Abstract
Extracts of the Floridian marine cyanobacterium Lyngbya cf. confervoides were found to deter feeding by reef fish and sea urchins (Diadema antillarum). This antifeedant activity may be a reflection of the secondary metabolite content, known to be comprised of many serine protease inhibitors. Further chemical and NMR spectroscopic investigation led us to isolate and structurally characterize a new serine protease inhibitor 1 that is formally derived from an intramolecular condensation of largamide D (2). The cyclization resulted in diminished activity, but to different extents against two serine proteases tested. This finding suggests that cyanobacteria can endogenously modulate the activity of their protease inhibitors.
Keywords: cyanobacteria, Lyngbya, antifeedant activity, serine protease inhibitors, cyclodepsipeptides
1. Introduction
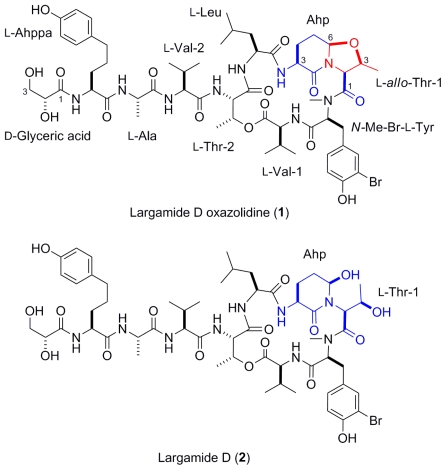
Both marine and freshwater cyanobacteria contain toxic secondary metabolites and are notorious for their ability to form extensive blooms. The compounds found in these bloom-forming cyanobacteria include the lyngbyatoxins and the hepatotoxic microcystins and cylindrospermopsin [1–6]. Several cyanobacterial extracts can reduce feeding by fish, which is thought to be driven by secondary metabolite diversity and abundance [6–9]. However, cyanobacteria also produce a range of protease inhibitors with little or no apparent cytotoxicity. One prevalent class found in marine [10–13] and freshwater [14–16] cyanobacteria is comprised of protease inhibitors with a cyclic depsipeptide scaffold that contains a 3-amino-6-hydroxy-2-piperidone (Ahp) moiety as a key feature for inhibition of certain serine proteases. Since many digestive enzymes such as trypsin and chymotrypsin are serine proteases and are inhibited by these compounds, these natural products could function as digestion inhibitors [17]. Serine protease inhibitors also co-occur with microcystins and are linked to an enhanced toxin activity [18] or thought to upregulate biosynthetic genes [19]. Here we investigate the antifeedant activity of a Lyngbya species identified as Lyngbya cf. confervoides (cf. = similar to but not the same as; compared to Lyngbya confervoides) [10,20] collected from reefs near Fort Lauderdale, Florida, towards natural assemblages of reef fish and the tropical sea urchin Diadema antillarum. This cyanobacterium produces a wide array of serine protease inhibitors including lyngbyastatins 4–6 [21,22], pompanopeptin A [23] and largamides D–G [24]. Extracts of this cyanobacterium deterred fish and urchin feeding, which may be linked to the production of these protease inhibitors, however, individual compounds have not been tested for their ability to deter feeding. This observed feeding deterrent activity led us to thoroughly investigate the natural product chemistry of this cyanobacterium. Through rigorous chemical investigation of the crude extracts we identified a new compound, largamide D oxazolidine (1), which is an intramolecular condensation product of the serine protease inhibitor largamide D (2) (Figure 1). Largamide D oxazolidine (1) exhibited a reduced and differential activity against chymotrypsin and elastase, providing evidence for an endogenous mode of deactivation or modulation of protease inhibitor activity in this marine cyanobacterium.
Figure 1.
Structures of largamide D oxazolidine (1) and the parent compound, largamide D (2).
2. Results and Discussion
2.1. Feeding Experiments
The extractions yielded a dry weight concentration of 21.15% for the non-polar extract and 6.95% for the polar extract. All feeding experiments were conducted at natural concentration by dry weight.
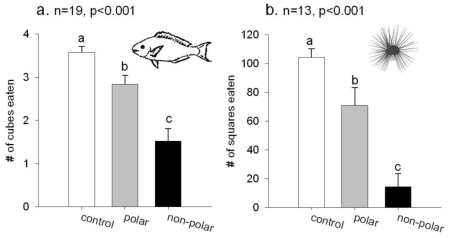
When tested against a natural assemblage of reef fish in field feeding experiments, where many individuals of diverse fish species would feed on food cubes containing cyanobacterial extracts, both the polar and non-polar extracts reduced feeding on agar food cubes. The fish consumed 3.58 ± 0.14 (mean ± SE) agar cubes of the control food, 2.84 ± 0.21 agar cubes of the food containing the polar extract, and 1.52 ± 0.29 of the agar cubes that contained the non-polar extract (Figure 2a). There were significant differences in the amount of food eaten (n = 19, Friedman’s random block, p < 0.001), with less of the polar extract eaten by the fish than the solvent controls and the least amount eaten of the food containing the non-polar extract (Student Newman Keuls post-hoc test, p < 0.05).
Figure 2.
Feeding experiments with the crude extracts of L. cf. confervoides. The bars represent the mean amount of food eaten and the error bars are +1 SE. Different letters above the bars represent means that are statistically different. (a). Feeding experiments with a natural assemblage of reef fish. (b). Feeding experiments with the sea urchin Diadema antillarum.
