Abstract
Eight marine cyanobacteria strains of the genera Cyanobium, Leptolyngbya, Oscillatoria, Phormidium, and Synechococcus were isolated from rocky beaches along the Atlantic Portuguese central coast and tested for ecotoxicity. Strains were identified by morphological characteristics and by the amplification and sequentiation of the 16S rDNA. Bioactivity of dichloromethane, methanol and aqueous extracts was assessed by the Artemia salina bioassay. Peptide toxin production was screened by matrix assisted laser desorption/ionization time of flight mass spectrometry. Molecular analysis of the genes involved in the production of known cyanotoxins such as microcystins, nodularins and cylindrospermopsin was also performed. Strains were toxic to the brine shrimp A. salina nauplii with aqueous extracts being more toxic than the organic ones. Although mass spectrometry analysis did not reveal the production of microcystins or other known toxic peptides, a positive result for the presence of mcyE gene was found in one Leptolyngbya strain and one Oscillatoria strain. The extensive brine shrimp mortality points to the involvement of other unknown toxins, and the presence of a fragment of genes involved in the cyanotoxin production highlight the potential risk of cyanobacteria occurrence on the Atlantic coast.
Keywords: marine cyanobacteria, cyanotoxins, ecotoxicology, Artemia salina, toxin genes
1. Introduction
Cyanobacteria are known for their diversity in terms of morphological, physiological and toxicological properties. Lately, the production of bioactive compounds with commercial and medical applications has also increased interest in these organisms [1]. In fact, together with the production of potent toxins, cyanobacteria produce many substances interesting in terms of antifungal, antibiotic and anticancerigenous activities [2,3].
The occurrence and ecotoxicology of cyanobacteria are particularly well documented in freshwater habitats [4,5]. In contrast, studies related to marine environments have not been performed to the same extent and are mainly from tropical regions, where cyanobacteria mass occurrences have been reported [6,7]. As described for cyanobacteria blooms in freshwater and brackish waters, the increase in cyanobacteria bloom formation registered in coastal areas has been attributed to factors such as high irradiation, high temperatures and increased nutrient loading, as a consequence of human population growth near these locations [8–11]. Similar to other littoral areas of the world, the Portuguese coast has been subjected to an increased human pressure and, consequently, coastal eutrophication. Eutrophication, powered by the phenomenon of global warming, increases the potential of marine cyanobacteria proliferation. As most coastal areas have a considerable importance for biodiversity, fish industry and tourism, it is of significant importance to also study the putative toxicity of cyanobacteria that occurs in this marine environment. Moreover, previous studies conducted with marine cyanobacteria isolated from north Portuguese rocky beaches, reported that strains of the genera Aphanotece, Oscillatoria, Phormidium, Synechococcus and Synechocystis caused toxic effects on mammals, marine invertebrates, growth inhibition of Gram-positive bacteria and cytotoxicity in human cell lines [12–15].
In this work we evaluated the potential toxicity of marine cyanobacteria isolated from the central coast of Portugal by means of biological, chemical and molecular biology analysis.
2. Materials and Methods
2.1. Cyanobacteria strains
Marine cyanobacteria strains were isolated from water samples and solid materials collected from rocky beaches on the central coastal area of Portugal [12] (Table 1). Strains were isolated and maintained in Z8 medium [16] supplemented with 20 g NaCl/L. Unicyanobacterial cultures were performed in 4 L Z8 medium at 25 ºC, irradiation of 10 μmol photons s−1 m−2 was provided by cool white fluorescent tubes and with a light/dark cycle of 14 h/10 h. Cells were harvested after one-month of growth by decantation for the non-floating strains, centrifugation for the floating strains and filtration through a 3 μm pore net for filamentous strains. Cyanobacterial biomass was washed repeatedly with deionized water to remove the excess of NaCl. Samples were frozen and freeze-dried and the material was stored at −20 ºC.
Table 1.
