to DNA, and the selective maker type have all been shown to influence transformation frequencies of other naturally competent organisms, such as A. calcoaceticus and Thermus thermophilus (17, 34). It is possible that many of these factors also influence transformation efficiency in Thermoanaerobacter and Thermoanaerobacterium strains and that the maximum transformation efficiencies remain to be determined. Nevertheless, with the efficiencies reported here, standard genetic manipulations, such as plasmid transformation, gene knockout, and gene integration, are easily performed, and transformation via linear DNA enables rapid PCR-based transformation strategies (12, 32).
The genome of T. saccharolyticum carries several genes that have homology to Gram-positive late-stage competence genes, including a 13-gene cluster with homology to type IV pilus (T4P) assembly genes which bind DNA during natural competence (6), comEA and comEC homologues, which are involved in DNA transport across the cell membrane (6), and cinA and recA homologues, which are thought to be involved in single-strand DNA protection and chromosomal integration after passage into the cytosol (2a). cinA, also referred to as colligrin or DNA damage/competence-induced protein, has been shown to mediate recA localization to the membrane when cells are in the competent state (30).
In T. saccharolyticum, homologues for T4P genes, comEA, and comEC are required for observable natural competence. This strongly suggests that natural competence occurs via a conserved Gram-positive mechanism involving these enzymes. Based on sequence similarity, the ΔT4P region (see Fig. S2 in the supplemental material) contains many T4P components, including putative traffic NTPases PilB and PilT (or1961 and or1960), pseudopilins PilE and PilV (or1958 and or1955), a prepilin processing peptidase, PilD (or1957), a polytopic membrane protein, PilG (or1959), and T4P or competence-associated proteins FimT, PilW, ComFB, PilM, PilN, and PilO (or1956, or1954, or1952, or1951, or1950, and or1949, respectively). The comEA and comEC genes of T. saccharolyticum are not located adjacent to other known competence genes, although the genetic organization at these two loci is conserved in other Thermoanaerobacter and Thermoanaerobacterium strains. The observed 250-fold drop in transformation efficiency of the ΔcinA ΔrecA strain suggests that CinA, RecA, or both also play a role during natural competence, as has been shown with B. subtilis and S. pneumoniae (30, 50).
Model organisms such as S. cerevisiae and E. coli are often considered for lignocellulosic biofuel and biochemical production due to the relative ease of genetic engineering, even though they lack one or more of the traits required of an ideal biocatalyst, such as hydrolytic capabilities, high productivities, or broad substrate utilization (1). Organisms such as Thermoanaerobacter and Thermoanaerobacterium bacteria have inherent advantages relative to these model organisms, such as the ability to rapidly hydrolyze and ferment low-cost polysaccharides and sugars (2, 27) and the ability to grow at temperatures above 50°C, which could improve process metrics, such as fermentation heat load, microbial contamination, substrate solubility, and product recovery (44). Still, the absence or rudimentary status of genetic systems for such thermophilic anaerobes constrained their development as biocatalysts. The simple and powerful transformation system described here, along with recent genomic sequencing projects for several Thermoanaerobacter and Thermoanaerobacterium strains (DOE Joint Genome Institute [http://www.jgi.doe.gov/]), should greatly accelerate the pace and extent to which genetic manipulations can be made in these biotechnologically relevant organisms.
Supplementary Material
Acknowledgments
This work was supported by Mascoma Corporation and grants from the Department of Energy under awards DE-FC36-07G017057 and DE-FG02-02-ER15350. L.R.L. was also supported by a grant from the DOE BioEnergy Science Center.
We thank Paul Weimer for generously providing Thermoanaerobacterium saccharolyticum B6A and Larry Feinberg, Nicholas Orem, and Justine Folden of the Mascoma Corporation for isolating and providing Thermoanaerobacterium thermosaccharolyticum strains M0523, M0524, and M0795.