When extracts of L. cf. confervoides were tested against the sea urchin Diadema antillarum, both the polar and non-polar extracts reduced urchin feeding. The urchins ate 104.3 ± 6.0 (mean ± SE) squares of the control food, 71.0 ± 12.3 squares of the food with the polar extract and 14.3 ± 9.3 squares of the food with the non-polar extract (Figure 2b). There were statistically different amounts of the food consumed (n = 13, Friedman’s random block, p < 0.001) with the solvent control the most consumed food, less eaten of the food with the polar extract and the least eaten of the food with the non-polar extract (Student Newman Keuls post-hoc test, p < 0.05).
Other benthic marine cyanobacteria have also been shown to be chemically defended from marine herbivores [6–9,25]. Ypaoamide, pitipeptolide A, malyngolide and malyngamides A and B are a few purified compounds from marine cyanobacteria known to reduce feeding by generalist herbivores such as fishes and sea urchins [7,26,27]. More specialized herbivores, such as the sea hare Stylocheilus striatus, are usually not deterred by cyanobacterial extracts or compounds [6,9,26], and extracts of L. cf. confervoides also did not deter this herbivore relative to controls [28]. However, Stylocheilus striatus grew more slowly on L. cf. confervoides relative to L. polychroa, which might be related to the presence of serine protease inhibitors in L. cf. confervoides that reduced conversion efficiency of this diet [28].
2.2. Isolation and Structure Determination of Largamide D Oxazolidine (1)
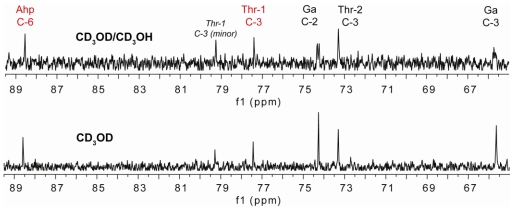
The organic extracts from separate samples of the marine cyanobacterium L. cf. confervoides collected from two different locations from reefs near Ft. Lauderdale (Florida, USA) were subjected to HP-20 fractionation and subsequent HPLC purification to yield either largamide D (2) [24] or a new intramolecular condensation product, largamide D oxazolidine (1) (Figure 1). The structure of 2 was established by comparison with reported data [24]. Compound 1 displayed a pseudomolecular ion [M + Na]+ peak at m/z 1236.4745 and an [M + 2 + Na]+ isotope peak of similar intensity characteristic for bromine, which was consistent with a molecular formula of C56H80BrN9NaO16. The 1H and 13C NMR spectra (Table 1), featuring typical signals for a peptide, were suggestive of a close analog of largamide D (2). A detailed analysis of the 2D NMR (COSY, HSQC, HMBC and TOCSY) spectral data of 1 in DMF-d7 clearly supported the structural fragments and enabled the connectivity of the depsipeptide along with the side chain, similar to 2. The major chemical shift differences between 1 and 2 were observed around the Ahp and Thr-1 units. The remaining 1H and 13C NMR data of 1 in MeOH-d4 (Table 1) were in close agreement with the reported data for 2 [24]. Most pronounced were the ~11 ppm downfield shifts relative to 2 of the signals for C-6 of the Ahp unit (δC 88.6) and C-3 of Thr-1 (δC 77.4), indicative of alkylation and, to be in agreement with the molecular formula, consistent with a formal condensation of Ahp and Thr-1 resulting in an oxazolidine and overall in a fused bicyclic system. A 13 A 13C NMR experiment carried out in a mixture of CD3OD/CD3OH clearly showed doubling of signals attributable to carbons attached to the hydroxyl groups in glyceric acid (Ga, δC 74.2 and 65.6). Singlets at δC 88.6 (C-6, Ahp) and 77.4 (C-3, Thr-1) supported the absence of exchangeable protons in both Ahp and Thr-1 where cyclization has occurred (Figure 3). A second set of resonances in a solvent-dependent ratio (1:0.3 in DMF-d7 and 1:0.7 in MeOH-d4) was observed in the NMR spectra, indicating the presence of conformers.
