Abstract
Members of the nitrite-oxidizing genus Nitrospira are most likely responsible for the second step of nitrification, the conversion of nitrite (NO2−) to nitrate (NO3−), within various sponges. We succeeded in obtaining an enrichment culture of Nitrospira derived from the mesohyl of the marine sponge Aplysina aerophoba using a traditional cultivation approach. Electron microscopy gave first evidence of the shape and ultrastructure of this novel marine Nitrospira-like bacterium (culture Aa01). We characterized these bacteria physiologically with regard to optimal incubation conditions, especially the temperature and substrate range in comparison to other Nitrospira cultures. Best growth was obtained at temperatures between 28°C and 30°C in mineral medium with 70% North Sea water and a substrate concentration of 0.5 mM nitrite under microaerophilic conditions. The Nitrospira culture Aa01 is very sensitive against nitrite, because concentrations higher than 1.5 mM resulted in a complete inhibition of growth. Sequence analyses of the 16S rRNA gene revealed that the novel Nitrospira-like bacterium is separated from the sponge-specific subcluster and falls together with an environmental clone from Mediterranean sediments (98.6% similarity). The next taxonomically described species Nitrospira marina is only distantly related, with 94.6% sequence similarity, and therefore the culture Aa01 represents a novel species of nitrite-oxidizing bacteria.
Numerous sponges have the capacity to accommodate large amounts of diverse microbes and represent significant sources for bioactive natural compounds (13). Many marine invertebrates excrete ammonium as a metabolic waste product (9), and the excretion of nitrite and nitrate has been taken as primary evidence that nitrifiers are active in these animals (10). By modulation of their pumping, sponges are a suitable habitat not only for aerobic microbes but also for anaerobic microbes. Accordingly, Hoffmann et al. (19) were able to detect major microbial pathways of the nitrogen cycle in the sponge Geodia barretti, including nitrification, the anammox process, and denitrification.
Nitrification involves the biological oxidation of ammonia (NH3) to nitrite (NO2−) and further to nitrate (NO3−) for energy purposes. It is of fundamental importance for the global nitrogen cycle in aquatic and terrestrial habitats. Nitrification is catalyzed by two phylogenetically distinct groups of microorganisms: in the first step, ammonia-oxidizing bacteria and archaea (AOB and AOA) take part in the oxidation of ammonia to nitrite, and in the second step nitrite-oxidizing bacteria (NOB) convert nitrite to nitrate (38).
Nitrite has a central position in the nitrogen cycle, connecting aerobic and anaerobic pathways. Nitrite-oxidizing bacteria play a major role in removing nitrite from the environment because it is toxic for living organisms (31). Based on morphological characteristics, NOB have been divided into five genera. This classification also reflects the phylogenetic diversity of NOB, which includes Nitrobacter and Nitrococcus (Alpha- and Gammaproteobacteria), Nitrospina (putative Deltaproteobacteria), and the candidate genus “Candidatus Nitrotoga” (Betaproteobacteria) (2). The genus Nitrospira is more distantly related to the other known NOB because it is part of its own deep-branching bacterial phylum Nitrospirae. Marine species are present in all genera of NOB except in the newly identified genus “Candidatus Nitrotoga.”
As all known nitrifying prokaryotes are slow growing and hard to maintain, their enrichment and isolation from environmental samples is difficult. Most physiological studies have been performed with pure cultures of a few “model” nitrifiers, in particular AOB related to the genus Nitrosomonas and NOB of the genus Nitrobacter. For the genus Nitrospira there are only four pure cultures available: the marine species Nitrospira marina (37), Nitrospira moscoviensis (12), “Candidatus Nitrospira bockiana” (25), and Nitrospira calida (E. Lebedeva, personal communication).
Sponges of the family Aplysinidae contain large amounts of bacteria embedded within the sponge tissue matrix (15). For example, the biomass of Aplysina aerophoba consists of up to 40% bacteria (36). These sponges are able to differentiate between food bacteria and their own bacterial symbionts (41). Investigations of the diversity of sponge-associated bacteria, including different genetic and also cultivation approaches, have been made with several specimens (15, 16, 39). In terms of nitrification, Hentschel et al. (17) gave first evidence for the presence of nitrite oxidizers, and it has been verified that sponges harbor AOB and AOA (8). Most of the recognized NOB in sponges are Nitrospira-like bacteria (17, 32, 35), although in the beginning, there were further hints to 16S rRNA sequences, which are most closely related to Nitrospina gracilis (17). However, as these sequences were found only once, it could be assumed that Nitrospira is the main nitrite oxidizer in this environment. Nitrospira-like bacteria are deemed to be recalcitrant and fastidious, and they are easily overgrown by other bacteria under suboptimal conditions. Despite these limitations in the laboratory, Nitrospira was determined to be the most important nitrite oxidizer during wastewater treatment (21, 33), in aquaculture biofilters (14) and in freshwater systems (20, 29).
