Abstract
Alexandrium catenella is widespread in western North America and produces a suite of potent neurotoxins that cause paralytic shellfish poisoning (PSP) in humans and have deleterious impacts on public health and economic resources. There are seasonal PSP-related closures of recreational and commercial shellfisheries in the Puget Sound, but the factors that influence cell distribution, abundance, and relationship to paralytic shellfish toxins (PSTs) in this system are poorly described. Here, a quantitative PCR assay was used to detect A. catenella cells in parallel with state shellfish toxicity testing during the 2006 bloom season at 41 sites from April through October. Over 500,000 A. catenella cells liter−1 were detected at several stations, with two main pulses of cells driving cell distribution, one in June and the other in August. PSTs over the closure limit of 80 μg of PST 100 per g of shellfish tissue were detected at 26 of the 41 sites. Comparison of cell numbers and PST data shows that shellfish toxicity is preceded by an increase in A. catenella cells in 71% of cases. However, cells were also observed in the absence of PSTs in shellfish, highlighting the complex relationship between A. catenella and the resulting shellfish toxicity. These data provide important information on the dynamics of A. catenella cells in the Puget Sound and are a first step toward assessing the utility of plankton monitoring to augment shellfish toxicity testing in this system.
Various species of the dinoflagellate genus Alexandrium, including members of the species complex comprising Alexandrium catenella, Alexandrium fundyense, and Alexandrium tamarense, produce saxitoxins and a number of related derivatives (1). Shellfish that ingest toxic Alexandrium cells accumulate these potent neurotoxins, which can then lead to paralytic shellfish poisoning (PSP) in human consumers of shellfish. As such, paralytic shellfish toxins (PSTs) pose a serious threat to both public health and economically important fisheries (16). Within the Alexandrium genus, A. catenella is widespread in the northwestern part of North America, including the Puget Sound, and is responsible for seasonal harmful algal blooms (HABs) in this region (17). In the Puget Sound, recreational shellfish harvesters collect nearly 2 million pounds of clams and oysters annually, and Washington is also a leading producer of farmed bivalve shellfish in the United States, generating an estimated $77 million in sales a year and supporting thousands of jobs (13).
PSTs are not a new problem in the Pacific Northwest; events have been documented as far back as the late 18th century (17). Currently, the Sentinel Monitoring Program of the Washington State Department of Health (WADOH) is in place to provide systematic early warning of harmful levels of PSTs, with caged mussels sampled at as many as 70 sites throughout all basins of Puget Sound at roughly 2-week intervals. Analysis of this long-term shellfish monitoring data indicates that maximum PST levels and PST-related closures have increased over the past 20 years, reaching >10,000 μg of PST per 100 g of shellfish tissue in multiple years and resulting in significant negative impacts on shellfisheries in the region (17).
To date, monitoring efforts in the Puget Sound have focused on measuring the level of PSTs present in shellfish tissue. Existing programs do not typically monitor for phytoplankton species composition or abundance. Information on A. catenella distribution and seasonal dynamics is limited for this region, despite its potential value for monitoring and understanding toxic A. catenella blooms and their impacts. Toward this end, we used a previously developed high-throughput quantitative PCR (qPCR) method (5, 6) to detect and enumerate A. catenella cells. We couple this specific and sensitive detection method for A. catenella with PST monitoring efforts to examine changes in A. catenella populations and accompanying shellfish toxicity in the Puget Sound. The data, collected from April through October, span nearly all of the 2006 A. catenella bloom season in the region. These results provide important information on the abundance and dynamics (e.g., possible source populations) of A. catenella cells during a bloom season and on their relationship to PSTs in shellfish. This effort represents a first step toward assessing the utility of plankton monitoring to augment shellfish toxicity testing in this region.
MATERIALS AND METHODS
Cultures.
A. catenella cultures (strains ACQH01 and ACQH02 isolated from Quartermaster Harbor, Puget Sound, WA) were obtained from D. Kulis of the Woods Hole Oceanographic Institution (WHOI). Cells were grown in f/2 medium at 15°C in cool white fluorescent light (about 200 μmol quanta m−2 s−1) on a 14-h light/10-h dark cycle to mimic typical Puget Sound summer conditions. The f/2 medium was made with 0.2-μm-pore-size filtered local seawater (salinity, ∼32 practical salinity units [psu]) autoclaved with inorganic f/2 nutrients (8). Filter-sterilized f/2 vitamins were added after autoclaving. Growth in the cultures was determined via cell counts of 1% Lugol's preserved samples.
Field sampling.
