Abstract
In contrast to most denitrifiers studied so far, Pseudomonas stutzeri TR2 produces low levels of nitrous oxide (N2O) even under aerobic conditions. We compared the denitrification activity of strain TR2 with those of various denitrifiers in an artificial medium that was derived from piggery wastewater. Strain TR2 exhibited strong denitrification activity and produced little N2O under all conditions tested. Its growth rate under denitrifying conditions was near comparable to that under aerobic conditions, showing a sharp contrast to the lower growth rates of other denitrifiers under denitrifying conditions. Strain TR2 was tolerant to toxic nitrite, even utilizing it as a good denitrification substrate. When both nitrite and N2O were present, strain TR2 reduced N2O in preference to nitrite as the denitrification substrate. This bacterial strain was readily able to adapt to denitrifying conditions by expressing the denitrification genes for cytochrome cd1 nitrite reductase (NiR) (nirS) and nitrous oxide reductase (NoS) (nosZ). Interestingly, nosZ was constitutively expressed even under nondenitrifying, aerobic conditions, consistent with our finding that strain TR2 preferred N2O to nitrite. These properties of strain TR2 concerning denitrification are in sharp contrast to those of well-characterized denitrifiers. These results demonstrate that some bacterial species, such as strain TR2, have adopted a strategy for survival by preferring denitrification to oxygen respiration. The bacterium was also shown to contain the potential to reduce N2O emissions when applied to sewage disposal fields.
Wastewater treatment processes produce one of the major greenhouse effect gases, nitrous oxide (N2O) (7, 25, 30). The global warming potential of N2O relative to that of carbon dioxide (CO2) is 298 for a 100-year time horizon, and its concentration in the atmosphere continues to increase by about 0.26% per year (9). Nitrogen removal in wastewater treatment plants is essentially based on the activities of nitrifying and denitrifying microorganisms, both of which are inhabitants of activated sludge. Nitrifying bacteria aerobically oxidize ammonium to nitrite (NO2−) and nitrate (NO3−), which are then reduced anaerobically by denitrifying bacteria to gaseous nitrogen forms, such as N2O and dinitrogen (N2). It has long been known that N2O can be produced during both nitrification and denitrification processes of wastewater treatment (3, 19, 23), but the cause of N2O emission during the nitrification process was not clear. We recently showed, however, using activated sludge grown under conditions that mimicked a piggery wastewater disposal, that N2O emission during the nitrification process depends on denitrification by ammonia-oxidizing bacteria (Nitrosomonas) (18). On the other hand, it is believed that denitrifying bacteria produce N2O as a by-product when anaerobiosis is insufficient during the denitrification process, because N2O reductase is the enzyme that is most sensitive to oxygen (6). Piggery wastewater, in particular, contains a high concentration of ammonia, and N2O emission tends to take place during the nitrogen removal process (5, 10). Experiments on the removal of ammonia and organic carbon by the aerobic denitrifier Pseudomonas stutzeri SU2 (24) and the heterotrophic nitrifier-aerobic denitrifier Alcaligenes faecalis no. 4 (16, 17) have been reported as examples of bioaugmentation in piggery wastewater treatment. Reduction of N2O emissions from pig manure compost by addition of nitrite-oxidizing bacteria has also been reported (11). However, there have been no reports of methods for reducing N2O emissions by bioaugmentation using aerobic denitrifying bacteria.
Takaya et al. isolated the aerobic denitrifying bacterium Pseudomonas stutzeri TR2 (26). The denitrification activity of strain TR2 was monitored in batch and continuous cultures, using denitrification and artificial wastewater media, and the strain was found to keep a distinct activity (producing N2 from NO3−) and to produce a very low level of N2O at a dissolved oxygen (O2) concentration of 1.25 mg liter−1. Therefore, strain TR2 should be useful in the future for reducing N2O emissions from wastewater treatment plants by bioaugmentation. To investigate the feasibility of using strain TR2 for future application to wastewater treatment processes, we examined its denitrification activity, N2O production, growth rate, and expression of denitrifying genes in batch cultures, using a medium that mimics the composition found in nitrogen removal wastewater plants. Comparison of the properties of strain TR2 with those of well-characterized denitrifying bacteria revealed characteristics of the strain that favor denitrification, although it can also respire oxygen.
MATERIALS AND METHODS
Strains.
P. stutzeri TR2 and Ralstonia pickettii K50 were isolated in a previous study (26). Paracoccus denitrificans JCM 20620 and Pseudomonas aeruginosa JCM 2412 were obtained from the Japan Collection of Microorganisms. Pseudomonas stutzeri ZoBell ATCC 14405 was obtained from the American Type Culture Collection.
Media.
Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, and 0.5% NaCl) was used for seed culture and preculture. In order to mimic the actual conditions of a sewage disposal field, the basal medium used was derived from a field where a swine herd is kept (Aichi, Japan). Piggery wastewater, comprising urine, washing water, and feces, was first digested by anaerobic methane fermentation, and the resulting supernatant after filtration was used as digestion liquid after methane fermentation of piggery wastewater (DLMF) that contained about 4,000 mg liter−1 NH4+-N. DLMF was further treated in a membrane sequencing batch bioreactor (MSBR) to remove nitrogen after supplementation with acetic acid, to give MSBR-treated water (TW). TW still contained 60 mg liter−1 NH4+-N and 160 mg liter−1 total N (pH 8.3). Mixed liquor medium (MLM) was prepared by mixing TW and DLMF at a ratio of 59:1, followed by supplementation with a trace element solution (1 ml liter−1). MLM was used as the basal medium throughout this work. The trace element stock solution consisted of 20% MgSO4·7H2O, 1% FeSO4·7H2O, 1% FeCl3·7H2O, 0.2% ZnSO4·7H2O, 0.4% CuSO4·5H2O, 0.05% NaMoO4·2H2O, 0.01% MnCl2·4H2O, 0.01% H3BO4, 0.03% Na2SeO3, and 1% citric acid. Before cultivation, the media were sterilized by filtration through a membrane filter (pore size, 0.2 μm).