Strain codes, substrates from which strains were isolated, genera identification (highest similarities of the sequence analysis in GenBank) and accession numbers of the cyanobacteria strains included in this study.
| Strain code | Substrate | Genus (16S rRNA highest match NCBI) | % similarity | Accession Nº |
|---|---|---|---|---|
| LEGE 06005 | Sea water | Synechococcus sp. UH7 | 92 | HM124558 |
| LEGE 06007 | Rock | Phormidium sp. HBC9 | 99 | HM124560 |
| LEGE 06008 | Rock | Cyanobium sp. NS01 | 99 | HM124561 |
| LEGE 06009 | Rock | Leptolyngbya sp. 0BB32S02 | 96 | HM124563 |
| LEGE 06010 | Rock | Leptolyngbya sp. 0BB32S02 | 98 | HM124564 |
| LEGE 06011 | Rock | Cyanobium sp. NS01 | 98 | HM124557 |
| LEGE 06015 | Patella sp. | Cyanobium sp. NS01 | 99 | HM124559 |
| LEGE 06018 | Rock | Oscillatoria sp. CCAP 1459/26 | 100 | HM124562 |
2.2. Identification of cyanobacteria isolates
Cyanobacteria isolates were identified by their morphology and size of cells and colonies and by a molecular approach. Phenotypic identification was based on the phycological and bacterial guides [17–21]. Molecular identification of the isolates was based on PCR amplification and sequencing of the 16S rDNA gene.
Molecular identification
Total genomic DNA was extracted from freeze dried biomass using the commercial DNA extraction kit AquaPure kit (Bio-Rad, Hercules, USA) and following the manufacturer’s instructions. The set of primers 27F/809R and 740F/1494R were used for PCR amplification of the cyanobacteria 16S rDNA gene [22,23] (Table 2). Thermal cycling program was based on the work by Jungblut et al. [23], with the exceptions that 35 cycles were preformed with a final phase extension of 72 ºC for 5 min. Reactions were carried out in a 100 μL reaction volume that consisted of 1X Tampon solution (Bioline, 10x NH4 Reaction Buffer), 2.5 μL dNTPs (Bioline, 2.5 mM), 2.5 mM MgCl2 (Bioline, 50 mM), 0.5 pmol of each primer (10 pM/μL), 2.5 U of Taq DNA polimerase (Bioline), 69.5 μl sterile ultra-pure water and 2.5 μL of DNA sample. PCR products were extracted from gel with the Jet Quick Spin Column Technique kit (Genomed) according to the manufacturer’s instructions and sent for direct sequencing to STABVIDA®. Automated sequencing was performed using the BigDye Terminator Kit by Applied Biosystems and the 96-capillary 3730xL DNA Analyzer by Applied Biosystems. Sequences were analyzed using the BLAST system (http://www.ncbi.nlm.nih.gov/BLAST/).
Table 2.
PCR primers used for amplification of 16S rRNA gene for cyanobacteria identification and for the amplification of genes related to cyanotoxins production. A—Individual annealing temperature, B—Reference annealing temperature, bp = base pairs.
| Primer | Sequence (5′→3′) | A | B | Size (bp) | Amplified gene | Reference |
|---|---|---|---|---|---|---|
| 27F | AGAGTTTGATCCTGGCTCAG | 52 | 60 | 780 | 16S rRNA | [22] |
| 809R | GCTTCGGCACGGCTCGGGTCGATA | 64 | [23] | |||
| 740F | GGCYRWAWCTGACACTSAGGGA | - | 50 | 754 | 16S rRNA | [22] |
| 1494R | TACGGCTACCTTGTTACGAC | 56 | ||||
| mcyA-Cd F | AAAATTAAAAGCCGTATCAAA | 51 | 59 | 297 | Microcystin synthetase | [28] |
| mcyA-Cd R | AAAAGTGTTTTATTAGCGGCTCAT | 43 | ||||
| HEPF | TTTGGGGTTAACTTTTTTGGGCATAGTC | 57 | 52 | 472 | Microcystin/nodularin synthetase | [23] |
| HEPR | AATTCTTGAGGCTGTAAATCGGGTTT | 55 | ||||
| PKS M4 | GAAGCTCTGGAATCCGGTAA | 52 | 55 | 650 | Cylindrospermopsin polyketide synthase | [29] |
| PKS M5 | AATCCTTACGGGATCCGGTGC | 56 | [29] | |||
| M13 | GGCAAATTGTGATAGCCACGAGC | 57 | 55 | 597 | Cylindrospermopsin peptide synthetase | [29] |
| M14 | GATGGAACATCGCTCACTGGTG | 57 | [29] | |||
2.3. Toxin genes detection
In order to determine the presence of genes implicated in the production of cyanotoxins, we applied a range of molecular primers that are currently used for detection of genes involved in the production of microcystins (MC), nodularins (NOD) and cylindrospermopsin (CYN) (Table 2). The MC gene cluster, mcy, comprises 10 genes in two transcribed operons, mcyA-C and mcyD-J [24]. The NOD gene cluster, nda, consists of nine open reading frames (ndaA-I) [25]. For the detection of genes involved in MC and NOD production we used the HEP primers set. This primers have as target sequences the aminotransferase (AMT) domain, which is located on the modules mcyE and ndaF of the MC and NOD synthetase enzyme complexes, respectively [24,26]. For the detection of other genes implied in the production of MC, we used the primers mcyA-C, which detect the mcyA, mcyB and mcyC genes [27,28]. For detection of genes involved in the CYN production we used the polyketide synthase PKS M4 and M5 primers and the peptide synthetase M13 and M14 primers designed by Schembri et al. [29] who demonstrate a direct link between the presence of the polyketide synthase and peptide synthetase genes and the ability of cyanobacteria to produce CYN.