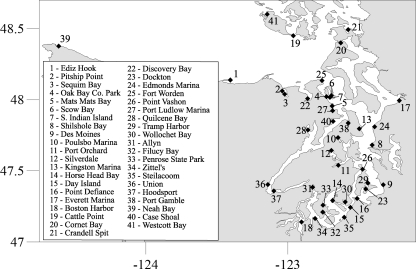
Field samples for qPCR assays and whole-cell counts were collected by volunteers recruited from the WADOH, local tribes, local health jurisdictions, and the general community. Samples were collected from late April 2006 through October 2006 at over 40 sites (Fig. 1) used in the Sentinel Monitoring Program. Volunteers were given a supply kit that included a small hand pump equipped with a Swinnex filter holder (Millipore, Billerica, MA) and were trained in sample collection. At each Sentinel Site, 1 liter of surface water was collected in a high-density polyethylene (HDPE) bottle. With a hand pump, the entire contents of the bottle were filtered through Nitex mesh (20-μm pore size) contained in the Swinnex holder. The Nitex filter circle was then transferred to a screw-cap tube containing 400 μl of buffer ATL (Qiagen, Valencia, CA). Additional near-surface water was collected (150-ml grab sample) and preserved in 1% Lugol's solution for whole-cell counts as needed. Samples were returned to the laboratory at the WADOH, and all filter and preserved samples were then shipped to WHOI for analysis. Shellfish were distributed, collected, and tested for PSTs as described elsewhere (17).
FIG. 1.
Map of station locations used in this study. The inset lists the station names. The station name and basin designation are also listed in Table 1.
DNA extractions.
All cultures (for standards) and field samples were extracted using a Generation Capture Column DNA Purification kit (Qiagen, Valencia CA). Briefly, 200 μl of well-mixed sample was removed from each sample tube and pipetted onto a DNA capture column. The samples were incubated at room temperature for 12 min. All samples were washed twice with the purification solution provided with the kit, and then DNA was eluted in 200 μl of elution solution. Samples were stored at −20°C until subsequent qPCR analysis.
Quantitative PCR.
PCR primers were designed to amplify a 183-bp sequence of the ribosomal large subunit (LSU) gene. The forward primer (5′-GCAAGTGCAACACTCCCACCAAGCAA-3′) was designed from an alignment of publicly available Alexandrium strain sequences. The reverse primer (5′-GCAAGTGCAACACTCCCACCAAGCAA-3′) was modified from a previously designed oligonucleotide (NA1 [2]). The NA1 oligonucleotide is specific for North American ribotypes of the A. catenella/A. fundyense/A. tamarense species complex (15). qPCR amplification of field samples, standards, and internal controls was performed in triplicate in a 96-well plate format using Full Velocity SYBR green qPCR master mix (Stratagene, La Jolla, CA) in an iCycler iQ real-time detection system (Bio-Rad, Hercules, CA). Reaction mixtures included 150 nM (each) forward and reverse primers and 5 μl of genomic DNA (gDNA) template (diluted 1:5) in a final volume of 25 μl. Internal controls and standards that ranged from 5 to 50,000 Alexandrium cells were included on each plate. The cycle conditions were as follows: 95°C for 5 min, 40 cycles of 95°C for 10 s and 63°C for 30 s, followed by 95°C for 1 min, and 80 cycles of 95°C for 10 s, decreasing by 0.5°C every 2 cycles for melt curve data collection. Melt curves were used to confirm single amplification products. The fluorescence threshold was set by the analytical software for the iCycler (Bio-Rad). The PCR cycle during which this threshold was crossed for each sample, standard, or internal control was designated the threshold cycle (CT). The reported CT was averaged for the standards, and a standard curve was generated for each plate of samples. Cell numbers for each field sample were determined through a comparison to the standard curve.
Standards were prepared in triplicate via dilution of a concentrated cell stock that was filtered onto 5-μm-pore-size Durapore filters (Millipore). Each standard was extracted separately to account for any variations in extraction efficiency at different cell numbers. Triplicate extracts for each cell number were then pooled and aliquoted for use during the entire sampling season. The standards included on each plate spanned 4 orders of magnitude to cover the most common range of cell densities encountered in the field. To account for potential variability in the target copy numbers between strains, all standards were made from a mixed stock of two A. catenella isolates from the Puget Sound, ACQH01 and ACQH02. PCR efficiency (E) was determined using the following equation: E = 10(−1/slope). Field samples analyzed by qPCR are potentially subject to interference by inhibitors present in the environment being sampled. To avoid inhibition field samples were routinely diluted 1:5 or more, and field samples that yielded a cell abundance of zero by qPCR were spiked with a known volume of A. catenella gDNA to confirm that negative results were not related to inhibition.
Microscope counts.
For comparison to the qPCR assay, whole-water samples were collected as needed by volunteers and fixed with 1% Lugol's solution as described above. The samples were poured into settling columns (Aquatic Research Instruments, Lemhi, ID). Generally, a 100-ml column was used; however, at certain times in the season when the concentration of Alexandrium cells was high, a 50-ml column was used. Alexandrium cells were counted in the whole-water samples at a magnification of ×200 with an inverted microscope by scanning the settling chamber. The area scanned depended on the number of Alexandrium cells observed and ranged from three transects of the settling chamber to the entire chamber.