Identification of sponge-associated microorganisms has been performed largely with culture-independent methods, which are 16S rRNA gene based (denaturing gradient gel electrophoresis [DGGE], terminal restriction fragment-length polymorphism [TRFLP]) or visual (fluorescence in situ hybridization [FISH], electron microscopy) (8, 11). Nevertheless, the cultivation of microorganisms is still essential for the investigation of their physiological potential and function in the environment. Information about physiological characteristics helps us to understand the metabolism and possible nutritional interactions of nitrifiers with the host sponge (8).
This is the first report about cultivation of nitrifying bacteria originating from a marine sponge. We obtained a nitrite-oxidizing enrichment culture of a Nitrospira-like bacterium derived from Aplysina aerophoba, characterized it phylogenetically, and analyzed the most important physiological features.
MATERIALS AND METHODS
Sponge preparation.
An Aplysina aerophoba sample was collected by scuba diving at depths from 3 to 15 m offshore (Banyuls sur Mer, France) in April 2003 by the research group of Ute Hentschel (University of Würzburg). The sample was sent to Hamburg in sterile polypropylene bags cooled at 4°C. Thin layers from the mesohyl were sliced with a scalpel and washed in calcium/magnesium-free seawater (CMFASW; 27.0 g NaCl, 1.00 g Na2SO4, 0.80 g KCl, 0.18 g Na2HCO3, deionized H2O 1 liter). The preparation of multiple extraction fractions was done by three centrifugation steps at the following different speeds: (i) 160 × g for 10 min, (ii) 7,000 × g for 4 min, and (iii) 7,000 × g for 15 min. The cell pellet retrieved after the third centrifugation step was used as 1% inoculum (vol/vol) in 300-ml Erlenmeyer flasks filled with 150 ml marine (70% seawater) medium.
Cultivation and physiology of Nitrospira-like bacteria.
Nitrospira-like bacteria were enriched in mineral salt medium (12) prepared with 70% seawater, which contained 0.3 mM NaNO2 as the energy source and no organic carbon in order to suppress the growth of heterotrophic bacteria. Incubation was performed at 22°C and 28°C in the dark without agitation. The presence of nitrite was regularly checked by the Griess-Ilosvay spot test (22) and replaced every 7 to 14 days in case of growth. Upscaling was performed as described by Spieck et al. (33) to obtain a sufficient amount of cells for further analysis. Cultures with a high cell density were grown in 3-liter flasks and used to separate Nitrospira-like bacteria from contaminants by Percoll density gradient centrifugation and subsequent serial dilutions (12). To enhance the growth of Nitrospira, a vitamin solution prepared according to Balch et al. (5) was added to the media. Closed bottles which were three-quarters full of liquid medium were used to produce a reduced oxygen content.
Growth experiments were performed at an incubation temperature of 28°C in 300-ml Erlenmeyer flasks filled with 150 ml medium. For the determination of the optimal temperature, incubations between 10 and 34°C were carried out. Nitrite tolerance was investigated with a nitrite concentration of 0.1 to 3.0 mM with precultures grown at 28°C.
Chemical analyses.
Concentrations of nitrite and nitrate were measured by high-performance liquid chromatography (HPLC) via ion-pair chromatography with a Hypersil ODS C18 column (5 μm; 125 by 4.6 mm) (30) and UV detection in an automated system (Kontron, Eching, Germany). Optimal temperature and nitrite tolerance were measured twice in three duplicates.
DNA extraction and phylogenetic and RFLP analysis.
Liquid culture samples were centrifuged at 15,000 × g for 15 min. Cell pellets smaller than a pinhead were used for DNA extraction, using the UltraClean DNA kit (MoBio Laboratories, Inc., Solana Beach, CA) according to the manufacturer's instructions.