The conditions of the PCR reactions were as those described for the amplification of the 16S rRNA gene. Concerning the cycling conditions, for mcyA-Cd genes the thermal cycling conditions were 1 cycle at 95 ºC for 2 min, 35 cycles at 95 ºC for 90 s, 56 ºC for 30 s and 72 ºC for 50 s and 1 cycle at 72 ºC for 7 min. For HEP genes, the thermal cycling conditions were 1 cycle at 95 ºC for 2 min, 35 cycles at 95 ºC for 90 s, 55 ºC for 30 s and 72 ºC for 1 min and 1 cycle at 72 ºC for 7 min. For CYL genes, the thermal cycling conditions were 1 cycle at 95 ºC for 2 min, 35 cycles at 95 ºC for 90 s, 55 ºC for 30 s and 72 ºC for 1 min and 1 cycle 72 ºC for 7 min [27,28]. PCR products were purified as described previously for the 16S rRNA gene amplification. As a positive control for MC and NOD, a Microcystis aeruginosa strain from Lagoa de Mira, Portugal (M6 strain), and for CYN a Cylindrospermopsis raciborskii strain from Queensland, Australia (AQS strain) were used.
2.4. Peptides screening by MALDI-TOF MS
The production of oligopeptides such as MC was screened by matrix assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). Freeze dried biomass from the marine strains was screened for the production of oligopeptides by MALDI-TOF MS as described in Welker et al. [30]. By this analysis, several hundreds of different oligopeptides of different types can potentially be detected and identified by post-source-decay fragmentation.
2.5. Cyanobacteria extracts for Artemia salina bioassay
Cyanobacteria freeze-dried biomass (100 mg/mL) was extracted with a diclorometane:methanol (50:50; v:v) solution (dichloromethane extract), a 80% methanol solution (methanolic extract) and deionised water (aqueous extract). After solvent addition, solutions were sonicated for 3 × 20 s on ice and kept in the dark for 12 h for the dichloromethane extract and for 1.5 h for the methanolic and aqueous extracts. Extracts were centrifuged at 23 000 × g for 10 min. Supernatants were collected, evaporated to dryness and ressuspended in filtered natural seawater. Extracts were diluted with filtered natural seawater (75, 50 and 25%) and then filtered through a 0.22 μm filter.
2.6. Artemia salina bioassay
A. salina bioassay was performed in 96 wells polystyrene plates [31]. Artemia cysts (available in aquaristic shops) were incubated in 1 L of natural filtered seawater for 24 h. Cysts were maintained at 25 ºC with continuous light and aeration. Ten to fifteen individuals were placed into 200 μL of test solution in triplicate. Control was performed with natural filtered seawater. Plates were incubated in the dark for 48 h, with very low agitation. After 24 and 48 h, dead nauplii were counted and by the end of the assay total numbers of individuals were registered. Results were expressed as percentage of mortality and LC50 was calculated using the statistic software 8.0 of StatSoft Inc. 1984–2007 version.