PSP impact factor.
PST results from the 41 Sentinel sites in 2006 were sorted into four categories based on PSP impact (4). WADOH categorizes the severity of the toxicity for a site as follows: none, PST level was less than 80 μg per 100 g of shellfish tissue (FDA action level); low, PST level ranged from 80 to 499 μg per 100 g of shellfish tissue; moderate, PST level ranged from 500 to 999 μg per 100 g of shellfish tissue; high, PST level was greater than 1,000 μg per 100 g of shellfish tissue. The sites were then ranked by calculating the annual PSP impact factor as follows: PSP impact factor = (TLOW)(wfLOW) + (TMOD)(wfMOD) + (THIGH)(wfHIGH), where TLOW is the number of PSP results in the low category, wfLOW is the “weighing factor” for low PSP results (set at 1), TMOD is the number of PSP results in the moderate category, wfMOD is the weighing factor for moderate PSP results (set at 2), THIGH is the number of PSP results in the high category, and wfHIGH is the weighing factor for high PSP results (set at 3).
RESULTS
Quantitative PCR assay.
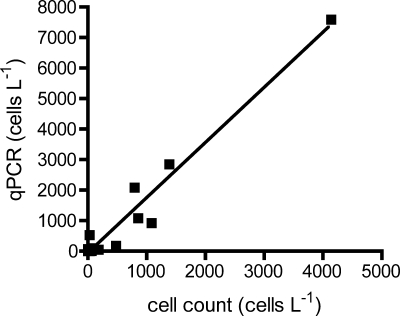
One set of standard samples was created at the start of the season using two different Puget Sound A. catenella isolates, and the same standards were used for the analysis of all samples during the year. Analyses of the mixed cell standards indicated that as few as 5 and as many as 500,000 cells liter−1 could be reliably detected. The standard curve was linear (r2 of ≥0.99) over 4 orders of magnitude and exhibited good efficiency (average E, 1.81; n = 25). All DNA extracts from field samples were diluted to mitigate potential PCR inhibition, and there was no evidence of inhibition in the diluted samples either in melt curves or template addition experiments or in comparison to the microscopic cell counts. For example, in selected cases where samples yielded a cell abundance of zero by qPCR, they were spiked with a known concentration of A. catenella gDNA. Upon reanalysis of these samples, they generated CT values consistent with the expected signal (data not shown). During 2006, roughly 500 near-surface samples were collected and assayed for A. catenella abundance at 41 stations using qPCR. Cells were detected at every sampling site at least once over the course of the field season, covering a wide range of cell numbers from roughly 5 cells liter−1 to at least 500,000 cells liter−1. At several stations in both 2006 and 2007, samples were taken and preserved for direct cell counts for comparison to a companion qPCR-assayed sample. There is a general correlation (slope, 1.809; r2 of 0.9572; P of <0.0001) between the cell densities determined by the two methods although the qPCR results tended to be about 2-fold higher than those from the Lugol's preserved and settled samples (Fig. 2).
FIG. 2.
A plot of A. catenella abundance data generated by qPCR or direct cell counts of fixed material for Puget Sound stations where both samples were analyzed. The qPCR assay generates cell numbers roughly two times (slope, 1.809 ± 0.08774) higher than microscopic cell counts (r2 = 0.9572; P < 0.0001).
Spatial and temporal distribution of cells and PSTs.
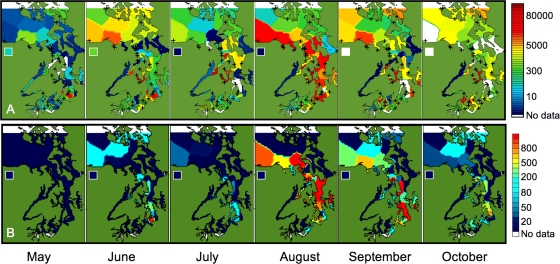
The 41 stations monitored during 2006 were examined for the spatial and monthly averaged temporal trends in A. catenella cells and PSTs (Fig. 3). In mid-May, high cell numbers were present in the southern Main Basin, but no PSTs were recorded. In June, A. catenella cells were fairly widespread (21 out of 35 stations), and PSTs were recorded at eight stations, including locations in the North Basin, throughout the Main Basin, and also at Ediz Hook (Fig. 3 and Fig. 4). No PSTs were reported from the Northwest or South Basins in June. July was generally quiet in terms of PSTs although A. catenella cells were still common throughout Puget Sound (Fig. 3). The period of August to October was very active as A. catenella cells and PSTs were detected throughout the Puget Sound (Fig. 3). In August, cells and PSTs were present in the Northwest and Main Basins as well as the eastern half of the South Basin (Fig. 3) and at Ediz Hook (Fig. 4). The North Basin had cells but no PSTs during that time. Late August was the only time that cells were observed in the Hood Canal, exceeding 2,000 cells liter−1 at Case Shoal (Fig. 4), just inside the entrance. Cells and PSTs were detected in those same areas during September, with both cell and PST distributions being quite patchy in the Main Basin (Fig. 3). In October, the last month of sampling, cells and PSTs continued to be detected in the North Basin but were generally absent in the Northwest Basin (Fig. 3). A few sites in the Main Basin had cells and/or PSTs during this time. In the South Basin, cells were found in both the eastern and western halves, but only one site had PSTs above the closure level.
