Summary
In mice, Lkb1 deletion and activation of KrasG12D results in lung tumors with a high penetrance of lymph node and distant metastases. We analyzed these primary and metastatic de novo lung cancers with integrated genomic and proteomic profiles and have identified gene and phosphoprotein signatures associated with Lkb1 loss and progression to invasive and metastatic lung tumors. These studies revealed that SRC is activated in Lkb1 deficient primary and metastatic lung tumors and that the combined inhibition of SRC, PI3K and MEK1/2 resulted in synergistic tumor regression. These studies demonstrate that integrated genomic and proteomic analyses can be used to identify signaling pathways that may be targeted for treatment.
Introduction
Lung cancer is the leading cause of cancer mortality worldwide. Genetic analyses and gene expression profiling of primary human lung tumors have identified several aberrant signaling pathways involved in the initiation of non-small cell lung cancer (NSCLC) (Bild et al., 2006). While many of these pathways are also likely to be involved in NSCLC progression and metastases, it is clear that the metastatic phenotype requires additional pathway perturbations that regulate cell motility, cell adhesion, the epithelial to mesenchymal transition (EMT), and extracellular matrix remodeling (Nguyen and Massague, 2007). As the development of metastases causes much of the morbidity and incurability of epithelial cancers, there is an urgent need to elucidate the events underlying this biological process.
The paucity of matched primary and metastatic human lung tumors has made it difficult to examine the genetic changes involved in the progression and metastasis of NSCLC. Studies using genetically engineered mice have demonstrated that the activation of certain combinations of oncogenes is sufficient for lung tumor initiation (Engelman et al., 2008; Jackson et al., 2001; Ji et al., 2006a; Ji et al., 2006b) and that additional genetic events are required for tumor progression (Iwanaga et al., 2008; Jackson et al., 2005) or metastases (Ji et al., 2007; Kim et al., 2009). We recently showed that the loss of the Lkb1 tumor suppressor gene in Kras-driven lung tumors results in metastases to lymph node and distant sites in mice (Ji et al., 2007). Moreover, we and others, have shown that LKB1 is inactivated by point mutations or deletions in at least 15-35% of NSCLC and concurrent KRAS and LKB1 mutation is observed in 4-10% of NSCLC (Carretero et al., 2004; Koivunen et al., 2008; Ji et al., 2007; Matsumoto et al., 2007; Sanchez-Cespedes et al., 2002).
In humans, germline mutation of LKB1 results in the Peutz-Jeghers syndrome, a familial cancer syndrome characterized by intestinal hamartomas and an increased risk for epithelial cancers, including lung cancers (Hearle et al., 2006). The LKB1 gene encodes a serine/threonine kinase that phosphorylates several substrates, many of which are also kinases (Alessi et al., 2006). However, identification of the pathways responsible for the pro-metastatic effects of Lkb1 loss has yet to be elucidated.
To identify altered signal transduction pathways involved in the progression and metastases of Lkb1 deficient lung tumors, we have performed an unbiased, integrated analysis of genomic and phosphoproteomic signatures of primary and metastatic mouse lung tumors.
Results
Lkb1 deficient metastatic lung tumors harbor unique gene expression changes
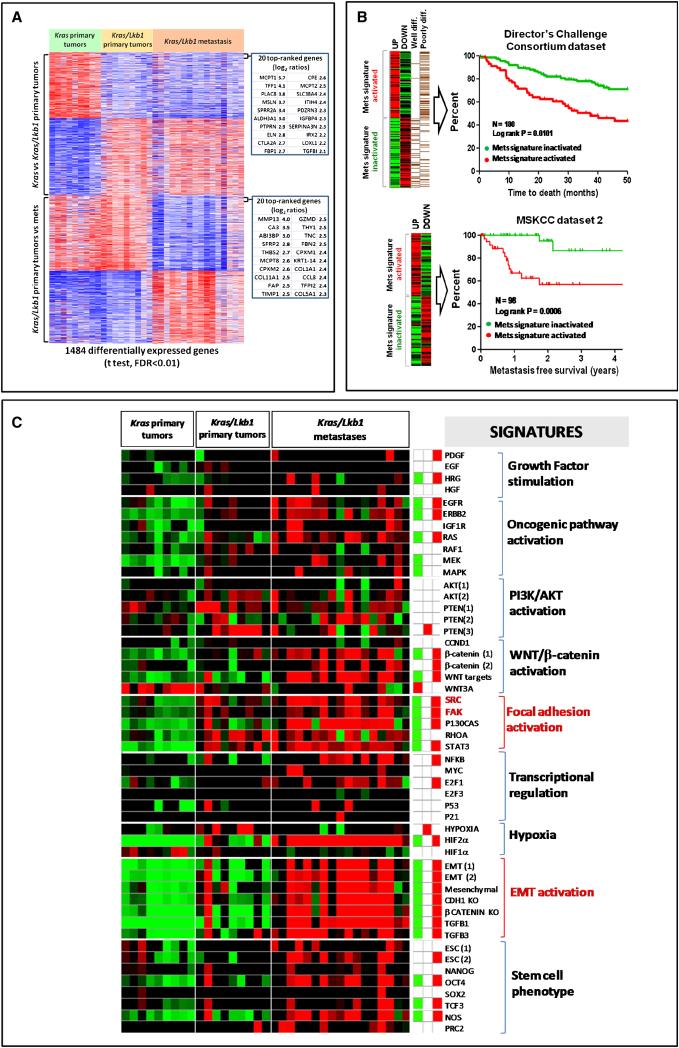
Lung specific activation of mutant KrasG12D results in bronchoalveolar hyperplasia and low grade adenocarcinomas (Jackson et al., 2001). We recently observed that concomitant loss of the Lkb1 tumor suppressor gene leads to a broadened histologic spectrum of lung tumors that have a high propensity for local invasion and metastases (Ji et al., 2007). To identify the gene expression changes associated with the invasive and metastatic progression of Lkb1-deficient mouse lung tumors, we compared RNA expression profiles from 9 Kras and 9 Kras/Lkb1 primary tumors as well as 16 Kras/Lkb1 metastases (lymph node and distant sites). Unsupervised hierarchical clustering of the samples revealed three distinct clusters corresponding to the following groups: 1) primary lung tumors with activated Kras G12D, 2) primary lung tumors with activated Kras G12D and Lkb1 loss, and 3) metastases from lung tumors with activated Kras G12D and Lkb1 loss (Figure S1A).