The 16S rRNA genes were amplified by the bacterial conserved primers 27F and 1492R (23). The PCR product was directly ligated into the pGEM-T vector cloning system (Promega, Mannheim, Germany) and transformed into competent cells as described in the manufacturer's instructions. For partial and near-complete sequencing of clone inserts, the plasmid primers SP6 and T7 were used to reamplify the insert. Bacterial conserved primers (341F, 907R) were additionally used for sequencing (23). 16S rRNA gene sequences were compared with those on publicly accessible databases by using the program Basic Local Alignment Search Tool (BLAST; NCBI) (4). By use of the program package ARB (26), a neighbor-joining tree was generated from an alignment of 16S rRNA gene sequences from selected Nitrospira-like bacterial clones.
For RFLP analysis, colony PCR analysis was performed on white clones using the Nitrospira-specific PCR primers Nsp60-kF (1) and 1158R (28). For colony PCR analysis, the primers T7 and SP6 were used and the annealing temperature was 45°C. Restriction digestion was performed with the enzyme HapII. Unique RFLP patterns were identified after separation on a 3% agarose gel.
Electron microscopy.
For observation by electron microscopy, cells were collected at 15,000 × g, washed in 2.0% NaCl, fixed with 2.5% (vol/vol) glutaraldehyde and 2% (wt/vol) osmium tetroxide, and embedded in Spurr medium (34). The embedded samples were sectioned and stained according to a previously published protocol (33). Examination was carried out with a transmission electron microscope (Zeiss model Leo 906E with a charge-coupled device [CCD] camera model 794). For negative staining, a few milliliters of the culture were centrifuged (15,000 × g) and the harvested cells were dropped on a copper grid and dried. The samples were stained with uranyl acetate (2%).
Nucleotide sequence accession numbers.
Nucleotide sequences were deposited in the GenBank database under accession numbers EU055608 (Aa01), HM131833 (F2), and HM131832 (S11).
RESULTS
Cultivation.
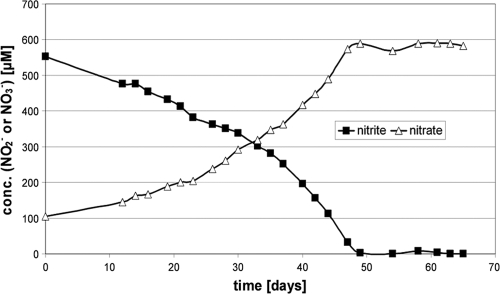
We succeeded in obtaining enrichment cultures of a Nitrospira-like bacterium derived from the mesohyl of Aplysina aerophoba after an initial lag phase of 10 months. The potential of the cultures to oxidize nitrite was proven by physiological investigations. Figure 1 shows the oxidation of about 0.5 mM nitrite within 50 days, attended by an equivalent increase of nitrate. After a period of nearly 3 years, the cultures began to oxidize up to 0.8 mM nitrite per week. In order to improve growth, we tested the influence of vitamins such as biotin, folic acid, and vitamin B12, but observed no positive effect. The best-growing enrichment culture was named Aa01. Several other enrichment cultures derived from the same material grown at temperatures of 22°C and 28°C, with or without enrichment by Percoll density gradient centrifugation, were phylogenetically identical.
FIG. 1.
Nitrite consumption by the enrichment culture of the Nitrospira-like bacterium Aa01 grown at 28°C in Erlenmeyer flasks plotted against nitrate production.
In contrast to Nitrospira marina (37), cells of the enrichment culture Aa01 did not grow in terrestrial mineral salt medium without seawater or in mineral salt medium with 70% Baltic Sea water. Growth was observed in artificial sea salt medium with salt concentrations similar to those of the Mediterranean Sea. The best growth was achieved with a mineral medium prepared with 70% North Sea water at pH 7.8 and 0.5 mM nitrite in 250-ml bottles at 28°C. In contrast to the Erlenmeyer flasks, the lower oxygen pressure associated with the use of bottles seems to enhance the growth of the novel Nitrospira culture. In Erlenmeyer flasks, a maximum nitrite consumption of around 25 μM per day was measured, whereas in bottles a consumption of almost 110 μM nitrite per day could be achieved (data not shown).
Morphology.