FIG. 3.
Contour maps from the 2006 field season with monthly averaged data for A. catenella abundance (cells liter−1) (A) and PSTs (μg of PST 100 g shellfish tissue−1) (B). Data for the Neah Bay station are shown in the inset. Symbols denote the station locations in the upper panel.
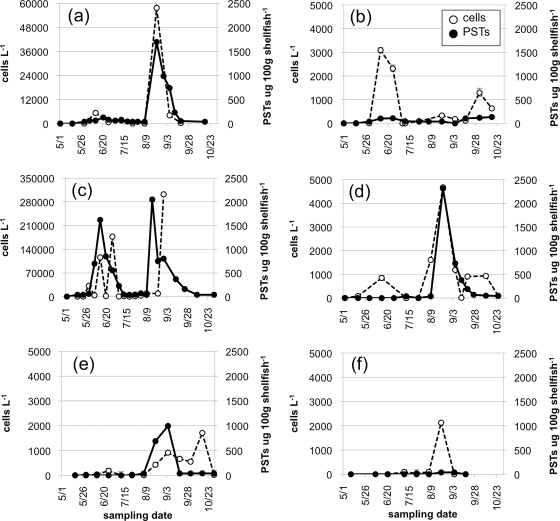
FIG. 4.
Example plots of A. catenella abundance (open circles and dashed line) and PST (solid circles and line) over time, representing different cell and PST patterns at a station from each Puget Sound basin. Ediz Hook (a) and Cattle Point (b) each had one instance of increased cell numbers prior to the appearance of PSTs above the regulatory limit and one instance of cell numbers and PST values increasing at the same sampling time. Dockton (c) and Fort Worden (d) showed increases in cell numbers prior to PSTs and increased cell numbers in the absence of PSTs. At Day Island (e), cells and PSTs increased at the same sampling time, and there was also an instance of elevated cell numbers in the absence of PSTs. Case Shoal (f) had one instance in which cell densities were >100 cells liter−1, yet no PSTs were detected. Dockton in the Main Basin contained both cells and PSTs for almost the entire sampling period; the time course illustrates the complex dynamics between cells and PSTs that can occur even within one station and one season. Error bars denote standard deviation, calculated from the mean of triplicate wells within a qPCR assay.
In general, the Hood Canal, Whidbey Basin, and the outer edge of the Strait of Juan de Fuca (Neah Bay) all had few cells and no PSTs from April to October 2006 (Fig. 3). Both A. catenella cells and PSTs were first observed early in the season (May) in the North Basin and the southern end of the Main Basin (Fig. 3). June cell and PST distributions were quite similar to those in October, with cells throughout the North Basin and along the western edge of the Main Basin (Fig. 3). In the Main and Northwest Basins, both cell numbers and PST values were highest during August but continued into September (Fig. 3). The North Basin was most active early (June) and late (October) in the season (Fig. 3). Average monthly cell numbers peaked somewhat later in the South Basin, with elevated cell abundances in September and October (Fig. 3). The Northwest Basin and Strait of Juan de Fuca had cells throughout the season. Likewise, three monitoring sites, Port Ludlow, Point Vashon, and Kingston, had both cells and closure levels of PSTs throughout virtually the entire sampling period.
PSP impact.
The WADOH monitored PST levels in shellfish from all 41 Sentinel Sites (Table 1). Record PST levels were measured throughout King, Pierce, Snohomish, and Kitsap counties, WA, in 2006. Approximately 70% of the sites sampled had PST values exceeding the FDA's regulatory limit. When the level of PST in a single sample of a particular shellfish species was determined to exceed the regulatory limit, WADOH closed commercial and recreational harvest areas for shellfish and initiated more frequent shellfish sampling. The areas were reopened only when continued monitoring ensured a return to safe conditions. PST results from the 41 Sentinel sites in 2006 were ranked by the annual PSP impact factor (Table 1).
TABLE 1.