3. Results
3.1. Cyanobacteria strains
Eight cyanobacteria strains were successfully isolated and studied (Table 1). Strains were isolated from seawater and solid material obtained from rock scrapings and Patella sp. shells. Taxonomic identification, based on morphology and on the sequencing of the 16S rRNA gene followed by a BLAST search, led to the coccoid Cyanobium and Synechococcus and to the filamentous Leptolyngbya, Oscillatoria and Phormidium. Strains code and the accession numbers of 16S rDNA sequences are presented in Table 1.
3.2. Detection of genes involved in toxin production
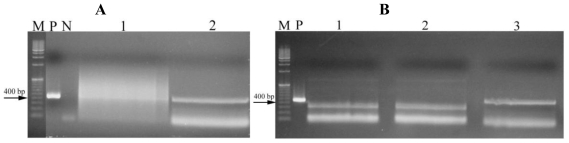
For most of the strains, no amplification of any of the toxin genes was obtained. Nevertheless, 439 and 431 base pair (bp) fragments were obtained with the HEP primers reaction for strains LEGE 06010 and LEGE 06018, Leptolyngbya sp. and Oscillatoria sp. respectively (Figure 1). In order to confirm the identity of the amplified fragments, the PCR products were sequenced. The sequences were compared with sequences from the GenBank and we found 99% similarity with the mcyE of the microcystin synthetase gene cluster from a Microcystis sp. CYN06 strain (Table 3).
Figure 1.
PCR products amplified from strains LEGE 06010 and LEGE 06018, Leptolyngbya sp. and Oscillatoria sp, respectively, with HEPF/HEPR primers. (A) Lane 1: strain LEGE 06025 (data not shown); Lane 2: strain LEGE 06010; M-1Kb; (B) Lanes 1 and 2: strain LEGE 06025 (data not shown); Lane 3: strain LEGE 06018. M: Marker; P: Positive control (M6 strain), N: Negative control.
Table 3.
Identification of toxin gene from the samples sequenced using the HEP primers for the strains LEGE 06010 and LEGE 06018. The highest similarities to the sequences in GenBank and Accession numbers are shown.
3.3. MALDI-TOF MS
Mass signals of known or putative peptides such as MC were not found for any of the eight tested strains. In all mass spectra, peaks at m/z 871 and m/z 593 for pheophytin and pheophorbide, respectively, were present, indicating that the extraction of the cells was efficient.
3.4. Artemia salina assay
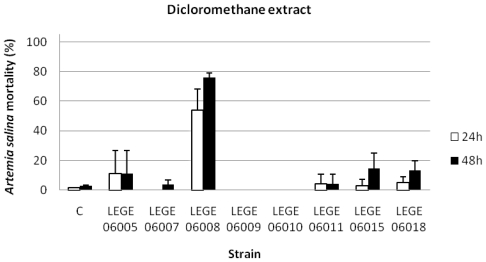
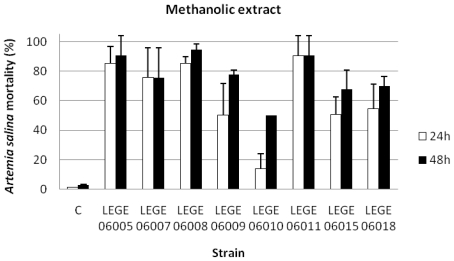
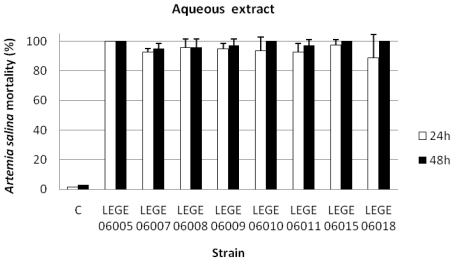
The results concerning the percentage of A. salina mortality exposed to the dichloromethane (Figure 2), the methanolic (Figure 3) and the aqueous extracts (Figure 4), showed that for the dichloromethane extract only the strain LEGE 06008 revealed mortality higher than 50%. Results concerning the methanolic and the aqueous extract were more pronounced for most of the strains, with strains LEGE 06005, LEGE 06010, LEGE 06015 and LEGE 06018 reaching 100% mortality with the aqueous extract. The average 24 h and 48 h LC50 for the methanolic extract were 50.8 mg/mL and 50.2 mg/mL, and the 24 h and 48 h LC50 for aqueous extract were on average 50.7 mg/mL and 50.5 mg/mL, respectively (Table 4).