Seed culture and preculture.
The denitrifying bacteria were inoculated from an LB agar plate to 5 ml of LB in a test tube, and the tube was shaken at 150 rpm at 30°C for 24 h (seed culture). Two milliliters of seed culture was transferred to 200 ml of LB medium in a 500-ml baffled Erlenmeyer flask, and the flask was shaken aerobically at 150 rpm at 30°C for 20 h (preculture). The preculture was collected by centrifugation, washed twice with MLM, suspended with an appropriate volume of MLM to adjust the optical density (OD) (resuspended preculture), and then transferred to the main culture.
Culture.
The growth of various bacterial strains under aerobic and denitrifying conditions was compared by culture in L-shaped test tubes (Monod tubes) (see Fig. 1). Nine microliters of the resuspended preculture, whose OD at 660 nm (OD660) was adjusted to 5.0, was inoculated into 9 ml MLM (pH 6.8) in an L-shaped test tube (27-ml volume). Thus, the initial OD660 of each culture was 0.005. Sodium acetate (10 mM) and the indicated amount of 15N-labeled NO3− or NO2− were added to the MLM. For aerobic culture, the test tube was plugged with an air-permeable foam plug and shaken at 120 rpm on a reciprocal shaker at 30°C. For denitrifying culture, the test tube was sealed with a double butyl rubber stopper after purging the headspace with argon gas and replacing its portion (1%) with O2 gas by use of a gas-tight syringe (Hamilton, Reno, NV).
FIG. 1.
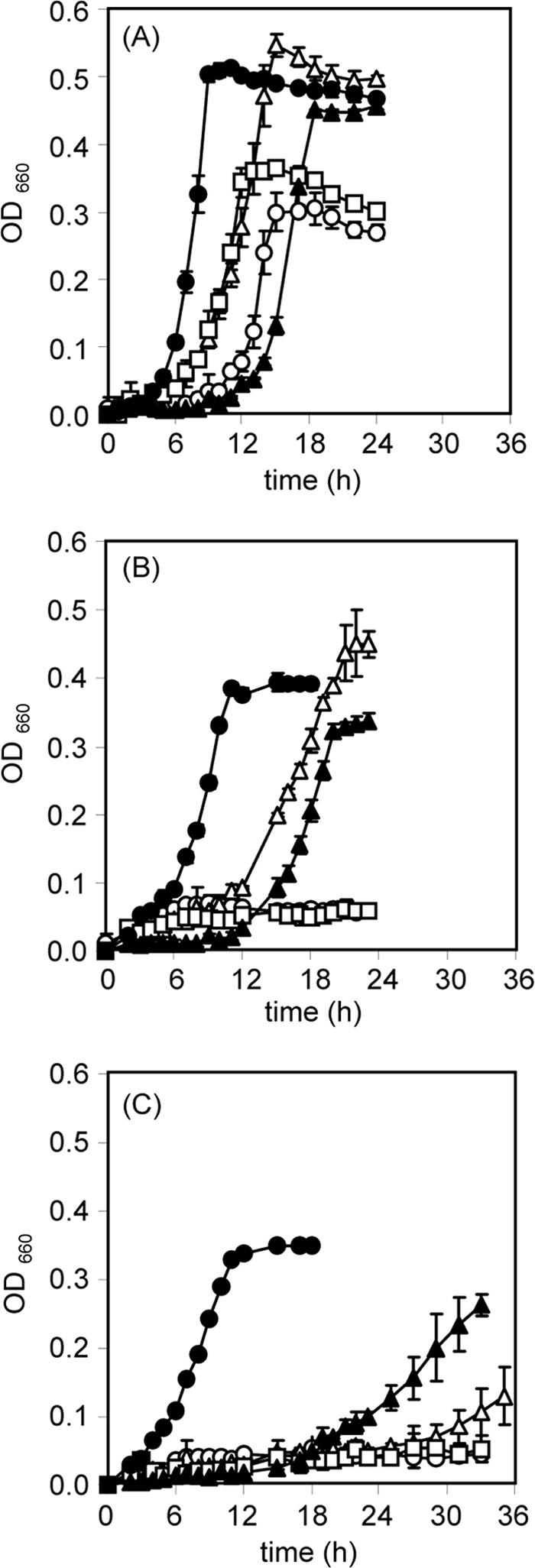
Time-dependent growth of TR2 and other denitrifiers under aerobic and denitrifying (initially 1% O2) conditions. Denitrifiers were cultured aerobically (A) or under denitrifying conditions in the presence of 20 mM Na15NO3 (B) or 20 mM Na15NO2 (C). Symbols (amount of inoculated cells [dry weight] per tube): open circles, P. aeruginosa (26 μg); open triangles, P. denitrificans (22 μg); open squares, R. pickettii K50 (35 μg); closed circles, strain TR2 (30 μg); closed triangles, P. stutzeri ZoBell (40 μg). Determinations were performed three times, and standard deviations (error bars) are indicated for each plot.