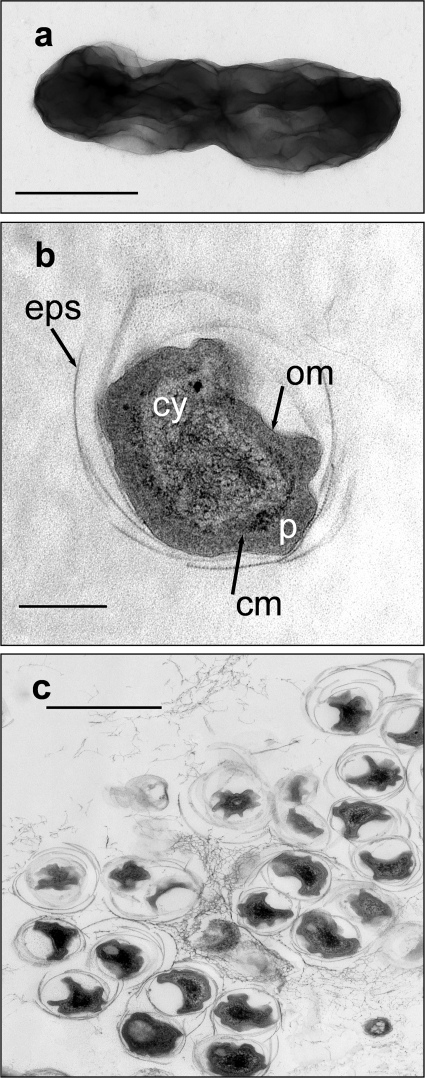
The shape of the Gram-negative Aa01 Nitrospira-like cells ranged from slightly curved to straight rods with 3 to 4 turns (Fig. 2 a). The size of the rod-shaped cells varied from 0.3 to 0.7 μm in width and 0.8 to 2.0 μm in length. The cells were surrounded by a multilayered cell envelope consisting of a cytoplasmic membrane and an outer membrane (Fig. 2b). The membranes were separated by an exceptionally wide periplasmic space (40 to 80 nm), which is typical for members of Nitrospira. No intracytoplasmic membranes or carboxysomes were found, but the cells often contained glycogen and polyphosphate-like deposits as storage compounds. The characteristic ultrastructure shown by electron microscopic investigations provided additional evidence that most of the enriched sponge-associated bacteria belonged to the genus Nitrospira (Fig. 2c).
FIG. 2.
Electron micrographs of the nitrite-oxidizing enrichment culture Aa01. (a) Overall shape of a cell stained with uranyl acetate. Bar = 500 nm. (b) Ultrathin section of a Nitrospira-like bacterium revealing the wide periplasmic space and extracellular polymeric substances. Cy, cytoplasm; cm, cytoplasmic membrane; om, outer membrane; p, periplasm; eps, extracellular polymeric substances. Bar = 200 nm. (c) Overview of the dominant cell type with a unique ultrastructure. Bar = 1.0 μm.
Optimal temperature.
Nitrospira-like bacteria are known to occur in environments with extreme temperatures, ranging from permafrost soils (6) to hot springs (24). As a consequence, their behavior in the laboratory reflects temperature-specific adaptations (Table 1). The temperature interval for growth spanned at least 15°C. For the enrichment culture of Aa01, growth took place over an interval of 13°C (17°C to 30°C), and optimum activity occurred between 28°C and 30°C. At a temperature of 22°C, 0.5 mM nitrite was oxidized in 4 months, whereas at 17°C, the same amount of substrate was oxidized only after 8 months. No nitrite consumption could be detected at temperatures below 10°C or above 32°C after 8 months of incubation.
TABLE 1.
Temperature range and tolerance to nitrite of pure and enrichment cultures of the genus Nitrospira
| Culture | Temperature range (°C) | Nitrite tolerance (mM) | Source or reference |
|---|---|---|---|
| Aa01, enrichment culture | 17-30 | 1.5 | This study |
| “Candidatus Nitrospira defluvii” | 10-32 | 20-25 | Unpublished result; 33 |
| Nitrospira moscoviensis | 28-43 | 15 | Unpublished result; 12 |
| “Candidatus Nitrospira bockiana” | 28-44 | 18 | 25 |
| Nitrospira calida | 35-58 | 6 | E. Lebedeva, personal communication |
| Nitrospira marina | 15-30 | 6 | 37 |
| Laptev S11 | 4-28 | NDa | 1 |
| Laptev F2 | 4-28 | ND | 1 |
ND, not determined.
Nitrite tolerance.