Figure 2.
Mortality of Artemia salina (%) after 24 and 48 h exposure to dichloromethane extracts at a concentration of 100 mg/mL (C: control).
Figure 3.
Mortality of Artemia salina (%)after 24 and 48 h exposure to the methanolic extracts at a concentration of 100 mg/mL (C: control).
Figure 4.
Mortality of Artemia salina (%) after 24 and 48 h exposure to the aqueous extracts at a concentration of 100 mg/mL (C: control).
Table 4.
24 and 48 h LC50 values (mg/mL), for the tested cyanobacteria strains and extracts (D: dichloromethane extract; M: methanol extract; A: aqueous extract).
| Strain |
LEGE | LEGE | LEGE | LEGE | LEGE | LEGE | LEGE | LEGE | |
|---|---|---|---|---|---|---|---|---|---|
| Extract | 6005 | 6007 | 6008 | 6009 | 60010 | 6011 | 6015 | 6018 | |
| 24h | D | - | - | 51.4 | - | - | - | - | - |
| M | 51 | 51.3 | 50.6 | 50.9 | - | 49.9 | 50.8 | 51.4 | |
| A | 49.5 | 51.1 | 51 | 51.3 | 51.5 | 50.7 | 51 | 49.6 | |
| 48h | D | - | - | 51.2 | - | - | - | - | - |
| M | 50.8 | 50.7 | 49.9 | 50 | - | 48.3 | 50.7 | 51.2 | |
| A | 49.2 | 50.9 | 50.8 | 51.1 | 51.5 | 50.2 | 51 | 49.6 | |
4. Discussion
Marine cyanobacteria blooms are mostly reported from tropical and subtropical regions in spite that cyanobacteria blooms in freshwater ecosystems are common worldwide. In the absence of large amounts of biomass from natural samples, the isolation of strains and cultivation under laboratory conditions remains essential to study the putative toxicity of strains and to infer about the potential threat to humans and to ecosystems. In this work, we found that aqueous and organic extracts of marine cyanobacteria from the coccoid genera Cyanobium and Synechococcus and the filamentous genera Leptolyngbya, Oscillatoria and Phormidium isolated from the Portuguese coast induced acute toxicity in nauplii of the brine shrimp A. salina. Toxicity revealed by the nauplii mortality was evident for the aqueous extracts of all the cyanobacteria strains studied (Figure 4). Although the MALDI-TOF MS analysis did not reveal the presence of known peptides, our results indicate the production of toxic compounds with diverse chemical natures, since toxic results were obtained with the dichloromethane, the methanolic and the aqueous extracts. Taken into account that the toxic effects on A. salina were more evident in the aqueous extract, the risk due to the occurrence of toxic cyanobacteria is high, due to the potential availability of water soluble toxins in the water column.
For toxins such as MC, NOD and CYN, the molecular machinery involved in their production is already elucidated and the presence of toxin-coding genes has been considered an important tool for the study of the potential toxicity of cyanobacteria [24]. All these toxins are produced by a wide range of cyanobacteria genera that have been involved in severe poisoning episodes all over the world [4]. For the strains LEGE 06010 and LEGE 06018, Leptolyngbya and Oscillatoria, respectively, we could amplify a fragment which showed 99% similarity with the mcyE gene of a Microcystis sp. strain (Table 2). However, no other genes that constitute the microcystin synthetase gene cluster were identified. The lack of all components of the gene cluster responsible for the production of MC can justify the fact that the toxins were not identified by MALDI-TOF MS since, as it was previously demonstrated that the synthesis of MC requires the presence of the entire mcy gene cluster [32]. Rantala et al. [33] suggested an ancient origin of MC and reported the loss of genes and, as a consequence, a loss in the ability of some strains to produce toxin. It is known that the transfer of fragments of DNA is common among prokaryotes [34] and, in the particular case of cyanobacteria, genes responsible for toxin production might be subjected to horizontal gene transfer, gene losses [35], transposition, mutagenesis, deletion and recombination [36]. In this case we can hypothesize that deletion events may have occurred resulting in the loss of genes. Besides a possible loss of the genes during evolution, we cannot exclude the possible occurrence of mutations during cultivation of strains. As described by Kaebernick et al. [37], mutations within the mcy gene cluster might occur during cultivation leading to a decrease in toxicity. Without involving molecular analysis, it was already reported a decrease in the toxicity of strains when maintained in culture. Cyanobacteria seem to be more toxic in the natural environment than under laboratory conditions [38].