Table 1.
NMR spectral data for the major conformer of largamide D oxazolidine (1) and largamide D (2) [24].
| Unita | 1 (DMF-d7) δH (J in Hz) |
δC | COSY | HMBC | 1 (methanol-d4) δH (J in Hz) |
δC | 2 (methanol-d4) δH (J in Hz) |
δC | |
|---|---|---|---|---|---|---|---|---|---|
| Val-1 | 1 | 171.4 | 173.6 | 174.8 | |||||
| 2 | 4.53, m | 56.8 | H-3, NH | 1, 3, 4, 5, 1 (N-Me-Br-Tyr) | 4.72, m | 58.1 | 4.49, d (7.4) | 58.9 | |
| 3 | 2.32, m | 31.1 | H-2, H3-4, H3-5 | 2, 4, 5 | 2.38, m | 32.7 | 2.15, m | 31.8 | |
| 4 | 0.92, d (7.0) | 19.0 | H-3 | 2, 3, 5 | 0.92, d (7.2) | 18.8 | 0.93, d (6.9) | 18.9 | |
| 5 | 0.86, d (7.0) | 17.5 | H-3 | 2, 3, 4 | 0.89, d (7.2) | 18.0 | 0.99, d (6.9) | 19.8 | |
| NH | 6.76, d (8.3) | H-2 | |||||||
| N-Me- Br-Tyr | 1 | 169.2 | 170.9 | 171.4 | |||||
| 2 | 5.71, dd (11.5, 2.5) | 62.5 | H-3a/3b | 1 | 5.75, dd (11.5, 2.4) | 63.9 | 5.06, dd (11.5, 3.1) | 62.6 | |
| 3a | 3.34, dd (−14.3, 11.5) | 32.4 | H-2, H-3b H-2, H-3a |
2, 4 | 3.28, dd (−14.4, 2.4) | 33.5 | 3.40, dd (−14.6, 3.1) | 33.8 | |
| 3b | 2.89, dd (−14.3, 2.5) | 2.84, dd (−14.4, 11.5) | 2.80, m | ||||||
| 4 | 130.5 | 131.4 | 130.4 | ||||||
| 5 | 7.44, d (1.9) | 133.8 | 4, 6, 7, 9 | 7.32, d (1.2) | 134.8 | 7.34, d (2.0) | 134.8 | ||
| 6 | 109.3 | 111.2 | 110.8 | ||||||
| 7 | 153.6 | 154.7 | 154.4 | ||||||
| 8 | 7.03, d (8.2) | 116.7 | H-9 | 4, 6, 7, 9 | 6.83, d (8.3) | 117.7 | 6.87, d (8.2) | 117.6 | |
| 9 | 7.13, dd (8.2, 1.9) | 129.9 | H-8 | 5, 8 | 7.02, dd (8.3, 1.2) | 130.8 | 7.17, dd (8.2, 2.0) | 130.7 | |
| N-Me | 2.92, s | 30.8 | 1, 2, 1 (Thr-1) | 2.90, s | 31.8 | 2.87, s | 31.2 | ||
| Thr-1a | 1 | 169.7 | 172.0 | 173.0 | |||||
| 2 | 4.04, d (3.4) | 59.5 | H-3 | 1, 3, 2 (Ahp), H-6 (Ahp) | 3.96, d (2.3) | 61.0 | 4.57, d (6.8) | 55.9 | |
| 3 | 4.36, qd (6.4, 3.4) | 76.4 | H3-4 | 1, 2, 4 | 4.38, dq (6.2, 2.3) | 77.4 | 3.74, m | 66.6 | |
| 4 | 0.71, d (6.4) | 19.4 | H-3 | 2, 3 | 0.70, d (6.2) | 20.5 | 0.57, d (6.2) | 19.6 | |
| Ahpa | 2 | 168.8 | 170.5 | 171.3 | |||||
| 3 | 4.63, ddd (12.4, 8.7, 4.9) | 50.3 | H-4a/4b, NH | 2, 4, 5, 6 | 4.63, dd (9.9, 5.3) | 51.7 | 4.64, dd (12.4, 6.4) | 50.7 | |
| 4a | 1.95, m | 24.6 | H-3, H-4b, H-5a/5b | 2, 3, 5, 6 | 1.97, m | 25.3 | 2.82, m | 22.0 | |
| 4b | 1.88, m | H-3, H-4a, H-5a/5b | 1.93, m | 1.88, m | |||||
| 5a | 2.38, m | 27.9 | H-4a/4b, H-5b, H-6 | 3, 4, 6 | 2.40, m | 29.2 | 2.04, m | 30.6 | |
| 5b | 1.68, m | H-4a/4b, H-5a, H-6 | 1.66, m | 1.87, m | |||||
| 6 | 5.01, dd (8.5, 6.2) | 87.4 | H-5a/5b | 2, 4, 5 | 5.03, dd (8.5, 6.5) | 88.6 | 5.33, brs | 76.9 | |
| NH | 7.22, d (8.7) | H-3 | 3, 1 (Leu) | ||||||
| Leu | 1 | 171.7 | 174.6 | 174.4 | |||||
| 2 | 4.49, m | 51.4 | H-3a/3b | 1, 3 | 4.58, dd (11.0, 3.0) | 52.6 | 4.56, dd (8.5, 3.3) | 52.9 | |
| 3a | 1.84, m | 40.6 | H-2, H-3b | 1, 2, 4 | 1.88, m | 41.3 | 1.98, ddd (12.6, 12.6, 3.3) | 40.2 | |
| 3b | 1.47, m | H-2, H-3a | 1.47, m | 1.55, m | |||||
| 4 | 1.61, m | 24.5 | H-3a/3b, H3-5, H3-6 | 2, 3, 5, 6 | 1.56, m | 25.8 | 1.65, m | 25.8 | |
| 5 | 0.87, d (6.6) | 22.9 | H-4 | 4 | 0.91, d (6.0) | 23.8 | 0.87, d (6.8) | 20.0 | |
| 6 | 0.82, d (6.6) | 20.8 | H-4 | 4 | 0.83, d (6.0) | 21.5 | 0.97, d (6.7) | 23.7 | |
| NH | 8.50, d (8.5) | H-2 | 2, 1 (Thr-2) | ||||||
| Thr-2 | 1 | 169.7 | 171.1 | 170.9 | |||||
| 2 | 4.92, dd (9.0, 1.7) | 55.4 | NH | 1, 3, 1 (Val-2) | 4.72, m | 56.7 | 4.78, d (1.2) | 56.3 | |
| 3 | 5.78, qd (6.0, 1.7) | 72.1 | H3-4 | 1, 2, 4, 1 (Val-1) | 5.64, qd (6.5, 1.4) | 73.2 | 5.59, qd (6.5, 1.2) | 73.4 | |
| 4 | 1.33, d (6.0) | 16.6 | H-3 | 2, 3 | 1.30, d (6.5) | 17.7 | 1.36, d (6.5) | 18.1 | |
| NH | 8.25, d (9.0) | H-2 | 2, 1 (Val-2) | ||||||