For characterization of denitrification by strain TR2 and P. denitrificans under various conditions, cultures in test tubes were employed (for the experiments in Fig. 2, 3, and 5). MLM (8.3 ml) supplemented with the indicated amounts (final concentrations) of sodium acetate and 15N-labeled NO3− or NO2− (pH 8.3) was placed in a 50-ml test tube, which was sealed with a double butyl rubber stopper after purging of its headspace with argon gas, with or without subsequent replacement of its proportion (3%) with O2 gas. To examine N2O utilization (see Fig. 3), 2.1 ml (2.5 mmol liter−1 of headspace gas; total, 84 μmol) N2O gas was also injected. Precultures of strain TR2 and P. denitrificans were harvested at late exponential phase (OD660 = 2.5 for strain TR2 and 5.0 for P. denitrificans), resuspended in MLM, injected using a gas-tight syringe to give the same bacterial concentrations as those in the cultures (OD660 = 2.5 and 5.0, respectively) and a final volume of 16.6 ml, and then cultured. For mixed culture with activated sludge grown in the MSBR (see Fig. 5), a resuspended preculture of strain TR2 was mixed with the sludge and injected in the same manner as that described above, to give the following final concentrations: strain TR2, 1 g liter−1 (dry weight); and sludge, indicated in the figure legends. The test tube for each culture (see Fig. 2, 3, and 5) was shaken at 120 rpm at 30°C, and the gas phase was monitored periodically by gas chromatography-mass spectrometry (GC-MS).
FIG. 2.
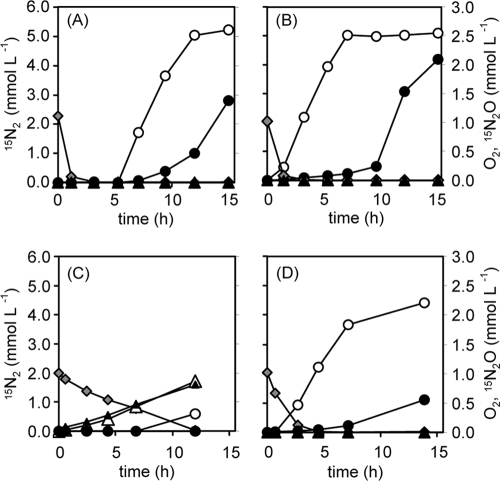
Denitrification of nitrate and nitrite by P. denitrificans and strain TR2. P. denitrificans (A and C) (34 mg cells [dry weight]) and strain TR2 (B and D) (16 mg cells) were incubated with 20 mM Na15NO3 (A and B) or 20 mM Na15NO2 (C and D) in the presence of 60 mM sodium acetate under initially microaerobic (3% O2) (open symbols) or anaerobic (argon-purged; closed symbols) conditions. Symbols: circles, 15N2; triangles, 15N2O; gray diamonds (for only initially microaerobic conditions), O2.
FIG. 3.
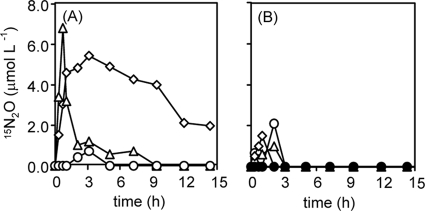
Denitrification in the presence of both 15N-labeled nitrite and nonlabeled N2O by P. denitrificans (A) and strain TR2 (B) under initially microaerobic (3% O2) conditions. Denitrifiers were incubated with 10 mM Na15NO2 and 2.5 mmol liter−1 14N2O. Symbols: closed triangles, 14N2O; open triangles, 15N2O; open circles, 15N2; gray circles, O2. Other conditions are the same as in Fig. 2.
FIG. 5.
Effects of strain TR2 on N2O production from culture of activated sludge. The indicated amount of sludge, in the absence (A) or presence (B) of strain TR2 (1 g liter−1 [dry weight]), was cultured in MLM (16.6 ml) containing 10 mM sodium acetate and 20 mM Na15NO2 in test tubes (50 ml) under denitrifying (initially 3% O2) conditions. Open symbols indicate culture of different amounts of activated sludge (dry weight). Circles, 12 g liter−1; triangles, 1.2 g liter−1; diamonds, 0.12 g liter−1. Closed circles in panel B indicate a control cultivation of strain TR2 without activated sludge.
When periodic extraction of culture broth (sampling) was required to monitor bacterial growth and gene expression, culture was performed on a large scale in a 500-ml baffled Erlenmeyer flask containing 200 ml MLM supplemented with 50 mM sodium acetate (pH 8.3) (see Fig. 4). The preculture was grown in a medium in which powder containing the ingredients for LB (tryptone, yeast extract, and NaCl) was dissolved in TW, pH 7.0. The preculture was inoculated into the flask so that the initial OD660 was adjusted to 0.25, giving dry weights of cells of 19 mg (P. denitrificans) and 28 mg (strain TR2). The flask was plugged with an air-permeable foam plug. Other details are given in the relevant figure legend.
FIG. 4.