Location, PSP impact factor, peak cell number, and peak PST concentration for each stationa
| Basin | Station name | PSP impact factor | Peak cell no. (cells/liter) | Peak PST (μg/100 g of shellfish tissue) |
|---|---|---|---|---|
| Main | Port Ludlow Marina | 35 | 31,800 | 17,000 |
| Main | Dockton | 15 | 302,200 | 2,000 |
| Main | Kingston Marina | 14 | 37,700 | 8,400 |
| Main | Point Vashon | 11 | 9,100 | 3,500 |
| Main | S. Indian Island | 11 | 6,300 | 4,800 |
| Strait of Juan de Fuca | Ediz Hook | 10 | 57,700 | 1,700 |
| Main | Tramp Harbor | 9 | 400 | 3,600 |
| South | Point Defiance | 9 | 36,900 | 1,100 |
| Main | Edmonds Marina | 7 | 140,100 | 5,500 |
| Northwest | Fort Worden | 7 | 4,700 | 2,300 |
| Main | Oak Bay County Park | 6 | 1,600 | 1,500 |
| Main | Port Orchard | 6 | 243,900 | 1,400 |
| Main | Silverdale | 6 | 526,900b | 1,300 |
| Northwest | Pitship Point | 6 | 14,300 | 1,000 |
| Main | Mats Mats Bay | 5 | 77,700 | 1,500 |
| Main | Poulsbo Marina | 5 | 812,500b | 1,000 |
| North | Cattle Point | 5 | 3,100 | 100 |
| Northwest | Sequim Bay | 5 | 33,700 | 900 |
| South | Des Moines | 5 | 17,700 | 2,700 |
| South | Horse Head Bay | 5 | 2,600 | 600 |
| Main | Shilshole Bay | 4 | 8,500 | 2,000 |
| South | Day Island | 4 | 1,700 | 1,000 |
| North | Crandell Spit | 3 | 10,800 | 200 |
| South | Steilacoom | 2 | 2,500 | 200 |
| Main | Port Gamble | 1 | 7,900 | 300 |
| Northwest | Discovery Bay | 1 | 264,100 | 200 |
| South | Wollochet Bay | 1 | 4,500 | 200 |
| Main | Scow Bay | 0 | 8,700 | 50 |
| Hood Canal | Case Shoal | 0 | 2,100 | <40 |
| Hood Canal | Hoodsport | 0 | 100 | NDc |
| Hood Canal | Quilcene Bay | 0 | 10 | ND |
| Hood Canal | Union | 0 | 10 | ND |
| North | Cornet Bay | 0 | 1,300 | <40 |
| North | Westcott Bay | 0 | 7,100 | <40 |
| South | Allyn | 0 | 600 | ND |
| South | Boston Harbor | 0 | 1,600 | ND |
| South | Filucy Bay | 0 | 7,600 | 40 |
| South | Penrose State Park | 0 | 160,200 | 70 |
| South | Zittel's | 0 | 900 | ND |
| Strait of Juan de Fuca | Neah Bay | 0 | 100 | <40 |
| Whidbey | Everett Marina | 0 | 30 | ND |
Stations are sorted by PSP impact factor.
Stations where the cell number is greater than 500,000 cell liter−1 are outside the range (5 to 500,000 cells liter−1) of cell numbers that can be confidently predicted from the qPCR standards.
ND, not detectable.
It is important to note that because the PSP impact factor accounts for the number of samples in a particular category, areas with closures due to PSTs may have higher impact factors because they are subject to more frequent sampling after the closure. The greatest impact factor occurred at Port Ludlow in the Main Basin, Jefferson County. Port Ludlow had the highest proportion (∼32%) of high PST levels (>1,000 μg 100 g shellfish tissue−1) and an annual PSP impact factor of 35 (Table 1). In general, sites in the Main and Northwest Basins had the highest PSP impact factors. In the Main Basin, only the Scow Bay site had PST levels that were always below the closure limit; no sites in the Northwest Basin were PST free (Table 1). Half of the four sites in the North Basin had no PST samples above the closure limit (Cornet Bay and Westcott Bay), and 5 out of 11 sites in the South Basin in Pierce County reported PSTs below the closure limit, including Allyn, Boston Harbor, Filucy Bay, Penrose Point, and Zittel's Marina (Table 1). All four of the sampling sites in Hood Canal—Case Shoal, Hoodsport, Quilcene Bay, and Union—were either PST free or were below the closure limit (Table 1). The easternmost site (Everett Marina, Whidbey Basin) and the westernmost site (Neah Bay, on the northwest corner of the Olympic peninsula) also had low PST levels, always below the closure limits (Table 1).
Relationship between cells and PSTs.
Maximum cell numbers ranged dramatically at any given station, from nondetectable to over 800,000 cells liter−1 (Table 1). Likewise, the PST values varied from below detection limits (32 μg 100 g shellfish tissue−1) to almost 17,000 μg per 100 g of shellfish tissue. The temporal association between elevated cell numbers (≥100 cells liter−1) and PST levels (≥80 μg 100 g shellfish tissue−1) varied between stations (Fig. 4). Because more than one instance of elevated cell and/or PST levels occurred at many of the sites, the temporal relationships varied even within the sampling period. To be considered distinct events or instances, the elevated cell concentrations or PST values must have been separated by at least one sample with values below the thresholds of 100 cells liter−1 and 80 μg of PST per 100 g of shellfish tissue, respectively. For the purposes of analysis and reporting, cells or PSTs were defined as “present” when values met or exceeded 100 cells liter−1 or 80 μg per 100 g of shellfish tissue, respectively.