| Val-2 | 1 | 171.7 | 173.8 | 173.9 | |||||
| 2 | 4.53, dd (8.6, 5.5) | 58.0 | H-3, NH | 1, 3, 4, 5 | 4.30, d (7.8) | 60.2 | 4.39, d (7.4) | 59.7 | |
| 3 | 2.17, m | 31.2 | H-2, H3-4, H3-5 | 2, 4, 5 | 2.09, dd (7.8, 6.8) | 32.0 | 2.17, m | 31.9 | |
| 4 | 0.90, d (7.0) | 18.5 | H-3 | 2, 3, 5 | 0.94, d (6.8) | 18.8 | 0.98, d (6.9) | 18.5 | |
| 5 | 0.87, d (7.0) | 17.5 | H-3 | 2, 3, 4 | 0.90, d (6.8) | 19.4 | 0.98, d (6.9) | 19.8 | |
| NH | 7.76, d (8.6) | H-2 | 2, 1 (Ala) | ||||||
| Ala | 1 | 172.0b | 174.8 | 174.4 | |||||
| 2 | 4.49, dq (9.0, 7.2) | 48.9 | H-3, NH | 1, 3 | 4.38, q (7.0) | 49.0 | 4.45, m | 49.9 | |
| 3 | 1.30, d (7.2) | 17.0 | H-2 | 1, 2 | 1.33, d (7.0) | 17.8 | 1.38, d (7.2) | 17.6 | |
| NH | 8.26, d (9.0) | H-2 | 2, 1 (Ahppa) | ||||||
| Ahppa | 1 | 171.9b | 174.1 | 173.5 | |||||
| 2 | 4.52, m | 52.6 | H-3a/3b, NH | 1, 3, 4, NH | 4.41, m | 54.4 | 4.46, m | 53.9 | |
| 3a | 1.89, m | 32.4 | H-2, H-3b, H-4a/4b | 2, 4, 5 | 1.87, m | 32.7 | 1.91, m | 32.6 | |
| 3b | 1.73, m | H-2, H-3a, H-4a/4b | 1.69, m | 1.73, m | |||||
| 4a | 1.68, m | 28.2 | H-3a/3b, H-4b | 2, 3, 5, 6 | 1.68, m | 29.0 | 1.70, m | 28.8 | |
| 4b | 1.62, m | H-3a/3b, H-4a | 1.63, m | 1.66, m | |||||
| 5 | 2.52, m | 34.2 | H-4a/4b | 3, 4, 6 | 2.54, m | 35.6 | 2.57, t (7.3) | 35.2 | |
| 6 | 132.6 | 134.0 | 133.6 | ||||||
| 7/11 | 7.03, d (8.3) | 129.8 | H-8/10 | 6, 8/10, 9 | 7.00, d (8.4) | 130.4 | 7.02, dd (8.2, 1.8) | 130.2 | |
| 8/10 | 6.76, d (8.3) | 115.2 | H-7/11 | 7/11, 9 | 6.67, d (8.4, 2.4) | 116.1 | 6.70, dd (8.2, 1.8) | 115.8 | |
| 9 | 156.0 | 7/11, 8/10 | 156.5 | 156.2 | |||||
| 9-OH | 9.36, s | 8/10, 9 | |||||||
| NH | 7.87, d (7.6) | H-2 | 1, 2, 1 (Ga) | ||||||
| Ga | 1 | 172.4 | 175.4 | 174.9 | |||||
| 2 | 4.11, m | 73.5 | H-3a/3b | 1, 3 | 4.11, d (3.5) | 74.2 | 4.13, t (3.8) | 73.9 | |
| 2-OH | c | ||||||||
| 3a | 3.76, dd (−10.9, 3.5) | 64.7 | H-2, H-3b | 1, 2 | 3.78, 2H, d (3.5) | 65.6 | 3.79, 2H, d (3.8) | 65.2 | |
| 3b | 3.70, dd (−10.9, 5.6) | H-2, H-3a |
The initial numbering from largamide D. (2) has been retained for comparison of data [24].
Can be interchanged.
Not observed.
Figure 3.
Expanded regions of the 13C NMR spectra (125 MHz) of 1 in CD3OD (bottom panel) and 1:1 CD3OD/CD3OH (top) showing peak splitting for carbons attached to the free hydroxyl groups.
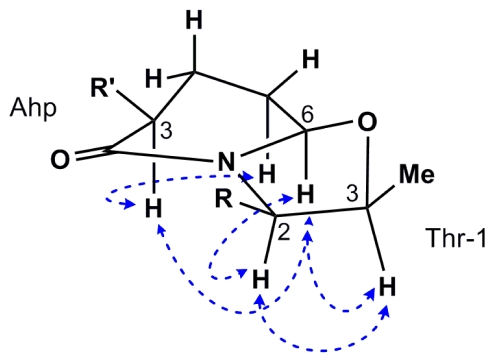
Based on the comparison of the 1H and 13C NMR data between 1 and 2 in methanol, we propose the same configuration for 1 as reported for largamide D (2) [24] for all unchanged residues. The oxazolidine heterocycle formation apparently forced the Ahp unit to adopt a twisted boat-like conformation (Figure 4) as evidenced by the ROESY correlations of H-3 with H-5b and H-6, all of which had to be in pseudo-axial position on the same face of the ring. This conclusion is consistent with the observed coupling constants for H-3 (12.4 and 4.9 Hz) and two relatively large coupling constants for H-6 (8.5 and 6.2 Hz) which may be rationalized in this conformation (Figure 4). Assuming 1 retained the same configuration at C-3 of the Ahp as in 2—since it is unaffected by cyclization—we propose a 3S,6R configuration for Ahp, which is identical to largamide D (2) [24]. Furthermore, ROESY correlations from Ahp H-6 to H-2 and H-3 of Thr-1 indicated that they are placed on the same side of the oxazolidine ring, providing evidence for a 2S,3S configuration for former Thr-1. Since the configuration of Thr-1 has been established as 2S,3R in 2, the configuration at C-3 is inverted in compound 1 and represents the only configurational change compared with largamide D (2).
Figure 4.
Selected key ROESY correlations among Ahp and Thr-1 for 1.