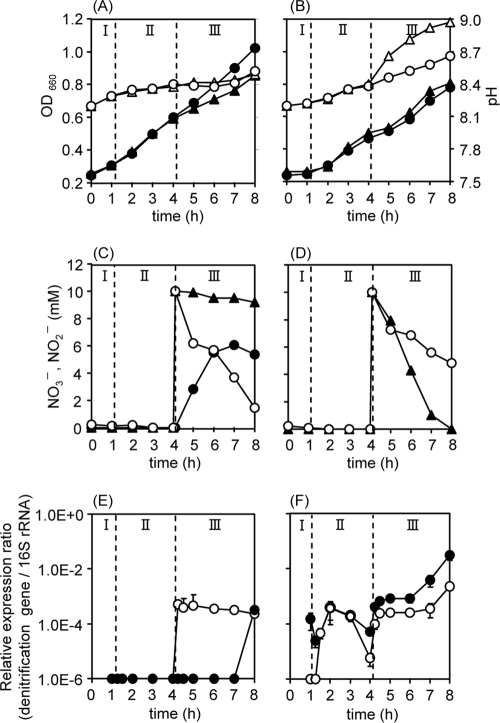
Growth of strain TR2 and of P. denitrificans under various conditions. P. denitrificans (A, C, and E) (123 mg cells [dry weight]) and strain TR2 (B, D, and F) (166 mg cells) were cultured first aerobically, with shaking at 150 rpm (phase I; DO = 5.0 mg liter−1), and then microaerobically, with a rotation speed of 75 rpm (phases II and III; DO = 0 mg liter−1). At 4 h, 10 mM NaNO3 or NaNO2 was added (phase III). Throughout the culture, time-dependent changes in the indicated items were monitored. (A and B) Closed circles, OD660 (cell growth) of the culture to which NO3− was added at phase III (upon NO3− addition); closed triangles, OD660 upon NO2− addition; open circles, pH upon NO3− addition; open triangles, pH upon NO2− addition. (C and D) Open circles, NO3− upon NO3− addition; closed circles, NO2− upon NO3− addition (not indicated in panel D, because all plots were at zero level); closed triangles, NO2− upon NO2− addition. NO3− was not detected upon nitrite addition (data not shown). (E and F) Gene expression upon NO3− addition. Closed circles, nosZ; open circles, nirS. Similar results were obtained with NO2− addition (data not shown).
The biological oxygen demand (BOD) of each culture was 540 (Fig. 1 and 5), 3,140 (Fig. 2 and 3), or 2,620 mg liter−1 (Fig. 4).
Three replicate experiments were performed for all results, but error bars are indicated only for Fig. 1, because a small difference in time lag made a large difference in gas evolution (see Fig. S1 in the supplemental material). Thus, a typical result was employed for the presentation.
Determinations.
Gas was monitored periodically by GC-MS with a Shimadzu GCMS-GP5050 instrument (Shimadzu, Kyoto, Japan) equipped with a CP-PoraPLOT-Q column (Varian, Palo Alto, CA); total flow was 118 ml min−1, the voltage was 0.9 V, and the temperature of the oven, injector, and detector was 50°C. Concentrations of NO2−, NO3−, and NH4+ ions were measured by ion chromatography with a Metrohm 861 Advanced Compact ion chromatograph (Metrohm, Zofingen, Switzerland). Bacterial growth was monitored by measuring the OD660 with a miniphoto 518R photometer (Taitec, Saitama, Japan) (see Fig. 1) and a UV-visible spectrophotometer (Jasco, Tokyo, Japan) (see Fig. 4). The dissolved oxygen (DO) concentration was measured by a DO meter (Able, Tokyo, Japan).
Quantitative real-time PCR analysis.
A primer set comprising EUB 338f (5′-ATC CCT ACG GGA GGC AGC-3′) (2) and EUB 907r (5′-CCG TCA ATT CMT TTR AGT T-3′) (29) was used to amplify the bacterial 16S rRNA gene. A primer set comprising nirS cd3aF and nirS R3cd and a set comprising nosZ-F and nosZ 1622R were used to amplify the cytochrome cd1 nitrite reductase (nirS) and nitrous oxide reductase (nosZ) genes, respectively (27). For real-time PCR analysis of denitrification genes, a 0.5-ml cell culture suspension was transferred to a 1.5-ml centrifuge tube containing 10 μl of 10-mg ml−1 rifampin. After centrifugation, the pellet was quick-frozen in liquid nitrogen and stored at −80°C before extraction of RNA. Total RNA was extracted using a Trizol reagent kit (Invitrogen, Carlsbad, CA). The sample was treated with DNase I (Takara, Ohtsu, Japan) and RNase inhibitor (Toyobo, Osaka, Japan), followed by phenol-chloroform extraction and ethanol precipitation. Reverse transcription (cDNA synthesis) was carried out using a ReverTra Ace-α kit (Toyobo) and random primers. DNA samples to generate the standard curve for real-time PCR analysis were prepared as follows. Genomic DNA was extracted from cells of denitrifying bacteria by phenol-chloroform treatment and amplified using KAPA2G Fast DNA polymerase (Kapa Biosystems, Woburn, MA). The PCR product was purified by extracting bands obtained by agarose gel electrophoresis, and a dilution series (50 μl of each dilution) was constructed (102 to 108 copies per μl) using an Easy Dilution kit (Takara). Real-time PCR was performed using Sybr Premix Ex Taq (Takara) and a LightCycler machine (Roche Diagnostics, Tokyo, Japan) under the following conditions: initial denaturation at 95°C for 10 s, followed by 30 amplification cycles, each of which consisted of denaturation for 5 s at 95°C, annealing for 20 s at 50°C (16S rRNA gene), 58°C (nirS), or 60°C (nosZ), and extension for 15 s at 72°C.