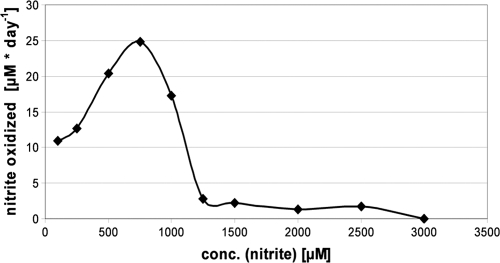
The substrate concentration has a great influence on the ecological distribution of nitrite oxidizers (7) and seems to differ within separate clusters of Nitrospira (28). In general, Nitrospira-like bacteria are known for their high sensitivity to their substrate nitrite, but large variances in this dependence have been noted in cultures from specialized habitats (Table 1). Culture Aa01 exhibited an extremely low tolerance to nitrite (1.5 mM), because all other representatives of Nitrospira, like N. marina, were able to grow in the presence of at least 6 mM nitrite (37). This species from the Atlantic Ocean as well as the novel thermophilic species N. calida (E. Lebedeva, personal communication) showed a relatively low tolerance toward nitrite, whereas the other described members of this genus were more resistant. The upper border of substrate tolerance was exhibited by “Candidatus Nitrospira defluvii” from a wastewater treatment plant, which could consume up to 25 mM nitrite. The optimal nitrite concentration for Aa01 was shown to lie between 0.5 and 0.75 mM (Fig. 3). The application of more than 1.5 mM nitrite resulted in an almost complete inhibition of growth.
FIG. 3.
Tolerance against nitrite of the Nitrospira-like culture Aa01. The maximum oxidation rate of nitrite per day is plotted against the nitrite concentration.
Phylogenetic affiliation.
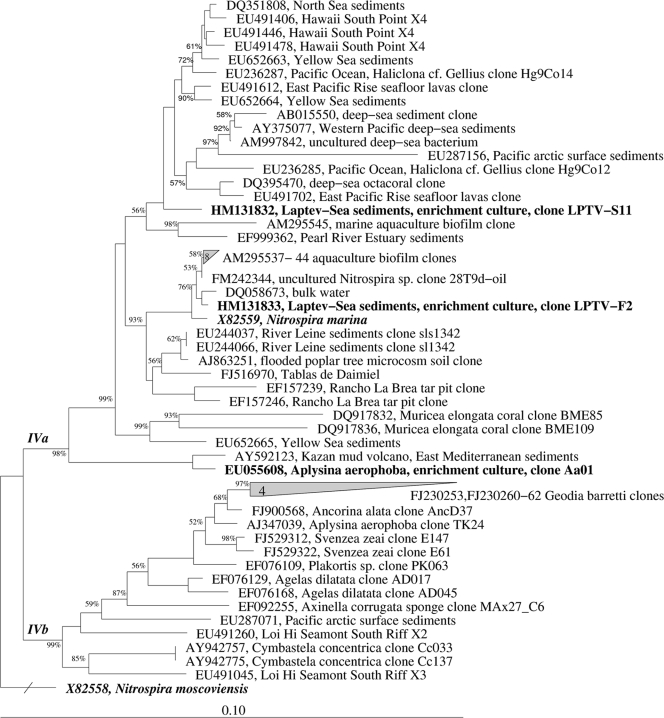
The 16S rRNA gene of the novel Nitrospira-like bacterium Aa01 was cloned, and the whole 16S rRNA gene sequence was compared to those in publicly accessible databases. A neighbor-joining tree was generated from an alignment of 16S rDNA sequences from selected Nitrospira-like bacteria (Fig. 4). Apart from the only described marine member, N. marina, two additional enrichment cultures of Nitrospira were included in the phylogenetic tree for comparison. Both cultures, F2 and S11, originated from marine sediments of the Laptev Sea in Siberia (1). Culture F2 is closely related to N. marina, whereas S11 represents a novel sublineage. The 16S rRNA sequence of the sponge culture does not belong to the monophyletic cluster of Nitrospira sequences found by molecular work inside sponges (17, 19, 27, 35). The closest relative of the sponge culture Aa01 is a sequence derived from an environmental clone from the Kazan mud volcano (East Mediterranean sediments; AY592123) with 98.6% similarity. The similarity to the only described species in cluster IV (N. marina) amounted to 94.6%.
FIG. 4.
Phylogenetic relationships between the cloned 16S rRNA sequence from the nitrite-oxidizing enrichment culture Aa01, representatives of marine Nitrospira-like bacteria, and sponge-associated environmental sequences. The tree was constructed using the neighbor-joining algorithm based on long sequences (≥1,000 nucleotides). Nodes supported by bootstrap values are indicated. Cultivated representatives are printed in boldface type. Bar = 10% sequence divergence.
Using the specific 16S rRNA gene-targeting primers Nsp60-kF (1) and 1158R (28), a 1,060-bp PCR product was obtained from the Aa01 enrichment culture. In total, 15 clones were compared by restriction fragment polymorphism analysis (RFLP). Two different restriction patterns were observed and sequenced. Both were identical to the Aa01 clone.