The inversion of C-3 configuration (Thr-1) suggests that the OH group of Ahp in 2 may have acted as a nucleophile to form the oxazolidine via nucleophilic substitution at C-3 of Thr-1 (rather than the opposite way), and this event could have been preceded by addition of an unknown leaving group at C-3 or the 3-OH group. Alternatively, Thr-1 could have been dehydrated to a 2-amino-2-butenoic acid (Abu) as encountered in lyngbyastatins, also produced by this cyanobacterium, which then served as a Michael acceptor, while the chiral environment induced the formation of a single diastereomer, compound 1. While it is possible that compound 1 is derived from 2 as an isolation artifact, it is tempting to postulate that 1 is a plausible biosynthetic product. In favor of this assumption we note that 1 was not found in the samples of L. cf. confervoides that yielded largamide D (2) and vice versa from the two different collection sites. To the best of our knowledge this fused oxazolidine-containing bicyclic system is unprecedented in cyanobacteria but is found in diterpene alkaloids from Spiraea japonica [29] and in several polyketide compounds from Actinomycete spp. [30–32].
2.3. Serine Protease Inhibition Study
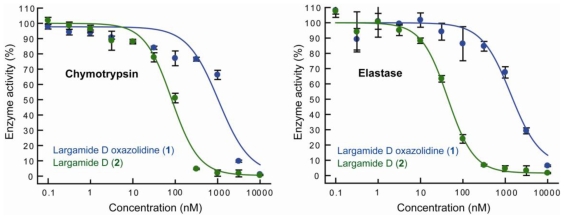
The presence of an Ahp unit is characteristic for many serine protease inhibitors, including lyngbyastatins. Largamide D (2) was previously reported to be a moderate chymotrypsin inhibitor [24]. We directly compared the activities of compound 1 and largamide D (2) against two serine proteases, chymotrypsin and porcine pancreatic elastase (Figure 5, Table 2). Compound 1 exhibited 11-fold and 33-fold reduced activity against chymotrypsin and elastase, respectively, indicating that the condensation of Ahp and Thr-1 considerably deactivated largamide D (2) and to different extents for both enzymes tested.
Figure 5.
Effect of compounds 1 and 2 on chymotrypsin and elastase activity.
Table 2.
Serine protease inhibitory activities (IC50, μM) of compounds 1 and 2.
| Chymotrypsin | Elastase | |
|---|---|---|
| Largamide D (2) | 0.083 ± 0.008 | 0.045 ± 0.003 |
| Largamide D oxazolidine (1) | 0.928 ± 0.093 | 1.52 ± 0.08 |
2.4. Conclusion
Marine cyanobacteria produce secondary metabolites that function as chemical defenses against consumers. Many of these compounds are serine protease inhibitors that could be responsible for the antifeedant activity observed for L. cf. confervoides from Fort Lauderdale reefs. Future studies should show the contribution of each serine protease inhibitor or other secondary metabolite produced by this particular collection of L. cf. confervoides to the observed antifeedant activity of the extracts. Furthermore, the isolation of compound 1 suggests that intramolecular condensation can modulate the inhibitory activity. The Ahp moiety is oftentimes critical for protease inhibition, and structural and conformational changes involving this unit are expected to affect activity. If indeed biosynthetically driven, this represents an endogenous pathway to modulate enzymatic activities. In turn, if this process is reversible it could unlock a more potent inhibitor.
3. Experimental Section
3.1. General Experimental Procedures
1H and 2D NMR spectra for largamide D oxazolidine were acquired in DMF-d7 or MeOH-d4 on a Bruker 600 MHz spectrometer using residual solvent signals as the internal standard (δH 8.02, δC 162.3 for DMF-d7 and δH 3.30 δC 49.0 for MeOH-d4). HSQC experiments were optimized for 1JCH = 145 Hz, and HMBC experiments were optimized for nJC,H = 7 Hz. 13C NMR experiments were performed on a Bruker 500 MHz spectrometer (5 mm probe). HRMS data were obtained using an Agilent LC-TOF mass spectrometer equipped with an APCI/ESI multimode ion source detector, and low resolution mass spectra were obtained on a A3200 Q TRAP LC/MS/MS (hybrid triple quadrupole linear ion trap mass spectrometer, Applied Biosystems, USA) with an electrospray ionization (ESI) interface operated in positive mode. HPLC–based compound isolation was performed on a Shimadzu LC-20AT prominence LC system with peak detection by a Shimadzu SPD-M20A prominence diode array detector.
3.2. Marine Cyanobacterial Samples
Largamide D oxazolidine (1) was isolated from L. cf. confervoides samples collected at approximately 15 m depth from reefs near the Port Everglades Inlet, Fort Lauderdale, Florida, USA (26°05.9902′N, 80°05.0184′W) in August 2004 and May and August 2005. Largamide D (2) was isolated from L. cf. confervoides samples collected off the coast of Broward County (Fort Lauderdale and Pompano Beach, Florida, USA) (26°01.1414′N, 80°05.9973′W; 26°15.134′N, 80°03.908′W) at a depth of 7–15 m in July 2004 and August 2005. S. Golubic identified the cyanobacterium [20] and its 16S rDNA gene sequence has been reported [10,20].
3.3. Feeding Experiments
The fresh L. cf. confervoides collected from Broward County was returned to the laboratory and frozen immediately and then freeze-dried. The freeze-dried material was weighed and extracted three times in 1:1 ethyl acetate–methanol (non-polar) and then three times in 1:1 ethanol–water (polar). Each solvent mixture was left on the cyanobacterium for 24 hours and then exchanged for fresh solvent. Each of the three extracts of the same solvents were pooled and dried down under vacuum.