This report was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
Footnotes
Published ahead of print on 14 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alper, H., and G. Stephanopoulos. 2009. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat. Rev. Microbiol. 7:715-723. [DOI] [PubMed] [Google Scholar]
- 2.Altaras, N. E., M. R. Etzel, and D. C. Cameron. 2001. Conversion of sugars to 1,2-propanediol by Thermoanaerobacterium thermosaccharolyticum HG-8. Biotechnol. Prog. 17:52-56. [DOI] [PubMed] [Google Scholar]
- 2a.Bergé, M., I. Mortier-Barrière, B. Martin, and J. P. Claverys. 2003. Transformation of streptococcus pneumoniae relies on DprA- and RecA- dependent protection of incoming DNA single strands. Mol. Microbiol. 50:527-536. [DOI] [PubMed] [Google Scholar]
- 3.Cameron, D. C., N. E. Altaras, M. L. Hoffman, and A. J. Shaw. 1998. Metabolic engineering of propanediol pathways. Biotechnol. Prog. 14:116-125. [DOI] [PubMed] [Google Scholar]
- 4.Carlier, J. P., I. Bonne, and M. Bedora-Faure. 2006. Isolation from canned foods of a novel Thermoanaerobacter species phylogenetically related to Thermoanaerobacter mathranii (Larsen 1997): emendation of the species description and proposal of Thermoanaerobacter mathranii subsp. Alimentarius subsp. nov. Anaerobe 12:153-159. [DOI] [PubMed] [Google Scholar]
- 5.Cayol, J. L., B. Ollivier, B. K. Patel, G. Ravot, M. Magot, E. Ageron, P. A. Grimont, and J. L. Garcia. 1995. Description of Thermoanaerobacter brockii subsp. lactiethylicus subsp. nov., isolated from a deep subsurface French oil well, a proposal to reclassify Thermoanaerobacter finnii as Thermoanaerobacter brockii subsp. finnii comb. nov., and an emended description of Thermoanaerobacter brockii. Int. J. Syst. Bacteriol. 45:783-789. [DOI] [PubMed] [Google Scholar]
- 6.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 7.Chen, I., R. Provvedi, and D. Dubnau. 2006. A macromolecular complex formed by a pilin-like protein in competent Bacillus subtilis. J. Biol. Chem. 281:21720-21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demain, A. L., M. Newcomb, and J. H. Wu. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai, S. G., M. L. Guerinot, and L. R. Lynd. 2004. Cloning of l-lactate dehydrogenase and elimination of lactic acid production via gene knockout in Thermoanaerobacterium saccharolyticum JW/SL-YS485. Appl. Microbiol. Biotechnol. 65:600-605. [DOI] [PubMed] [Google Scholar]
- 10.Dotzauer, C., M. A. Ehrmann, and R. F. Vogel. 2002. Occurrence and detection of Thermoanaerobacterium and Thermoanaerobacter in canned food. Food Technol. Biotechnol. 40:21-26. [Google Scholar]
- 11.Dubnau, D. 1993. Genetic exchange and homologous recombination, p. 555-584. In A. L. Sonenshein, T. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: physiology, biochemistry, and molecular genetics. American Society for Microbiology, Washington, DC.