Among the mcy genes, a region of the mcyE has been used as a reliable molecular marker for the detection of MC producers [39]. This gene is related to enzymes that are involved in the synthesis of Adda and addiction of D-glutamate, which are both essential amino acids for the toxicity of MC. In phylogenetic studies it was shown that mcyE sequences from different producer genera form their own clusters and remain excluded from horizontal gene transfer [39]. This fact may explain the prevalence of this gene in the strains and the loss of others. Nevertheless, we should point out that we got in our two positive mcyE gene isolates, 99% similarity with the mcyE of the microcystin synthetase gene cluster from a Microcystis sp. CYN06 strain that belongs to a different order. This will be further investigated. The lack of positive results for NOD highlight the production of these toxins only by species of the genus Nodularia since this toxin seem to be produced only by species of this genus [36].
Although no peptides such as MC, NOD or other partially known peptides were identified by MALDI-TOF MS, we cannot exclude the presence of other toxins, since ecotoxicological assays were positive for many of the strains and extract types. Existing records on cyanotoxins production are mainly related to freshwater and brackish cyanobacteria [4], but there are some reports about their production by cyanobacteria in marine environments. Some authors reported the production of MC by marine strains of Synechocystis and Synechococcus, and by filamentous forms of Oscillatoria, Geitlerinema, Leptolyngbya, Phormidium, Pseudoanabaena and Spirulina respectively [12,40,41].
The increasing occurrence of cyanobacteria in large densities at maritime shores resulting from eutrophication and global warming poses a serious threat to humans and ecosystems. Clearly there is a need to increase our knowledge about toxin production by marine cyanobacteria strains, in order to prevent possible adverse effects of its occurrence.
Acknowledgements
This work was partially funded by the FCT project PTDC/MAR/102258/2008 and Atlantox (Interreg). Martin Welker (TU Berlin, Germany) is thanked for MALDI-TOF MS analyses of strain material.
Footnotes
Samples Availability: Available from the authors.
References
- 1.Tan LT. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry. 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Dittmann E, Neilan BA, Borner T. Molecular biology of peptide and polyketide biosynthesis in cyanobacteria. Appl. Microbiol. Biotechnol. 2001;57:467–473. doi: 10.1007/s002530100810. [DOI] [PubMed] [Google Scholar]
- 3.Volk RB. Screening of microalgae for species excreting norharmane, a manifold biologically active indole alkaloid. Microbiol. Res. 2008;163:307–313. doi: 10.1016/j.micres.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Codd GA, Morrison LF, Metcalf JS. Cyanobacterial toxins: risk management for health protection. Toxicol. Appl. Pharmacol. 2005;203:264–272. doi: 10.1016/j.taap.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Wiegand C, Pflugmacher S. Ecotoxicological effects of selected cyanobacterial secondary metabolites: a short review. Toxicol. Appl. Pharmacol. 2005;203:201–218. doi: 10.1016/j.taap.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Albert S, O’Neil JM, Udy JW, Ahern KS, O’Sullivan CM, Dennison WC. Blooms of the cyanobacterium Lyngbya majuscula in coastal Queensland, Australia: disparate sites, common factors. Mar. Pollut. Bull. 2005;51:428–437. doi: 10.1016/j.marpolbul.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Pittman SJ, Pittman KM. Short-term consequences of a benthic cyanobacterial bloom (Lyngbya majuscula Gomont) for fish and penaeid prawns in Moreton Bay (Queensland, Australia) Estuar. Coast. Shelf Sci. 2005;63:619–632. [Google Scholar]