Fish feeding experiments were conducted with a natural assemblage of reef fish in the field at Golden Reef (7 m depth) in Belize (GPS: N 16° 48.575, W 088° 05.138) as previously described [25]. This site was selected because the reef fish assemblages were representative of Florida and Caribbean coral reefs, our methods were well worked out at this site and large numbers of tropical reef fishes would feed during the assay. Feeding experiments were conducted with agar-based food, composed of 5 g of fish food (Kent Platinum Reef Herbivore), 1.25 g of agar, 1.25 g of carrageenan, and mixed with 100 mL of boiling water. This mixture was poured warm into 1 cm3 molds and allowed to cool. The two treatment foods were made by incorporating a natural concentration of the extract (polar or non-polar) dissolved in 5 mL of ethanol into the agar mixture just before it was poured. Solvent only (5 mL of ethanol) was added to the control food. Most of the ethanol evaporated as the agar cooled and gelled. Four agar food cubes were attached with safety pins to each polypropylene rope and offered to fish in groups of three ropes (two treatments (one with nonpolar extract and one with polar extract incorporated at natural dry mass concentrations) and one control). Nineteen (19) replicate groups of three ropes were distributed around the reef. Fish including Thalassoma bifasciatum, Scarus iserti, and Acanthurus chirurgus were observed feeding on the food cubes. In the field the number of agar cubes eaten was recorded. Results were analyzed with Friedman’s random block test followed by a post-hoc multiple comparisons using the Student Newman Keuls method.
Feeding experiments with the sea urchin Diadema antillarum were conducted in the laboratory at Carrie Bow Cay in Belize. Individuals of D. antillarum were collected from the reef flat adjacent to the island and maintained individually in 5 gallon buckets with flow through seawater delivered to each bucket. The urchins were fed a small amount of Padina sp. before the experiment to ensure that they were not starved. The urchins were offered L. cf. confervoides extracts incorporated into artificial food that consisted of 2 g of dried and powdered Gracilaria and 1 g of agar. The dried ingredients were mixed with 40 mL of boiling water and either the polar or non-polar extract dissolved in 5 mL of ethanol or the ethanol alone was added to the agar mixture while it was cooling. The artificial food was poured as 2 × 35 cm strips on a sheet of window screen. The window screen was cut so that each urchin was offered a 2 × 3 cm strip of each food type, each of which covered 120 squares of the screen. The food was left with each urchin for approximately 24 hours. At the end of the experiment the number of window screen squares that were no longer covered by the agar food was counted. If the food was completely consumed or not consumed at all, that replicate urchin was not included in the statistical analysis. The number of squares eaten was recorded for each food type and was statistically compared using a Friedman’s random block test, followed by a post-hoc multiple comparisons using the Student Newman Keuls method.
3.4. Extraction and Isolation
The extraction and initial fractionation procedures of L. cf. confervoides samples collected from Port Everglades Inlet, Fort Lauderdale, Florida, are described in the isolation of tiglicamides [33]. The tiglicamide-rich fractions [33] were collected and subjected to repeated semi-preparative reversed-phase HPLC (Phenomenex Synergi 4 μm Hydro-RP, 250 × 10 mm, 2.0 mL/min; PDA detection at 200–400 nm) using two sequential linear gradients of MeOH in 0.05% aqueous TFA (60–90% over 25 min, 90–100% over 10 min) to furnish mixtures containing 1 and tiglicamides A–C. The final purification of 1 was achieved by using a Phenomenex Luna Phenyl-hexyl column 250 × 10 mm while maintaining the same HPLC conditions described above (1, tR 25.5 min; 6.7 mg). The extraction and isolation of L. cf. confervoides samples collected from Broward County and Pompano Beach, Florida, are described in the isolation of lynbyastatin 4 [21]. The fraction eluting with 25% aqueous MeOH off a C18 Alltech SPE cartridge [21] was then purified by semi-preparative reversed-phase HPLC (YMC-Pack ODS-AQ, 250 × 10 mm, 2.0 mL/min; UV detection at 220 and 240 nm) using a MeOH–H2O linear gradient (20–100% over 70 min and then 100% MeOH for 10 min), to furnish largamide D (2, tR 53.0 min; 2 mg).
3.5. Largamide D oxazolidine (1)
Colorless amorphous solid; [α]20D –35 (c 0.1, MeOH), UV (MeOH) λmax (log ɛ) 204 (4.23), 214 (sh) (4.02), 230 (sh) (3.71), 280 (3.22) nm; NMR data, see Table 1; HRESI/APCIMS m/z [M + Na]+ 1236.4745, 1238.4742 (ratio 1:1.2, calcd for C56H8079BrN9NaO16, 1236.4804; C56H8081BrN9NaO16, 1238.4784).
3.6. Protease Inhibition Assays
Elastase assay was assessed using high-purity porcine pancreatic elastase (Elastin Products Company, EC134, 135 units/mg). The assay buffer used was 1M Tris-HCl (pH 8.0). Assay buffer (79 μL), elastase solution (75 μg/mL in assay buffer, 5 μL) and various concentrations of 1 and 2 (1 μL, dissolved in DMSO) were pre-incubated for 15 min at room temperature in a microtiter plate. After this time, 15 μL of substrate solution (2 mM N-succinyl-Ala-Ala-Ala-p-nitroanilide in assay buffer) was added to each well and the reaction was followed by measuring the absorbance at 405 nm every 30 s. Inhibition of chymotrypsin was determined using the α-chymotrypsin from bovine pancreas (Sigma, C4129, 55 units/mg). The assay buffer was 50 mM Tris-HCl, 100 mM NaCl and 1 mM CaCl2 (pH 7.8). Assay buffer (39 μL), enzyme solution (100 μg/mL in assay buffer, 10 μL), and various concentrations of 1 and 2 (1 μL, dissolved in DMSO) were pre incubated for 30 min at room temperature before substrate solution (1.5 mM N-succinyl-Gly-Gly-Phe-p-nitroanilide in assay buffer) was added. The reaction was followed by measuring the absorbance at 405 nm every 30 s for 30 min. For each assay, enzyme activity of each well was calculated using the initial slope of each progress curve, expressed as a percentage of the slope of uninhibited reaction. All assays were carried out in triplicate at ambient temperature (T = 29 ºC). Molassamide, which also causes complete enzyme inhibition, was used as a positive control for inhibition of elastase (IC50 32 nM) and chymotrypsin (IC50 234 nM) [12]. Dose–response curve fitting was done using Xlfit Excel, MathlQ Version 2.2.2 (IDBS Ltd.).