RESULTS
Comparative analyses of growth of strain TR2 and other denitrifiers under aerobic and denitrifying conditions.
In all experiments of this study, we performed batch cultures of denitrifying bacteria by using MLM as a medium that mimics the sewage from a piggery. For the growth experiments (Fig. 1), MLM was supplemented with a low concentration of acetate (10 mM) to give a low C/N ratio of 0.85. This mimics the actual sewage disposal field, in which carbon sources are always limited. Five denitrifiers were compared with respect to growth under aerobic and initially microaerobic (denitrifying) conditions and with various electron acceptors (O2, NO2−, and NO3−) (Fig. 1). Strain TR2 grew the most rapidly under all conditions examined, with the shortest lag periods. The other denitrifiers showed little growth or began to grow after a long lag period under denitrifying conditions. Strain K50 and P. aeruginosa did not grow at all during the incubation time of 36 h under both denitrifying conditions (in the presence of NO2− or NO3−) (Fig. 1B and C). P. denitrificans and P. stutzeri ZoBell grew well in the presence of NO3−, but only after long lag periods (Fig. 1B). They showed poorer growth with a longer lag period in the NO2−-containing medium (Fig. 1C). In contrast, strain TR2 grew well, without a lag period, even under denitrifying conditions, with the same rate (0.32 h−1) for growth in the NO3−- and NO2−-containing media. The growth rate was near comparable to those for aerobic growth of the same strain (0.56 h−1) and other denitrifiers (P. stutzeri ZoBell, 0.41 h−1; P. aeruginosa, 0.39 h−1; strain K50, 0.36 h−1; and P. denitrificans, 0.32 h−1). Among the denitrifiers other than strain TR2, P. denitrificans exhibited the best adaptation to MLM. Therefore, we compared strain TR2 with P. denitrificans in the following experiments.
Strain TR2 produces a low level of N2O from nitrite.
Time-dependent denitrification of 15N-labeled nitrate or nitrite by strain TR2 and P. denitrificans was examined after aerobically grown cells were moved to these conditions (Fig. 2). The initial O2 concentration was set at 3% for the microaerobic culture because FNR-regulated genes show half-maximum expression at O2 concentrations of around 0.5 to 2.4% (31). In order to investigate denitrification ability without carbon limitation, an adequate carbon source (60 mM sodium acetate) was added. Under microaerobic conditions, denitrification of nitrate by P. denitrificans showed a lag period of 5 h (Fig. 2A). On the other hand, denitrification by strain TR2 started at 1 h, preceding the complete consumption of O2, and completed within 7 h (100% turnover of 15N of nitrate into N2) (Fig. 2B). Both denitrifiers did not produce N2O from nitrate. When nitrite was used, O2 consumption by P. denitrificans was significantly decelerated, and slight but continuous N2O production was observed (Fig. 2C). On the other hand, O2 consumption and 15N2 production by strain TR2 did not decelerate substantially compared with the rate of denitrification of nitrate, and N2O production was not detected (Fig. 2D). Under anaerobic conditions in which the headspace of the test tube was purged with argon gas, the start of denitrification by both strain TR2 and P. denitrificans was significantly delayed (closed symbols in Fig. 2). P. aeruginosa, P. stutzeri ZoBell, and R. pickettii K50 also exhibited a delay in denitrification under anaerobic conditions (data not shown). The results show that denitrification from nitrite under anaerobic conditions is the most severe to occur. Even under such conditions, strain TR2 did not produce N2O, in contrast to the continuous emission of N2O by P. denitrificans (Fig. 2C and D).
Denitrification in the presence of two electron accepters, nitrite and N2O.
Next, we examined denitrification by P. denitrificans and strain TR2 in the presence of both nitrite and N2O, mimicking the composition of sewage disposal fields producing N2O (Fig. 3). 15N-labeled nitrite and nonlabeled N2O were initially added to the liquid and gas phases, respectively, and the initial O2 concentration was set at 3%. With regard to the culture of P. denitrificans, 14N2O in the gas phase decreased during the first 1 h but remained constant for several hours subsequently (Fig. 3A). The first decrease in N2O should depend on dissolution of N2O into the liquid phase. All oxygen was consumed within 3 h; however, 14N2O did not decrease during this period. For incubation times of 5 to 6 h, a decrease in 14N2O, a temporary appearance of 15N2O, and production of 15N2 started, showing that these phenomena relevant to denitrification were started almost simultaneously.
On the other hand, strain TR2 started to consume 14N2O before the complete consumption of O2, and it almost exhausted it within 3 h (Fig. 3B). Production of 15N2O was not observed. Production of 15N2 started at about 2.5 h and finished within 5 h. Under these conditions, the rates of denitrification of both N2O (14N2O decrease) and nitrite (increase in 15N2) by strain TR2 were significantly higher than those by P. denitrificans. The rate of 15N2 production reached its maximum at around 3.5 h, clearly lagging behind the stage at which 14N2O consumption reached its maximum (around 2.5 h). This indicates that strain TR2 first used N2O rather than nitrite as an electron acceptor, demonstrating that strain TR2 prefers N2O to nitrite under aerobic conditions.
Effects of nitrate and nitrite on aerobic cultures of P. denitrificans and strain TR2.