DISCUSSION
In general, Nitrospira-like bacteria grow very slowly, with generation times of up to 90 h for Nitrospira marina (37). However, the initial 10-month lag phase of the sponge culture Aa01 seems to be extraordinarily long, even for Nitrospira. One reason for this behavior could be the low cell density of Nitrospira-like bacteria in the mesohyl of the sponge A. aerophoba combined with the normally slow growth rate. The most relevant reasons are probably the lack of knowledge about optimal growth conditions, such as the correct composition of different salts in the media as well as the loss of interactions with adjoining microorganisms. Although it was possible to cultivate a sponge-associated nitrifying bacterium for the first time, the 16S rRNA sequence was not sponge-specific, and it is not clear if this bacterium is a real sponge inhabitant. As revealed by RFPL analysis, the enrichment culture Aa01 contains only one member of Nitrospira. This finding was further confirmed by DGGE (using a eubacterial primer set) and cloning of different cultures (semispecific PCR analysis for Nitrospira) (not shown).
Growth of the Nitrospira culture Aa01 was enhanced when the oxygen pressure was lowered. This is surprising, because the nitrification process is generally thought to be strictly aerobic. However, in freshwater sediments members of Nitrospira are known to exist in anoxic microhabitats (3), which are also documented within sponges (18, 19).
In comparison to other Nitrospira-like bacteria, the enrichment culture Aa01 has a small temperature range of active growth (Table 1). This finding correlates with studies of the temperature thresholds for bacterial symbiosis in sponges, which have revealed a rapid response to elevated temperatures (40). A temperature shift from 27°C to 33°C resulted in a significant community shift, and Nitrospira-like sequences were no more detectable. This upper limit of temperature agrees well with that found in our laboratory culture. Furthermore, culture Aa01 exhibited a very low tolerance to nitrite in comparison with other pure and enrichment cultures of Nitrospira-like bacteria and seems to be adapted to the low quantity of nitrite inside the mesohyl.
In laboratory experiments, the enrichment culture Aa01 oxidized up to 110 μM nitrite per day. A net nitrification rate in the sponge Geodia barretti of 566 nmol N cm−3 sponge per day was calculated by Hoffman and colleagues (19). The nitrate excretion rate in sponges of Aplysina aerophoba reached values of up to 344 nmol g−1 dry weight per hour, increasing to 1,325 nmol g−1 dry weight per hour when additional ammonium was supplied (8). In comparison to these nitrification rates within whole sponges, the enrichment culture Aa01 is not able to reach this amount of nitrite consumption.
The rapidly increasing phylogenetic diversity within the genus Nitrospira is supplemented by the 16S rRNA sequence of the enrichment culture Aa01. Members of the Nitrospira are ubiquitously distributed in a wide range of aquatic and terrestrial ecosystems, but in sublineage IV all clones and cultures were derived from marine habitats and invertebrates. As expected, the 16S rRNA gene sequence of Aa01 was affiliated with cluster IV (Fig. 4). This sequence is located between cluster IVa and IVb (27) but actually belongs to IVa. In cluster IVb, most of the sponge-associated Nitrospira-like bacteria are grouped, whereas the bacteria in cluster IVa originate from different marine habitats. The similarity to environmental clones of Nitrospira-like bacteria detected in the sponge Aplysina aerophoba is relatively low, with a maximum similarity of 91.1% (AJ347039; clone TK24). However, this classification will remain provisional until more full sequences of marine Nitrospira cultures are available.
Within the last decade, it was realized that Nitrospira is the key organism of nitrite oxidation in moderate and extreme habitats. These microorganisms dominate the guild of NOB in natural and artificial environments varying in factors like temperature and nitrogen charge. Accordingly, an increasing diversity of Nitrospira-derived 16S rRNA sequences can be found in public databases, but simultaneously it was possible to take several representatives into laboratory culture, which reflected a physiological behavior in adaptation to their special origin. As shown above, the most susceptible culture of Nitrospira originated from a marine sponge, whereas the highly resistant “Candidatus Nitrospira defluvii” was retrieved from activated sludge, revealing a substrate tolerance of 25 mM sodium nitrite, which is in the range of Nitrobacter. Further on, different Nitrospira isolates span a temperature interval of 4°C to 58°C in growth, providing the potential for colonization of different ecological niches.