- 8.Camargo JA, Alonso A. Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: A global assessment. Environ. Int. 2006;32:831–849. doi: 10.1016/j.envint.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Ahern KS, Ahern CR, Udy JW. In situ field experiment shows Lyngbya majuscula (cyanobacterium) growth stimulated by added iron, phosphorus and nitrogen. Harmful Algae. 2007;7:389–404. [Google Scholar]
- 10.Hakanson L, Bryhn AC, Hytteborn JK. On the issue of limiting nutrient and predictions of cyanobacteria in aquatic systems. Sci. Total Environ. 2007;379:89–108. doi: 10.1016/j.scitotenv.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Pliński M, Mazur-Marzec H, Jóźwiak T, Kobos J. The potential causes of cyanobacterial blooms in Baltic Sea estuaries. Oceanol. Hydrobiol. Stud. 2007;36:125–137. [Google Scholar]
- 12.Martins R, Pereira P, Welker M, Fastner J, Vasconcelos VM. Toxicity of culturable cyanobacteria strains isolated from the Portuguese coast. Toxicon. 2005;46:454–464. doi: 10.1016/j.toxicon.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Martins R, Fernandez N, Beiras R, Vasconcelos V. Toxicity assessment of crude and partially purified extracts of marine Synechocystis and Synechococcus cyanobacterial strains in marine invertebrates. Toxicon. 2007;50:791–799. doi: 10.1016/j.toxicon.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Selheim F, Herfindal L, Martins R, Vasconcelos V, Doskeland SO. Neuro-apoptogenic and blood platelet targeting toxins in benthic marine cyanobacteria from the Portuguese coast. Aquat. Toxicol. 2005;74:294–306. doi: 10.1016/j.aquatox.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Martins RF, Ramos MF, Herfindal L, Sousa JA, Skaerven K, Vasconcelos VM. Antimicrobial and cytotoxic assessment of marine cyanobacteria - Synechocystis and Synechococcus. Mar. Drugs. 2008;6:1–11. [PMC free article] [PubMed] [Google Scholar]
- 16.Kotai J. Instructions for Preparation of Modified Nutrient Solution Z8 for Algae. Norwegian Institute for Water Research B-11769; Blindern, Oslo, Norway: 1972. p. 5. [Google Scholar]
- 17.Baker P. Identification of Common Noxious Cyanobacteria, Part II Chroococcales, Oscillatoriales. Urban Water Research Association of Australia; Melbourne, Australia: 1992. [Google Scholar]
- 18.Rippka R. Recognition and Identification of Cyanobacteria, Methods in Enzymology. Academic press Inc; New York, NY, USA: 1998. pp. 28–67. [Google Scholar]
- 19.Komárek J, Anagnostidis K. Cyanoprokariota. 1. Teil: Chroococcales. Gustav Fischer; Berlin, Germany: 1999. [Google Scholar]
- 20.Castenholz RW, Boone DR. The Archae and the Deeply Branching and Phototrophic Bacteria. In: Garrity GM, editor. Bergey’s Manual of Systematic Bacteriology. Springer; New York, NY, USA: 2001. [Google Scholar]
- 21.Cronberg G, Annadotter H. Manual on Aquatic Cyanobacteria-A Photo Guide and a Synopsis of Their Toxicology. Internatinal Society for the Study of Harmful Algae, United Nations Educational, Scientific and Cultural Organization; Denmark: 2006. [Google Scholar]
- 22.Neilan BA, Jacobs D, Del Dot T, Blackall LL, Hawkins PR, Cox PT, Goodman AE. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Bacteriol. 1997;47:693–697. doi: 10.1099/00207713-47-3-693. [DOI] [PubMed] [Google Scholar]
- 23.Jungblut AD, Hawes I, Mountfort D, Hitzfeld B, Dietrich DR, Burns BP, Neilan BA. Diversity within cyanobacterial mat communities in variable salinity meltwater ponds of McMurdo Ice Shelf, Antarctica. Environ. Microbiol. 2005;7:519–529. doi: 10.1111/j.1462-2920.2005.00717.x. [DOI] [PubMed] [Google Scholar]
- 24.Pearson LA, Neilan BA. The molecular genetics of cyanobacterial toxicity as a basis for monitoring water quality and public health risk. Curr. Opin. Biotechnol. 2008;19:281–288. doi: 10.1016/j.copbio.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Moffitt MC, Neilan BA. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl. Env. Microbiol. 2004;70:6353–6362. doi: 10.1128/AEM.70.11.6353-6362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jungblut AD, Neilan BA. Molecular identification and evolution of the cyclic peptide hepatotoxins, microcystin and nodularin, synthetase genes in three orders of cyanobacteria. Arch. Microbiol. 2006;185:107–114. doi: 10.1007/s00203-005-0073-5. [DOI] [PubMed] [Google Scholar]