Both bacterial strains were precultured in TW medium (see Materials and Methods) to accustom them to TW and then were cultured under changing conditions in order to investigate their response to nitrate or nitrite (Fig. 4). For the first 1 h, they were incubated aerobically without the denitrification substrate (phase I; DO = 5 mg liter−1), and then aeration was decreased by lowering the rotation speed (phase II; DO = 0 mg liter−1). Nitrate or nitrite was added at 4 h (phase III; DO = 0 mg liter−1). Although the DO concentration was zero in phases II and III, the aerating conditions should still have been microaerobic, since the baffled flask continued to be aerated (see Materials and Methods).
The growth (OD660) of P. denitrificans was faster in this case than that of strain TR2 under aerobic conditions (phases I and II) (Fig. 4A and B). Addition of nitrate or nitrite affected the growth of the strains to various extents (phase III). The growth rate of P. denitrificans was not affected much by the addition of nitrate, whereas it was lowered by nitrite. The growth of strain TR2 was lowered at the initial stage of phase III, but a high growth rate was attained in the latter stage (Fig. 4B). The increase in pH was slow for the culture of P. denitrificans, whereas that for strain TR2 was more rapid, in particular after addition of nitrite in phase III. This suggests that the cells of strain TR2 were denitrifying at this stage. Nitrate added at phase III was immediately consumed by P. denitrificans, and most of the consumed nitrate was excreted as nitrite into the medium. However, the excreted or newly added nitrite was not utilized any more during the incubation period (Fig. 4C), showing that the bacterium does not denitrify nitrite under microaerobic conditions or that it takes more time to induce the denitrifying system in this bacterium. In contrast, strain TR2 immediately consumed nitrite (Fig. 4D), with concomitant rapid growth and increase in pH (Fig. 4B), suggesting that the strain was denitrifying nitrite in phase III. Strain TR2 also utilized nitrate (Fig. 4D, open circles) without secreting nitrite. The rate of nitrate consumption was slower than that by P. denitrificans or the rate of nitrite consumption by itself.
The profile of gene expression during the cultures (at phases II and III) was examined (Fig. 4E and F). Expression of the nirS gene in P. denitrificans was initiated soon after the addition of nitrate, whereas that of the nosZ gene in the bacterium was observed after a long time lag (4 h) after the addition of nitrate (Fig. 4E, phase III). Similar results were obtained upon addition of nitrite. In spite of the immediate response of nirS gene expression to nitrate (or nitrite), nitrite added to the medium or formed from nitrate decreased only slowly (Fig. 4C). This suggests that another level of regulation was working for utilization of nitrite. In contrast, expression of nirS was attained in strain TR2 immediately after the transition to low-aeration conditions (phase II) (Fig. 4F), demonstrating that the gene's expression responded to less aeration but not to nitrite. Interestingly, the nosZ gene was expressed in the strain from the beginning of phase II, indicating that the gene was already expressed during aerobic culture in the absence of nitrate or nitrite (phase I; DO = 5 mg liter−1). The constitutive expression of nosZ is consistent with the immediate consumption of N2O under microaerobic conditions (Fig. 2B).
Ability of strain TR2 to reduce N2O production from activated sludge.
The results above suggest the potential ability of strain TR2 to prevent N2O emission when the bacterium is applied to sewage disposal fields. Here we employed a model system that mimics sewage disposal fields by employing activated sludge grown in an MSBR. It is known that N2O emission is active when carbon sources are limited and the denitrification substrate is nitrite (1, 15). Under such extreme conditions, the system evolved N2O (Fig. 5 A), especially when the sludge concentration was low. This means that a heavy load on activated sludge causes N2O emission. Addition of strain TR2 was markedly effective in reducing N2O emissions under such conditions (Fig. 5B). The results demonstrate that strain TR2 has the potential ability to augment the denitrification system in wastewater treatment via the nitrite pathway (20).
DISCUSSION
The present results characterize strain TR2 as an expert of denitrification. The strain adapted rapidly to microaerobic denitrifying conditions (Fig. 1 and 2), in sharp contrast to other denitrifiers, which could not grow or showed a long time lag for growth under the same conditions. The rapid response of strain TR2 to denitrification is consistent with the constitutive or rapid expression of denitrification genes (nosZ and nirS) (Fig. 4). The nosZ gene was constitutively expressed even under high-aeration conditions (DO = 5 mg liter−1) in the absence of nitrate or nitrite, and the nirS gene responded only to low DO, not to nitrate or nitrite. The rate of TR2 growth by denitrification was even comparable or close to the rate of aerobic growth of other denitrifiers (Fig. 1). The aerobic growth of strain TR2 was also the fastest among the denitrifiers examined (Fig. 1A), indicating that the MLM derived from piggery wastewater fits its growth well. The present results also show that rapid induction of denitrification requires an adequate amount of O2 (Fig. 2). Among the enzymes involved in denitrification, cytochrome-type NiR and nitric oxide reductase (NoR) require heme for their activity (8). Under oxygen-limiting conditions, heme biosynthesis, which requires oxygen (13), may have been delayed. P. denitrificans could denitrify nitrite when its concentration was lower (10 mM) (Fig. 3A; cf. Fig. 1C and 2C [20 mM]), suggesting that a toxic effect of nitrite at a higher concentration lowered the growth of P. denitrificans and other denitrifiers, except for strain TR2.