Acknowledgements
This work was funded by NIGMS grant P41GM086210. Research conducted in Belize was supported by the Smithsonian Marine Science Network. We thank A. Erickson, G. Harrison, C. Ross, K. Arthur, S. Reed, and A. Capper for assistance with collections and extraction of the cyanobacterium over many years, which allowed us to thoroughly investigate its natural products chemistry. This is contribution #820 from the Smithsonian Marine Station at Fort Pierce and contribution #883 from the Caribbean Coral Reef Ecosystems Program.
Footnotes
Supplementary Material Available: NMR spectra of compound 1.
References
- 1.Cardellina JH, Marner FJ, Moore RE. Seaweed dermatitis: structure of lyngbyatoxin A. Science. 1979;204:193–195. doi: 10.1126/science.107586. [DOI] [PubMed] [Google Scholar]
- 2.Botes DP, Kruger H, Viljoen CC. Isolation and characterization of four toxins from the blue-green alga, Microcystis aeruginosa. Toxicon. 1982;20:945–954. doi: 10.1016/0041-0101(82)90097-6. [DOI] [PubMed] [Google Scholar]
- 3.Ohtani I, Moore Moore RE, Maria TC. Cylindrospermopsin: a potent hepatotoxin from the blue-green alga Cylindrospermopsis raciborskii. J. Am. Chem. Soc. 1992;114:7941–7942. [Google Scholar]
- 4.Carmichael WW. Cyanobacteria secondary metabolites–The cyanotoxins. J. Appl. Bacteriol. 1992;72:445–459. doi: 10.1111/j.1365-2672.1992.tb01858.x. [DOI] [PubMed] [Google Scholar]
- 5.Humpage A. Toxin types, toxicokinetics and toxicodynamics. In: Hudnell HK, editor. Cyanobacterial Harmful Algal Blooms: State of the Science and Research Needs, Advances in Experimental Medicine and Biology. Vol. 619. Springer; New York, NY, USA: 2008. pp. 383–415. [DOI] [PubMed] [Google Scholar]
- 6.Paul VJ, Arthur KE, Ritson-Williams R, Ross C, Sharp K. Chemical defenses: from compounds to communities. Biol. Bull. 2007;213:226–251. doi: 10.2307/25066642. [DOI] [PubMed] [Google Scholar]
- 7.Nagle DG, Paul VJ. Chemical defense of a marine cyanobacterial bloom. J. Exp. Mar. Biol. Ecol. 1998;225:29–38. [Google Scholar]
- 8.Nagle DG, Paul VJ. Production of secondary metabolites by filamentous tropical marine cyanobacteria: ecological functions of the compounds. J. Phycol. 1999;35:1412–1421. [Google Scholar]
- 9.Capper A, Cruz-Rivera E, Paul VJ, Tibbetts IR. Chemical deterrence of a marine cyanobacterium against sympatric and non-sympatric consumers. Hydrobiologia. 2006;553:319–326. [Google Scholar]
- 10.Sharp K, Arthur KE, Gu L, Ross C, Harrison G, Gunasekera SP, Meickle T, Matthew S, Luesch H, Thacker RW, Sherman DH, Paul VJ. Phylogenetic and chemical diversity of three chemotypes of bloom-forming Lyngbya species (cyanobacteria: Oscillatoriales) from reefs of southeastern Florida. Appl. Environ. Microbiol. 2009;75:2879–2888. doi: 10.1128/AEM.02656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linington RG, Edwards DJ, Shuman CF, McPhail KL, Matainaho T, Gerwick WH. Symplocamide A, a potent cytotoxin and chymotrypsin inhibitor from the marine cyanobacterium Symploca sp. J. Nat. Prod. 2008;71:22–27. doi: 10.1021/np070280x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunasekera SP, Miller MW, Kwan JC, Luesch H, Paul VJ. Molassamide, a depsipeptide serine protease inhibitor from the marine cyanobacterium Dichothrix utahensis. J. Nat. Prod. 2010;73:459–462. doi: 10.1021/np900603f. [DOI] [PubMed] [Google Scholar]
- 13.Kwan JC, Taori K, Paul VJ, Luesch H. Lyngbyastatins 8–10, elastase inhibitors with cyclic depsipeptide scaffolds isolated from the marine cyanobacterium Lyngbya semiplena. Mar. Drugs. 2009;7:528–538. doi: 10.3390/md7040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banker R, Carmeli S. Inhibitors of serine proteases from a waterbloom of the cyanobacterium Microcystis sp. Tetrahedron. 1999;55:1083–10844. [Google Scholar]
- 15.Grach-Pogrebinsky O, Sedmak BC. Protease inhibitors from a Slovenian Lake Bled toxic waterbloom of the cyanobacterium Planktothrix rubescens. Tetrahedron. 2003;59:8329–8336. [Google Scholar]
- 16.Radau G. Cyanopeptides: A new and nearly inexhaustible natural resource for the design and structure-activity relationship studies of the new inhibitors of trypsin-like serine proteases. Curr. Enz. Inhib. 2005;1:295–307. [Google Scholar]