- 12.Fabret, C., S. D. Ehrlich, and N. Philippe. 2002. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol. 46:25-36. [DOI] [PubMed] [Google Scholar]
- 13.Fardeau, M. L., M. Bonilla Salinas, S. L'Haridon, C. Jeanthon, F. Verhe, J. L. Cayol, B. K. Patel, J. L. Garcia, and B. Ollivier. 2004. Isolation from oil reservoirs of novel thermophilic anaerobes phylogenetically related to Thermoanaerobacter subterraneus: reassignment of T. subterraneus, Thermoanaerobacter yonseiensis, Thermoanaerobacter tengcongensis and Carboxydibrachium pacificum to Caldanaerobacter subterraneus gen. nov., sp. nov., comb. nov. as four novel subspecies. Int. J. Syst. Evol. Microbiol. 54:467-474. [DOI] [PubMed] [Google Scholar]
- 14.Georgieva, T., and B. Ahring. 2007. Evaluation of continuous ethanol fermentation of dilute-acid corn stover hydrolysate using thermophilic anaerobic bacterium Thermoanaerobacter BG1L1. Appl. Microbiol. Biotechnol. 77:61-68. [DOI] [PubMed] [Google Scholar]
- 15.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnsborg, O., V. Eldholm, and L. S. Håvarstein. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158:767-778. [DOI] [PubMed] [Google Scholar]
- 17.Koyama, Y., T. Hoshino, N. Tomizuka, and K. Furukawa. 1986. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacis, L. S., and H. G. Lawford. 1991. Thermoanaerobacter ethanolicus growth and product yield from elevated levels of xylose or glucose in continuous cultures. Appl. Environ. Microbiol. 57:579-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang, E., K. Haugen, B. Fleckenstein, H. Homberset, S. A. Frye, O. H. Ambur, and T. Tonjum. 2009. Identification of neisserial DNA binding components. Microbiology 155:852-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen, L., P. Nielsen, and B. K. Ahring. 1997. Thermoanaerobacter mathranii sp. nov., an ethanol-producing, extremely thermophilic anaerobic bacterium from a hot spring in Iceland. Arch. Microbiol. 168:114-119. [DOI] [PubMed] [Google Scholar]
- 21.Lee, Y. E., M. K. Jain, C. Lee, S. E. Lowe, and J. G. Zeikus. 1993. Taxonomic distinction of saccharolytic thermophilic anaerobes: description of Thermoanaerobacterium xylanolyticum gen. nov., sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov.; reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and Clostridium thermohydrosulfuricum E100-69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov., and Thermoanaerobacter thermohydrosulfuricus comb. nov., respectively; and transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus. Int. J. Syst. Bacteriol. 43:41-51. [Google Scholar]
- 22.Liu, S. Y., F. A. Rainey, H. W. Morgan, F. Mayer, and J. Wiegel. 1996. Thermoanaerobacterium aotearoense sp. nov., a slightly acidophilic, anaerobic thermophile isolated from various hot springs in New Zealand, and emendation of the genus Thermoanaerobacterium. Int. J. Syst. Bacteriol. 46:388-396. [Google Scholar]
- 23.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Mol. Biol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe, S. E., M. K. Jain, and J. G. Zeikus. 1993. Biology, ecology, and biotechnological applications of anaerobic-bacteria adapted to environmental stresses in temperature, pH, salinity, or substrates. Microbiol. Mol. Biol. Rev. 57:451-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynd, L. R., H. J. Ahn, G. Anderson, P. Hill, D. S. Kersey, and T. Klapatch. 1991. Thermophilic ethanol-production—investigation of ethanol yield and tolerance in continuous culture. Appl. Biochem. Biotechnol. 28-29:549-570. [Google Scholar]
- 26.Lynd, L. R., W. H. van Zyl, J. E. McBride, and M. Laser. 2005. Consolidated bioprocessing of cellulosic biomass: an update. Curr. Opin. Biotechnol. 16:577-583. [DOI] [PubMed] [Google Scholar]
- 27.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mai, V., W. W. Lorenz, and J. Wiegel. 1997. Transformation of Thermoanaerobacterium sp. strain JW/SL-YS485 with plasmid pIKM1 conferring kanamycin resistance. FEMS Microbiol. Lett. 148:163-167. [Google Scholar]
- 29.Marraffini, L. A., and E. J. Sontheimer. 2008. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 322:1843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masure, H. R., B. J. Pierce, H. Shio, and B. Spellerberg. 1998. Membrane targeting of RecA during genetic transformation. Mol. Microbiol. 27:845-852. [DOI] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Metzgar, D., J. M. Bacher, V. Pezo, J. Reader, V. Doring, P. Schimmel, P. Marliere, and V. de Crecy-Lagard. 2004. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmen, R., P. Buijsman, and K. J. Hellingwerf. 1994. Physiological regulation of competence induction for natural transformation in Acinetobacter calcoaceticus. Arch. Microbiol. 162:344-351. [Google Scholar]