It is known that the proton motive forces yielded by the four reducing steps in denitrification, catalyzed by nitrate reductase (NaR), NiR, NoR, and NoS, are the same (2 H+ ions translocated per 2 electrons) (4). This means that these reducing steps theoretically contribute equally to ATP synthesis by respiration. It is therefore intriguing that denitrifiers give preference to a specific step over the others, even if they contain all of the reducing steps, and that the preferred step varies depending on the respective denitrifier. Strain TR2 preferred the last step, by NoS, to the step by NiR (Fig. 3). The priority of N2O over other denitrification substrates as the electron acceptor is supported by the constitutive expression of the nosZ gene. On the other hand, P. denitrificans showed a preference for the first step, by NaR, over the second step, by NiR (Fig. 1 and 4). The immediate response of P. denitrificans to nitrate under microaerobic conditions (Fig. 4C) suggests that NaR was already expressed prior to the addition of nitrate. It therefore appears that P. denitrificans first reduces nitrate to nitrite under denitrifying conditions and then, after reducing most of the nitrate, initiates the subsequent reduction from nitrite to N2. This is why the denitrification of nitrate by this bacterium showed a time lag for initiation (Fig. 2A). In contrast, strain TR2 quickly responded not only to N2O (Fig. 3B) and nitrite (Fig. 4B, D, and F) but also to nitrate (Fig. 4D), showing that the full process of denitrification is ready to respond quickly in this strain. It is thus possible to classify bacterial denitrifiers by their preference for a specific step, e.g., nitrate denitrifiers, nitrite denitrifiers, N2O denitrifiers, full denitrifiers, and so on (21). Adoption of a particular reducing step(s) as a preference might have diversified bacterial denitrifiers, which could be a strategy to gain a niche for survival under various environmental conditions.
In a previous study (26), we demonstrated that strain TR2 exhibits aerobic denitrification that produces a low level of N2O in artificial media. In the present study, we demonstrated suitable and similar characteristics of strain TR2 for nitrogen removal in MLM, which mimics the composition of the sewage disposal field. The characteristics of strain TR2 should be advantageous when applied to sequencing batch nitrogen removal systems in which nitrification and denitrification are continuously repeated. The denitrifying system of strain TR2 can quickly respond to various conditions. Not only the constitutive gene expression of nosZ but also the high oxygen tolerance of the gene product, N2O reductase (T. Toya et al., unpublished data), may be the major factors that give strain TR2 the ability for aerobic denitrification in which a low level of N2O is emitted even under aerobic conditions.
Recently, various novel biological nitrogen removal processes, such as anaerobic ammonium oxidation (anammox), have been developed (28). Partial nitrification by ammonia-oxidizing bacteria to accumulate nitrite is a key procedure for implementing those novel processes (22). Among the new microbial nitrogen removal processes, nitrification and denitrification via the nitrite pathway have been investigated with a view to introducing the process into wastewater plants (12, 20). However, the process produces a significant amount of N2O, probably because of the toxicity of nitrite (14). The results of this study showing that strain TR2 can rapidly denitrify nitrite even under aerobic conditions (Fig. 5B) suggest the possibility for the strain to be applied effectively to novel wastewater treatment processes for preventing N2O emission.
Supplementary Material
Acknowledgments
This study was supported by the Research and Development Program for New Bio-Industry Initiatives and by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (grant 20248009 to H.S.).
Footnotes
Published ahead of print on 21 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alinsafi, A., N. Adouani, F. Beline, T. Lendormi, L. Limousy, and O. Sire. 2008. Nitrite effect on nitrous oxide emission from denitrifying activated sludge. Process Biochem. 43:683-689. [Google Scholar]
- 2.Bachoon, D. S., R. E. Hodson, and R. Araujo. 2001. Microbial community assessment in oil-impacted salt marsh sediment microcosms by traditional and nucleic acid-based indices. J. Microbiol. Methods 46:37-49. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, B., M. Snozzi, A. J. Zehnder, and J. R. Van Der Meer. 1996. Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J. Bacteriol. 178:4367-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berks, B. C., S. J. Ferguson, J. W. Moir, and D. J. Richardson. 1995. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232:97-173. [DOI] [PubMed] [Google Scholar]
- 5.Bernet, N., N. Delgenes, and R. Moletta. 1996. Denitrification by anaerobic sludge in piggery wastewater. Environ. Technol. 17:293-300. [Google Scholar]
- 6.Bonin, P., M. Gilewicz, and J. C. Bertrand. 1989. Effects of oxygen on each step of denitrification on Pseudomonas nautica. Can. J. Microbiol. 35:1061-1064. [Google Scholar]
- 7.Czepiel, P., P. Crill, and R. Harriss. 1995. Nitrous oxide emissions from municipal wastewater treatment. Environ. Sci. Technol. 29:2352-2356. [DOI] [PubMed] [Google Scholar]
- 8.Einsle, O., and P. M. Kroneck. 2004. Structural basis of denitrification. Biol. Chem. 385:875-883. [DOI] [PubMed] [Google Scholar]
- 9.Forster, P., V. Ramaswamy, P. Artaxo, T. Berntsen, R. Betts, D. W. Fahey, J. Haywood, J. Lean, D. C. Lowe, G. Myhre, J. Nganga, R. Prinn, G. Raga, M. Schulz, and R. van Dorland. 2007. Changes in atmospheric constituents and in radiative forcing, p. 129-234. In S. Solomon, D. Qin, M. Manning, Z. Chen, M. Marquis, K. B. Averyt, M. Tingor, and H. L. Miller (ed.), Climate change 2007: the physical science basis. Contribution of working group I to the fourth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom.