Future research might aim to examine if the novel Nitrospira-like bacterium is a permanent member of the mesohyl microbiota. The supply of biomass is essential to check the specificity of newly designed probes for fluorescence in situ hybridization as well as novel PCR primers. Additional experiments are required to assess if the novel Nitrospira bacterium has a symbiotic function inside sponges.
Acknowledgments
This research was funded by the German Research Foundation (DFG; project Nitrospira, number SP 667/3-1) and the Federal Ministry of Education and Research (BMBF; project Laptev Sea System).
We are grateful to Markus Wehrl, University of Würzburg, for supplying the sponge sample. Britta Soltau is acknowledged for laboratory help. We thank Elke Woelken for excellent technical assistance in electron microscopy.
Footnotes
Published ahead of print on 28 May 2010.
REFERENCES
- 1.Alawi, M. 2007. Diversität Nitrit oxidierender Bakterien in Böden des nordsibirischen Permafrostes und Sedimenten der Laptev-See. Ph.D. dissertation. University of Hamburg, Hamburg, Germany.
- 2.Alawi, M., A. Lipski, T. Sanders, E.-M. Pfeiffer, and E. Spieck. 2007. Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J. 1:256-264. [DOI] [PubMed] [Google Scholar]
- 3.Altmann, D., P. Stief, R. Amann, D. De Beer, and A. Schramm. 2003. In situ distribution and activity of nitrifying bacteria in freshwater sediment. Environ. Microbiol. 5:798-803. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Wyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, S., C. Hartwig, E. Spieck, and E. Bock. 2002. Immunological detection of Nitrospira-like bacteria in various soils. Microb. Ecol. 43:26-33. [DOI] [PubMed] [Google Scholar]
- 7.Bartosch, S., I. Wolgast, E. Spieck, and E. Bock. 1999. Identification of nitrite-oxidizing bacteria with monoclonal antibodies recognizing the nitrite oxidoreductase. Appl. Environ. Microbiol. 65:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayer, K., S. Schmitt, and U. Hentschel. 2008. Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ. Microbiol. 10:2942-2955. [DOI] [PubMed] [Google Scholar]
- 9.Brusca, R. C., and G. J. Brusca. 1990. Phylum Porifera: the sponges, p. 181-210. In A. D. Sinauer (ed.), Invertebrates. Sinauer Press, Sunderland, MA.
- 10.Corredor, J. E., C. R. Wilkinson, V. P. Vicente, J. M. Morell, and E. Otero. 1988. Nitrate release by Caribbean reef sponges. Limnol. Oceanogr. 33:114-120. [Google Scholar]
- 11.Diaz, M. C., D. Akob, and C. S. Cary. 2004. Denaturing gradient gel electrophoresis of nitrifying microbes associated with tropical sponges. Boll. Mus. Ist. Biol. Univ. Genova 68:279-289. [Google Scholar]
- 12.Ehrich, S., D. Behrens, E. Lebedeva, W. Ludwig, and E. Bock. 1995. A new obligately chemolithoautotrophic, nitrite oxidizing bacterium, Nitrospira moscoviensis sp. nov., and its phylogenetic relationship. Arch. Microbiol. 164:16-23. [DOI] [PubMed] [Google Scholar]
- 13.Faulkner, D. J. 2000. Marine natural products. Nat. Prod. Rep. 17:7-55. [DOI] [PubMed] [Google Scholar]
- 14.Foesel, B. U., A. Gieseke, C. Schwermer, P. Stief, L. Koch, E. Cytryn, J. R. de la Torré, J. van Rijn, D. Minz, H. L. Drake, and A. Schramm. 2008. Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol. Ecol. 63:192-204. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich, A. B., H. Merkert, T. Fendert, J. Hacker, P. Proksch, and U. Hentschel. 1999. Microbial diversity in the marine sponge Aplysina cavernicola (formerly Verongia cavernicola) analyzed by fluorescence in situ hybridization (FISH). Mar. Biol. 134:461-470. [Google Scholar]
- 16.Friedrich, A. B., I. Fischer, P. Proksch, J. Hacker, and U. Hentschel. 2001. Temporal variation of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38:105-113. [Google Scholar]
- 17.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann, F., O. Larsen, V. Thiel, H. T. Rapp, T. Pape, W. Michaelis, and J. Reitner. 2005. An anaerobic world in sponges. Geomicrobiol. J. 22:1-10. [Google Scholar]
- 19.Hoffmann, F., R. Radax, D. Woebken, M. Holtappels, G. Lavik, H. T. Rapp, M. L. Schläppy, C. Schleper, and M. M. Kuypers. 2009. Complex nitrogen cycling in the sponge Geodia barretti. Environ. Microbiol. 11:2228-2243. [DOI] [PubMed] [Google Scholar]
- 20.Hovanec, T. A., L. T. Taylor, A. Blakis, and E. DeLong. 1998. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl. Environ. Microbiol. 64:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juretschko, S., G. Timmermann, M. Schmid, K.-H. Schleifer, A. Pommerening-Röser, H.-P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keeney, D. R., and D. W. Nelson. 1982. Nitrogen—inorganic forms, p. 643-693. In A. Page (ed.), Methods of soil analysis, part 2. Chemical and microbiological properties. American Society of Agronomy, Inc., Crop Science Society of America, Inc., and Soil Science Society of America, Inc., Madison, WI.