- 27.Fergusson KM, Saint CP. Multiplex PCR assay for Cylindrospermopsis raciborskii and cylindrospermopsin-producing cyanobacteria. Environ. Toxicol. 2003;18:120–125. doi: 10.1002/tox.10108. [DOI] [PubMed] [Google Scholar]
- 28.Hisbergues M, Christiansen G, Rouhiainen L, Sivonen K, Borner T. PCR-based identification of microcystin-producing genotypes of different cyanobacterial genera. Arch. Microbiol. 2003;180:402–410. doi: 10.1007/s00203-003-0605-9. [DOI] [PubMed] [Google Scholar]
- 29.Schembri MA, Neilan BA, Saint CP. Identification of genes implicated in toxin production in the cyanobacterium Cylindrospermopsis raciborskii. Environ. Toxicol. 2001;16:413–421. doi: 10.1002/tox.1051. [DOI] [PubMed] [Google Scholar]
- 30.Welker M, Marsalek B, Sejnohova L, von Dohren H. Detection and identification of oligopeptides in Microcystis (cyanobacteria) colonies: toward an understanding of metabolic diversity. Peptides. 2006;27:2090–2103. doi: 10.1016/j.peptides.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Carballo JL, Hernández-Inda ZL, Pérez P, García-Grávalos MD. A comparison between two brine shrimp assays to detect in vitro cytotoxicity in marine natural products. BMC Biotechnology. 2002;2:17. doi: 10.1186/1472-6750-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christiansen G, Fastner J, Erhard M, Borner T, Dittmann E. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J. Bacteriol. 2003;185:564–572. doi: 10.1128/JB.185.2.564-572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rantala A, Fewer DP, Hisbergues M, Rouhiainen L, Vaitomaa J, Borner T, Sivonen K. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA. 2004;101:568–573. doi: 10.1073/pnas.0304489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuge K, Akiyama T, Shoda M. Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 2001;183:6265–6273. doi: 10.1128/JB.183.21.6265-6273.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikalsen B, Boison G, Skulberg OM, Fastner J, Davies W, Gabrielsen TM, Rudi K, Jakobsen KS. Natural variation in the synthase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 2003;185:2774–2785. doi: 10.1128/JB.185.9.2774-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moffitt MC, Neilan BA. On the presence of peptide synthetase and polyketide synthase genes in the cyanobacterial genus Nodularia. FEMS Microbiol. Lett. 2001;196:207–214. doi: 10.1111/j.1574-6968.2001.tb10566.x. [DOI] [PubMed] [Google Scholar]
- 37.Kaebernick M, Rohrlack T, Christoffersen K, Neilan BA. A spontaneous mutant of microcystin biosynthesis: genetic characterization and effect on Daphnia. Environ. Microbiol. 2001;3:669–679. doi: 10.1046/j.1462-2920.2001.00241.x. [DOI] [PubMed] [Google Scholar]
- 38.Prati M, Molteni M, Pomati F, Rossetti C, Bernardini G. Biological effect of the Planktothrix sp. FP1 cyanobacterial extract. Toxicon. 2002;40:267–272. doi: 10.1016/s0041-0101(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 39.Rantala A, Rajaniemi-Wacklin P, Lyra C, Lepistö L, Rintala J, Mankiewicz-Boczek J, Sivonen K. Detection of microcystin-producing cyanobacteria in Finnish lakes with genus-specific microcystin synthetase gene E (mcyE) PCR and associations with environmental factors. Appl. Environ. Microbiol. 2006;72:6101–6110. doi: 10.1128/AEM.01058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson LL, Sekar R, Myers JL, Gantar M, Voss JD, Kaczmarsky L, Remily ER, Boyer GL, Zimba PV. The presence of the cyanobacterial toxin microcystin in black band disease of corals. FEMS Microbiol. Lett. 2007;272:182–187. doi: 10.1111/j.1574-6968.2007.00751.x. [DOI] [PubMed] [Google Scholar]
- 41.Gantar M, Sekar R, Richardson LL. Cyanotoxins from black band disease of corals and from other coral reef environments. Microb. Ecol. 2009;58:856–864. doi: 10.1007/s00248-009-9540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]