Each station was examined to identify the instances of cells before PSTs, cells and PSTs together, and cells but no PSTs. Of the 41 stations, six had insufficient data to assess these categories: Discovery Bay, Port Vashon, Port Gamble, Tramp Harbor which had data for PST levels but little or no cell count data, and Hoodsport and Everett Marina, which had data for A. catenella but not PSTs. Of the 33 stations with sufficient data, 23 (70%) experienced at least one PST event. A total of 28 separate PST events were recorded: 18 stations had one event, and 5 stations had two PST events. The majority of these events (17/28, or 57%) occurred in the Main Basin. Of the 28 PST events, 20 of them (71%) were preceded by elevated A. catenella numbers while the remainder had cells and PSTs appearing concurrently. A majority of stations (27/33, or 82%) had at least one instance of elevated cell numbers without any accompanying PSTs. It was also common for sites to have both a PST event accompanied by cells and a separate instance of A. catenella cells but no PSTs (17/33 sites). Ten of the stations (30%) had elevated cell numbers at least once during the sampling period but never developed PSTs, and half of these were in the South Basin. The cell number thresholds, i.e., the numbers of cells before which PSTs were not detected above the FDA's regulatory limit, differed substantially between sites and basins. The apparent threshold is ∼400 cells liter−1 in the South Basin, ∼80 cells liter−1 in the Main Basin, and ∼3,000 and ∼4,500 cells liter−1 in the North and Northwest Basins, respectively.
In a comparison of the entire 41-station data set, there was no correlation between the cell numbers observed prior to or concurrent with PST level determination in shellfish (data not shown). However, the cell numbers observed prior to PST levels in shellfish were correlated at six individual stations (Ediz Hook, r2 = 0.8732 and P < 0.0001; Horse Head Bay, r2 = 0.9574 and P < 0.0001; Kingston Marina, r2 = 0.8366 and P < 0.0015; Point Defiance, r2 = 0.2839 and P < 0.0498; Poulsbo Marina, r2 = 0.8449 and P < 0.0005; and Silverdale, r2 = 0.9175 and P < 0.0001).
DISCUSSION
For the first time in this system, qPCR-based plankton monitoring was paired with PST testing to assess the concentrations and distributions of A. catenella and the relationship between cell numbers and PST levels in the Puget Sound. The application of qPCR to enumerate A. catenella cells made this study uniquely tractable, providing novel insight into bloom dynamics and identifying multiple source populations and the relationships between cells and shellfish PSTs. The data suggest that phytoplankton monitoring could be important in a management context, providing an early warning of shellfish PSTs.
Plankton monitoring with qPCR.
Plankton monitoring does not currently accompany shellfish PST testing in the Puget Sound, in part because of the time and expertise required to enumerate toxic species in a mixed water column assemblage. Although there are many methods for enumerating Alexandrium species (e.g., fluorescent in situ hybridization [2]), the qPCR method was chosen for this study primarily because of its sensitivity, speed, and high throughput. The qPCR assay enables a minimum of 24 samples to be extracted and analyzed in an 8-h day, more than any microscopy-based method. This allows the analysis of samples in near-real time, an important benefit in a management context. However, equipment and supply costs for different techniques are variable and must be weighed against the time and expertise required to perform the method. Here, a subset of samples was assayed with both qPCR and microscopy counts. There was a positive relationship between the cell numbers obtained from the two methods although the qPCR values were about 2-fold higher than those obtained by microscopy. Overestimation by qPCR has been previously reported (7) although the difference observed here is far less than the 5- to 10-fold differences reported by other authors. The overestimation may be compounded by the tendency for the microscopy method used here to underestimate cell numbers at higher densities and in the presence of cooccurring organisms (7). Taken together, qPCR shows promise as a viable alternative to microscopy for plankton monitoring in a management context.
Spatial and temporal patterns of A. catenella and PSTs in Puget Sound.
This is the first comprehensive data on A. catenella dynamics in the Puget Sound. The concurrent emergence of A. catenella cells at geographically distant sites (e.g., the Northwest, Main, and South Basins) in late May and early June suggests that there are multiple source populations in Puget Sound. In the Northwest Basin, Cox et al. (3) analyzed sedimentary A. catenella cyst distributions in Sequim Bay and concluded that its dense cyst population could initiate A. catenella blooms (3), consistent with water column observations in the present study. Trainer et al. (17) hypothesized that cells from this area traveled into the North Basin to initiate blooms there (17). That could be the case with the cells observed in that region in June; however, the first cells in the North Basin appeared at the eastern side, at Crandell Spit, before cells were detected in Sequim Bay. Horner et al. (9) found close to 100 cysts ml−1 of sediment in Bellingham Bay, north of Crandell Spit, and similar densities at East Sound on Orcas Island in the western part of the North Basin (9). Thus, the cells in the southern part of the North Basin could be derived from a local population or via currents or historical transport from the Northwest Basin.