- 34.Palmen, R., B. Vosman, P. Buijsman, C. K. D. Breek, and K. J. Hellingwerf. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol. 139:295-305. [DOI] [PubMed] [Google Scholar]
- 35.Roh, Y., S. V. Liu, G. Li, H. Huang, T. J. Phelps, and J. Zhou. 2002. Isolation and characterization of metal-reducing Thermoanaerobacter strains from deep subsurface environments of the Piceance Basin, Colorado. Appl. Environ. Microbiol. 68:6013-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudel, T., D. Facius, R. Barten, I. Scheuerpflug, E. Nonnenmacher, and T. F. Meyer. 1995. Role of pili and the phase-variable PilC protein in natural competence for transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 92:7986-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanks, R. M. Q., N. C. Caiazza, S. M. Hinsa, C. M. Toutain, and G. A. O'Toole. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl. Environ. Microbiol. 72:5027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao, W., S. DeBlois, and J. Wiegel. 1995. A high-molecular-weight, cell-associated xylanase isolated from exponentially growing Thermoanaerobacterium sp. strain JW/SL-YS485. Appl. Environ. Microbiol. 61:937-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw, A. J., D. A. Hogsett, and L. R. Lynd. 2009. Identification of the [FeFe]-hydrogenase responsible for hydrogen generation in Thermoanaerobacterium saccharolyticum and demonstration of increased ethanol yield via hydrogenase knockout. J. Bacteriol. 191:6457-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw, A. J., K. K. Podkaminer, S. G. Desai, J. S. Bardsley, S. R. Rogers, P. G. Thorne, D. A. Hogsett, and L. R. Lynd. 2008. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc. Natl. Acad. Sci. U. S. A. 105:13769-13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommer, P., T. Georgieva, and B. K. Ahring. 2004. Potential for using thermophilic anaerobic bacteria for bioethanol production from hemicellulose. Biochem. Soc. Trans. 32:283-289. [DOI] [PubMed] [Google Scholar]
- 42.Stewart, G. J., and C. A. Carlson. 1986. The biology of natural transformation. Annu. Rev. Microbiol. 40:211-231. [DOI] [PubMed] [Google Scholar]
- 43.Suihko, M. L., L. Partanen, T. Mattila-Sandholm, and L. Raaska. 2005. Occurrence and molecular characterization of cultivable mesophilic and thermophilic obligate anaerobic bacteria isolated from paper mills. Syst. Appl. Microbiol. 28:555-561. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, M. P., K. L. Eley, S. Martin, M. I. Tuffin, S. G. Burton, and D. A. Cowan. 2009. Thermophilic ethanologenesis: future prospects for second-generation bioethanol production. Trends Biotechnol. 27:398-405. [DOI] [PubMed] [Google Scholar]
- 45.Tyurin, M. V., C. R. Sullivan, and L. R. Lynd. 2005. Role of spontaneous current oscillations during high-efficiency electrotransformation of thermophilic anaerobes. Appl. Environ. Microbiol. 71:8069-8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van de Werken, H. J. G., M. R. A. Verhaart, A. L. VanFossen, K. Willquist, D. L. Lewis, J. D. Nichols, H. P. Goorissen, E. F. Mongodin, K. E. Nelson, E. W. J. van Niel, A. J. M. Stams, D. E. Ward, W. M. de Vos, J. van der Oost, R. M. Kelly, and S. W. M. Kengen. 2008. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl. Environ. Microbiol. 74:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weimer, P. J., L. W. Wagner, S. Knowlton, and T. K. Ng. 1984. Thermophilic anaerobic bacteria which ferment hemicellulose: characterization of organisms and identification of plasmids. Arch. Microbiol. 138:31-36. [DOI] [PubMed] [Google Scholar]
- 48.Wiegel, J., L. G. Ljungdahl, and J. R. Rawson. 1979. Isolation from soil and properties of the extreme thermophile Clostridium thermohydrosulfuricum. J. Bacteriol. 139:800-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiegel, J., C. P. Mothershed, and J. Puls. 1985. Differences in xylan degradation by various noncellulolytic thermophilic anaerobes and Clostridium thermocellum. Appl. Environ. Microbiol. 49:656-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasbin, R. E., D. L. Cheo, and K. W. Bayles. 1992. Inducible DNA repair and differentiation in Bacillus subtilis: interactions between global regulons. Mol. Microbiol. 6:1263-1270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.