- 23.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, London, United Kingdom.
- 24.Lebedeva, E. V., M. Alawi, C. Fiencke, B. Namsaraev, E. Bock, and E. Spieck. 2005. Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiol. Ecol. 54:297-306. [DOI] [PubMed] [Google Scholar]
- 25.Lebedeva, E. V., M. Alawi, F. Maixner, P. G. Jozsa, H. Daims, and E. Spieck. 2008. Physiological and phylogenetical characterization of a new lithoautotrophic nitrite-oxidizing bacterium ‘Candidatus Nitrospira bockiana’ sp. nov. Int. J. Syst. Evol. Microbiol. 58:242-250. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, K. Yadhu, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maixner, F. 2009. The ecophysiology of nitrite-oxidizing bacteria in the genus Nitrospira: novel aspects and unique features. Ph.D. thesis. Universität Wien, Vienna, Austria.
- 28.Maixner, F., D. Noguera, B. Anneser, K. Stoecker, G. Wegl, M. Wagner, and H. Daims. 2006. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ. Microbiol. 8:1487-1495. [DOI] [PubMed] [Google Scholar]
- 29.Martiny, A. C., H.-J. Albrechtsen, E. Arvin, and S. Molin. 2005. Identification of bacteria in biofilm and bulk water samples from a nonchlorinated model drinking water distribution system: detection of a large nitrite-oxidizing population associated with Nitrospira spp. Appl. Environ. Microbiol. 71:8611-8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meincke, M., E. Bock, D. Kastrau, and P. M. H. Kroneck. 1992. Nitrite oxidoreductase from Nitrobacter hamburgensis: redox centers and their catalytic role. Arch. Microbiol. 158:127-131. [Google Scholar]
- 31.Philips, S., H. J. Laanbroek, and W. Verstraete. 2002. Origin, causes, and effects of increased nitrite concentrations in aquatic environments. Rev. Environ. Sci. Biotechnol. 1:115-141. [Google Scholar]
- 32.Schmitt, S., J. B. Weisz, N. Lindquist, and U. Hentschel. 2007. Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl. Environ. Microbiol. 73:2067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spieck, E., C. Hartwig, I. McCormack, F. Maixner, M. Wagner, A. Lipski, and H. Daims. 2006. Selective enrichment and molecular characterisation of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ. Microbiol. 8:405-415. [DOI] [PubMed] [Google Scholar]
- 34.Spurr, A. R. 1969. A low viscosity epoxy resin embedding medium of electron microscopy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, M. W., R. Radax, D. Steger, and M. Wagner. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vacelet, J., and C. Donadey. 1977. Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Ecol. 30:301-314. [Google Scholar]
- 37.Watson, S. W., E. Bock, F. W. Valois, J. B. Waterbury, and U. Schlosser. 1986. Nitrospira marina gen. nov. sp. nov: a chemolithotrophic nitrite-oxidizing bacterium. Arch. Microbiol. 144:1-7. [Google Scholar]
- 38.Watson, S. W., E. Bock, H. Harms, H. P. Koops, and A. B. Hooper. 1989. Nitrifying bacteria, p. 1808-1834. In J. T. Stanley (ed.), Bergey's manual of systematic bacteriology, vol. 3. Williams & Wilkins, Baltimore, MD.
- 39.Webster, N. S., K. J. Wilson, L. L. Blackall, and R. T. Hill. 2001. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 67:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webster, N. S., R. S. Cobb, and A. P. Negri. 2008. Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2:830-842. [DOI] [PubMed] [Google Scholar]
- 41.Wehrl, M., M. Steinert, and U. Hentschel. 2007. Bacterial uptake by the marine sponge Aplysina aerophoba. Microb. Ecol. 53:355-365. [DOI] [PubMed] [Google Scholar]