In the Main Basin, Nishitani and Chew (12) hypothesized that blooms were initiated in Quartermaster Harbor (Dockton) on the southern end of Vashon Island (12), where there is a high concentration of cysts in the sediments (>70,000 cysts/ml sediment) (9). The data from this water column study support Dockton as a source population to the Main Basin. However, cells were also observed very early at other disparate stations in this area, suggesting that there may be multiple source populations of cells in the Main Basin.
Cox et al. (3) concluded, based on the paucity of cysts in Carr Inlet, that the South Basin was likely impacted by blooms transported into the region from other sites in the Sound (3). However, the early presence of cells in the South Basin (e.g., Allyn) suggests a potential A. catenella source in the South Basin as well. In September and October, cell numbers increased in the eastern part of the South Basin, near the sill at the Tacoma Narrows. These increases paralleled those occurring in the southern portion of the Main Basin and, most likely, were due to transport of cells into the South Basin. This is also supported by the lag in peak cell numbers observed in the South Basin, suggesting that both transport and resident populations were factors in A. catenella blooms in the South Basin in 2006.
There was a second pulse of cells in August at many stations, regardless of basin. This could be due to either further transport of cells between stations and basins or a second wave of cyst germination. Under the transport scenario, cells from the central Main Basin would have spread throughout the Main Basin during July and both northward and southward during August because cells generally disappeared from the North Basin and southern Main Basin during July. Surface waters in Puget Sound flow north, whereas deeper waters flow toward the south. This could potentially transport cells, especially germinating cells, in both directions. If the blooms in August are due to a second wave of germinating cells, this would suggest the presence of two different physiological populations, or ecotypes, of A. catenella that have different germination triggers and environmental tolerances. In brief, patterns of A. catenella abundance and distribution suggest that multiple independent source populations of cells exist in the Puget Sound that, combined with physical transport and environmental conditions, affect the initiation and spread of A. catenella blooms. Many of the uncertainties regarding bloom initiation, transport, and the presence of multiple distinct A. catenella populations could be addressed through further seasonal sampling and population genetic studies (14).
On a broad scale, the spatial and temporal distributions of PSTs tracked the A. catenella dynamics. The highest PSP impact factors were in the Main Basin, with sustained periods of elevated PST levels, while the lowest PSP impact factors were in the Whidbey Basin and the Hood Canal. There is considerable historical data for PST distribution in the Puget Sound, and the spatial and temporal patterns of PST levels observed during 2006 were consistent with the historical distribution of PSTs in the different Puget Sound basins, as well as in the Hood Canal (11, 17).
A. catenella and PST comparison.
In 29% of the PST events, cells and PST levels increased concurrently. However, the majority (71%) of the 28 separate PST events recorded in 2006 were predicted by an increase in cell abundance one sampling period in advance of the increase in PSTs (e.g., Kingston Marina). This was a significant correlation for six stations. Some of the lag between increases in cell numbers and detection of PSTs is likely driven by the timing of the sampling; most sites were sampled every other week, but several were sampled only monthly. However, even with this limited resolution, these data suggest that increases in cell number can be an indicator for PSTs. This kind of advance warning would be extremely beneficial to shellfish growers and local health departments, allowing management actions, such as focused monitoring of specific sites or relocating shellfish to safe areas. Routine tracking of Alexandrium species in other systems, like the Gulf of Maine, has become a powerful complement to shellfish management efforts, providing early warning and predictive power (10). Further A. catenella monitoring in the Puget Sound would be similarly valuable.
While PST increases were always associated with increased cell numbers, either before or concurrently, the reverse was not true. Most of the stations (82%) had at least one case of increased A. catenella cell numbers that was not accompanied by an increase in PSTs. Almost one-third of the stations had no PSTs despite the presence of A. catenella cells. At this point, it is also not possible to use absolute cell numbers as an indicator of PST levels or of the PSP impact factor as there is not a significant correlation between cell number and the concentration of PSTs in shellfish across that data set as a whole; rather, there is a correlation only for certain stations. These observations are likely a function of several factors, including, but not limited to, local water and cell residence times, the potential presence of ecotypes with variable toxicity, shellfish feeding rate, and detoxification kinetics. However, no sites were found to have PST in tested shellfish before the detection of A. catenella cells by qPCR.
This study encompasses only one bloom season, yet it has raised numerous questions about the sources, movement, and toxicity of A. catenella cells in the Puget Sound region. In 2006, PSTs in shellfish were always associated with increases in A. catenella cell numbers. However, it was common to have cells present at a site with no development of PSTs, highlighting the complex relationship between A. catenella cells and PSTs in the Puget Sound. More detailed seasonal monitoring of A. catenella in this system would help elucidate patterns of bloom dynamics and potentially contribute to an early warning system of shellfish PST.
Acknowledgments
We thank all of the extraordinary volunteers that comprise PSAMP and Dick Dyhrman, who provided field assistance.