- 17.Baumann HI, Keller S, Wolter FE, Nicholson GJ, Jung G, Süssmuth RD, Friedrich J. Planktocyclin, a cyclooctapeptide protease inhibitor produced by the freshwater cyanobacterium Planktothrix rubescens. J. Nat. Prod. 2007;70:1611–1615. doi: 10.1021/np0700873. [DOI] [PubMed] [Google Scholar]
- 18.Nakano Y, Shirai M, Mori N, Nakano M. Neutralization of microcystin shock in mice by tumor necrosis factor alpha antiserum. Appl. Environ. Microbiol. 1991;57:327–330. doi: 10.1128/aem.57.1.327-330.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schatz D, Keren Y, Vardl A, Sukenlk A, Carmeli S, Börner T, Dittmann E, Kaplan A. Towards clarification of the biological role of microcystins, a family of cyanobacterial toxins. Environ. Microbiol. 2007;9:965–970. doi: 10.1111/j.1462-2920.2006.01218.x. [DOI] [PubMed] [Google Scholar]
- 20.Paul VJ, Thacker RW, Banks K, Golubic S. Benthic cyanobacterial bloom impacts the reefs of South Florida (Broward county, USA) Coral Reefs. 2005;24:693–697. [Google Scholar]
- 21.Matthew S, Ross C, James RR, Paul VJ, Luesch H. Lyngbyastatin 4, a dolastatin 13 analogue with elastase and chymotrypsin inhibitory activity from the marine cyanobacterium Lyngbya confervoides. J. Nat. Prod. 2007;70:124–127. doi: 10.1021/np060471k. [DOI] [PubMed] [Google Scholar]
- 22.Taori K, Matthew S, Ross C, James RR, Paul VJ, Luesch H. Lyngbyastatin 5–7, potent elastase inhibitors from Floridian marine cyanobacteria, Lyngbya spp. J. Nat. Prod. 2007;70:1593–1600. doi: 10.1021/np0702436. [DOI] [PubMed] [Google Scholar]
- 23.Matthew S, Ross C, Paul VJ, Luesch H. Pompanopeptins A and B, new cyclic peptides from the marine cyanobacterium Lyngbya confervoides. Tetrahedron. 2008;64:4081–4089. [Google Scholar]
- 24.Plaza A, Bewley CA. Largamides A–H, Unusual cyclic peptides from the marine cyanobacterium Oscillatoria sp. J. Org. Chem. 2006;71:6898–6907. doi: 10.1021/jo061044e. [DOI] [PubMed] [Google Scholar]
- 25.Gunasekera SP, Ritson-Williams R, Paul VJ. Carriebowmide, a new cyclodepsipeptide from the marine cyanobacterium Lyngbya polychroa. J. Nat. Prod. 2008;71:2060–2063. doi: 10.1021/np800453t. [DOI] [PubMed] [Google Scholar]
- 26.Cruz-Rivera E, Paul VJ. Chemical deterrence of a cyanobacterial metabolite against generalized and specialized grazers. J. Chem. Ecol. 2007;33:213–217. doi: 10.1007/s10886-006-9212-y. [DOI] [PubMed] [Google Scholar]
- 27.Thacker RW, Nagle DG, Paul VJ. Effects of repeated exposures to marine cyanobacterial secondary metabolites on feeding by juvenile rabbitfish and parrotfish. Mar. Ecol. Prog. Ser. 1997;147:21–29. [Google Scholar]
- 28.Capper A, Paul VJ. Grazer interactions with four species of Lyngbya in southeast Florida. Harmful Algae. 2008;7:717–728. [Google Scholar]
- 29.He H-P, Shen Y-M, Zhang J-X, Zuo G-Y, Hao X-J. New diterpene alkaloids from the roots of Spiraea japonica. J. Nat. Prod. 2001;64:379–380. doi: 10.1021/np0004911. [DOI] [PubMed] [Google Scholar]
- 30.Kunimoto S, Lu J, Esumi H, Yamazaki Y, Kinoshita N, Honma Y, Hamada M, Ohsono M, Ishizuka M, Takeuchi T. Kigamicins, novel antitumor antibiotics. I. Taxonomy, isolation, physico-chemical properties and biological activities. J. Antibiot. 2003;56:1004–1011. doi: 10.7164/antibiotics.56.1004. [DOI] [PubMed] [Google Scholar]
- 31.Hopp DC, Milanowski DJ, Rhea J, Jacobsen D, Rabenstein J, Smith C, Romari K, Clarke M, Francis L, Irigoyen M, Luche M, Carr GJ, Mocek U. Citreamicins with potent gram-positive activity. J. Nat. Prod. 2008;71:2032–2035. doi: 10.1021/np800503z. [DOI] [PubMed] [Google Scholar]
- 32.Asai Y, Nonaka N, Suzuki S-I, Nishio M, Takahashi K, Shima H, Ohmori K, Ohnuki T, Komatsubara S. TMC-66, a new endothelin converting enzyme inhibitor produced by Streptomyces sp. A5008. J. Antibiot. 1999;52:607–612. doi: 10.7164/antibiotics.52.607. [DOI] [PubMed] [Google Scholar]
- 33.Matthew S, Paul VJ, Luesch H. Tiglicamides A–C, Cyclodepsipeptides from the marine cyanobacterium Lyngbya confervoides. Phytochemistry. 2009;70:2058–2063. doi: 10.1016/j.phytochem.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]