- 10.Fukumoto, Y., and T. Osada. 2003. Patterns and quantities of NH3, N2O and CH4 emissions during swine manure composting without forced aeration—effect of compost pile scale. Bioresour. Technol. 89:109-114. [DOI] [PubMed] [Google Scholar]
- 11.Fukumoto, Y., K. Suzuki, T. Osada, K. Kuroda, D. Hanajima, T. Yasuda, and K. Haga. 2006. Reduction of nitrous oxide emission from pig manure composting by addition of nitrite-oxidizing bacteria. Environ. Sci. Technol. 40:6787-6791. [DOI] [PubMed] [Google Scholar]
- 12.Guo, J., Y. Peng, S. Wang, Y. Zheng, H. Huang, and Z. Wang. 2009. Long-term effect of dissolved oxygen on partial nitrification performance and microbial community structure. Bioresour. Technol. 100:2796-2802. [DOI] [PubMed] [Google Scholar]
- 13.Heinemann, I. U., M. Jahn, and D. Jahn. 2008. The biochemistry of heme biosynthesis. Arch. Biochem. Biophys. 474:238-251. [DOI] [PubMed] [Google Scholar]
- 14.Hwang, S., K. Jang, H. Jang, J. Song, and W. Bae. 2006. Factors affecting nitrous oxide production: a comparison of biological nitrogen removal processes with partial and complete nitrification. Biodegradation 17:19-29. [DOI] [PubMed] [Google Scholar]
- 15.Itokawa, H., K. Hanaki, and T. Matsuo. 2001. Nitrous oxide production in high-loading biological nitrogen removal process under low COD/N ratio condition. Water Res. 35:657-664. [DOI] [PubMed] [Google Scholar]
- 16.Joo, H. S., M. Hirai, and M. Shoda. 2005. Characteristics of ammonium removal by heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis no. 4. J. Biosci. Bioeng. 100:184-191. [DOI] [PubMed] [Google Scholar]
- 17.Joo, H. S., M. Hirai, and M. Shoda. 2006. Piggery wastewater treatment using Alcaligenes faecalis strain no. 4 with heterotrophic nitrification and aerobic denitrification. Water Res. 40:3029-3036. [DOI] [PubMed] [Google Scholar]
- 18.Kim, S. W., M. Miyahara, S. Fushinobu, T. Wakagi, and H. Shoun. 2010. Nitrous oxide emission from nitrifying activated sludge dependent on denitrification by ammonia-oxidizing bacteria. Bioresour. Technol. 101:3958-3963. [DOI] [PubMed] [Google Scholar]
- 19.Otte, S., N. G. Grobben, L. A. Robertson, M. S. Jetten, and J. G. Kuenen. 1996. Nitrous oxide production by Alcaligenes faecalis under transient and dynamic aerobic and anaerobic conditions. Appl. Environ. Microbiol. 62:2421-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng, Y., and G. Zhu. 2006. Biological nitrogen removal with nitrification and denitrification via nitrite pathway. Appl. Microbiol. Biotechnol. 73:15-26. [DOI] [PubMed] [Google Scholar]
- 21.Philippot, L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 22.Philips, S., H. J. Laanbrock, and W. Verstraete. 2002. Origin, causes and effects of increased nitrite concentrations in aquatic environments. Rev. Environ. Sci. Biotechnol. 1:115-141. [Google Scholar]
- 23.Robertson, L. A., E. W. van Niel, R. A. Torremans, and J. G. Kuenen. 1988. Simultaneous nitrification and denitrification in aerobic chemostat cultures of Thiosphaera pantotropha. Appl. Environ. Microbiol. 54:2812-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su, J. J., B. Y. Liu, and Y. C. Chang. 2001. Identifying an interfering factor on chemical oxygen demand (COD) determination in piggery wastewater and eliminating the factor by an indigenous Pseudomonas stutzeri strain. Lett. Appl. Microbiol. 33:440-444. [DOI] [PubMed] [Google Scholar]
- 25.Sümer, E., A. Weiske, G. Benckiser, and J. C. G. Ottow. 1995. Influence of environmental conditions on the amount of N2O released from activated sludge in a domestic waste water treatment plant. Experientia 51:419-422. [Google Scholar]
- 26.Takaya, N., M. A. Catalan-Sakairi, Y. Sakaguchi, I. Kato, Z. Zhou, and H. Shoun. 2003. Aerobic denitrifying bacteria that produce low levels of nitrous oxide. Appl. Environ. Microbiol. 69:3152-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Throbäck, I. N., K. Enwall, Å. Jarvis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-407. [DOI] [PubMed] [Google Scholar]
- 28.Verstraete, W., and S. Philips. 1998. Nitrification-denitrification processes and technologies in new contexts. Environ. Pollut. 102:717-726. [Google Scholar]
- 29.Yamada, T., K. Miyauchi, H. Ueda, Y. Ueda, H. Sugawara, Y. Nakai, and G. Endo. 2007. Composting cattle dung wastes by using a hyperthermophilic pre-treatment process: characterization by physicochemical and molecular biological analysis. J. Biosci. Bioeng. 104:408-415. [DOI] [PubMed] [Google Scholar]
- 30.Yoshinari, T., and M. Wahlen. 1985. Oxygen isotope ratios in N2O from nitrification at a wastewater treatment facility. Nature 317:349-350. [Google Scholar]
- 31.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.