This work was supported by NOAA MERHAB Program grant NA05NOS4781224.
Footnotes
Published ahead of print on 21 May 2010.
REFERENCES
- 1.Anderson, D. M., D. Kulis, J. Sullivan, S. Hall, and C. Lee. 1990. Dynamics and physiology of saxitoxin production by the dinoflagellates Alexandrium spp. Mar. Biol. 104:511-524. [Google Scholar]
- 2.Anderson, D. M., D. M. Kulis, B. A. Keafer, and E. Berdalet. 1999. Detection of the toxic dinoflagellate Alexandrium fundyense (Dinophyceae) with oligonucleotide and antibody probes: variability in labeling intensity with physiological condition. J. Phycol. 35:870-883. [Google Scholar]
- 3.Cox, A. M., D. H. Shull, and R. A. Horner. 2008. Profiles of Alexandrium catenella cysts in Puget Sound sediments and the relationship to paralytic shellfish poisoning events. Harmful Algae 7:379-388. [Google Scholar]
- 4.Determan, T. 2003. Paralytic shellfish poisoning (PSP) patterns in Puget Sound shellfish in 2001—a report for the Puget Sound Ambient Monitoring Program. Office of Food Safety and Shelfish Programs, Washington State Department of Health, Olympia, WA. http://www.doh.wa/gov/ehp/sf/Pubs/PSPtrend2001.pdf.
- 5.Dyhrman, S. T., D. Erdner, J. La Du, M. Galac, and D. M. Anderson. 2006. Molecular quantification of toxic Alexandrium fundyense in the Gulf of Maine using real-time PCR. Harmful Algae 5:242-250. [DOI] [PubMed] [Google Scholar]
- 6.Erdner, D. L., L. Percy, B. A. Keafer, J. M. Lewis, and D. M. Anderson. 2010. A quantitative real-time PCR assay for the identification and enumeration of Alexandrium cysts in marine sediments. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 57:279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godhe, A., C. Cusack, J. Pedersen, P. Andersen, D. M. Anderson, E. Bresnan, A. Cembella, E. Dahl, S. Diercks, M. Elbrachter, L. Edler, L. Galluzzi, C. Gescher, M. Gladstone, B. Karlson, D. Kulis, M. LeGresley, O. Lindahl, R. Marin, G. McDermott, L. K. Medlin, L. J. Naustvoll, A. Penna, and K. Tobe. 2007. Intercalibration of classical and molecular techniques for identification of Alexandrium fundyense (Dinophyceae) and estimation of cell densities. Harmful Algae 6:56-72. [Google Scholar]
- 8.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 29-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum, New York, NY.
- 9.Horner, R. A., C. L. Greengrove, J. R. Postel, J. E. Gawel, K. S. Davies-Vollum, A. M. Cox, S. Hoffer, K. Sorenson, J. Hubert, J. Neville, and B. W. Frost. 2008. Alexandrium cysts in Puget Sound, Washington, USA, poster 16-07. Abstr. XII Int. Conf. Harmful Algae, Copenhagen, Denmark, 4 to 8 September 2006.
- 10.Li, Y. Z., R. Y. He, D. J. McGillicuddy, D. M. Anderson, and B. A. Keafer. 2009. Investigation of the 2006 Alexandrium fundyense bloom in the Gulf of Maine: in-situ observations and numerical modeling. Cont. Shelf Res. 29:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore, S. K., N. J. Mantua, B. M. Hickey, and V. L. Trainer. 2009. Recent trends in paralytic shellfish toxins in Puget Sound, relationships to climate, and capacity for prediction of toxic events. Harmful Algae 8:463-477. [Google Scholar]
- 12.Nishitani, L., and K. K. Chew. 1984. Recent developments in paralytic shellfish poisoning research. Aquaculture 39:317-329. [Google Scholar]
- 13.Puget Sound Water Quality Action Team. 2003. Shellfish economy: treasures of the tidelands. Puget Sound Partnership, Olympia, WA. http://www.psparchives.com/publications.htm.
- 14.Rynearson, T. A., and E. V. Armbrust. 2004. Genetic differentiation among populations of the planktonic marine diatom Ditylum brightwellii (Bacillariophyceae). J. Phycol. 40:34-43. [Google Scholar]
- 15.Scholin, C. A., and D. M. Anderson. 1994. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). 1. RFLP analysis of SSU rRNA genes. J. Phycol. 30:744-754. [Google Scholar]
- 16.Shumway, S. S. 1988. Toxic algal blooms: hazards to the shellfish industry. J. Shellfish Res. 7:587-705. [Google Scholar]
- 17.Trainer, V. L., B. L. Eberhart, J. C. Wekell, N. G. Adams, L. Hanson, F. Cox, and J. Dowell. 2003. Paralytic shellfish toxins in Puget Sound, Washington State. J. Shellfish Res. 22:213-223. [Google Scholar]