To better characterize the molecular alterations associated with Lkb1 loss and metastasis we derived expression signatures from our mouse lung tumors that reflected the gene expression changes induced by Lkb1 loss in primary tumors (Lkb1) by comparing primary Kras tumors to primary Kras/Lkb1 tumors and metastases (Mets) by comparing Kras/Lkb1 primary tumors to Kras/Lkb1 metastases. We used a two-class unpaired differential expression analysis and a false discovery rate (FDR) of <0.01. The resulting ‘Lkb1’ and ‘Mets’ gene signatures contain 727 and 757 differentially expressed genes respectively (Figure 1A). In aggregate, these results indicate that Lkb1 loss induces significant transcriptional changes in primary cancers and that there are further gene expression alterations that characterize the progression to lymph node and distant metastasis.
Figure 1. A gene expression signature of metastasis derived from a Kras-driven, Lkb1-deficient mouse lung cancer model predicts survival in human NSCLC.
A. Generation of an expression signature of metastasis. Kras/Lkb1 primary tumors were compared to Kras/Lkb1 metastases using two-class unpaired differential expression analysis and a false discovery rate (FDR) of <0.01. A total of 757 unique transcripts were significantly differentially deregulated and used to define a metastatic gene signature (Mets).
B. Kaplan-Meier analysis of the Director's Challenge Consortium dataset (N=180) and Memorial Sloan Kettering Cancer Center dataset 2 (N=98) of human lung tumors comparing the overall survival between tumors showing activation (red line) or inactivation (green line) of our mouse derived Mets signature. Red line = Mets signature activated. Green line = Mets signature inactivated.
C. Heat map of relative enrichment scores derived by gene set enrichment analysis. Tumors are represented in columns and genes signatures are represented in rows (red = significant enrichment of overexpressed genes; green, significant enrichment of under-expressed genes; black, not significant; p<0.05). The right panel shows the aggregate enrichment scores for each of the three classes of tumors (Kras, Kras/Lkb1 primary tumors and Kras/Lkb1 metastases).
See also Table S1 and Figure S1.
A cross species comparison across mouse and human datasets validates an expression signature of metastases as prognostic in human NSCLC
We next sought to determine whether this Mets signature predicted clinical outcome in human lung cancer patients. Using the human orthologs of the murine genes in the Mets signature, we interrogated two publicly available gene expression datasets of primary NSCLCs, annotated for either overall survival and histologic grade (Director's Challenge Consortium dataset, Shedden et al., 2008) or metastasis free recurrence (MSKCC dataset 2, Nguyen et al., 2009). The gene expression profiles of these human NSCLCs were queried for the presence or absence of our Mets expression signature as defined by coordinate upregulation and downregulation of overexpressed or under-expressed genes from the Mets signature.
Consistent with the notion that our Mets gene signature correlates with a proclivity towards metastasis, patients with tumors expressing the Mets signature had a significantly worse survival and increased occurrence of metastasis than did patients with tumors without Mets signature expression in these two datasets (Figure 1B). In contrast, the Lkb1 signature, generated by comparing Kras tumors to Kras/Lkb1 primary tumors, had no prognostic value (data not shown). These data suggest that the same gene expression changes induced during progression of Lkb1 deficient tumors to metastases in this murine model faithfully recapitulate those associated with the development of advanced incurable disease in human primary NSCLC and might be useful to prognosticate patients with early stage lung cancer. The disparity in the ability of the Lkb1 signature and the Mets signature in predicting survival suggest that either there is a subset of cells in the primary tumor that is destined for metastases which can not be discerned from expression analyses of the whole tumor or that additional expression changes developed at the site of metastases from cancer-stromal cell interactions. Lastly, and as shown on Figure S1B, the repressed component of the Mets signature showed a strong and significant correlation with the human samples that are mutant for both KRAS and LKB1 in the University of North Carolina dataset (Ji et al., 2007).
Lkb1 deficient metastases have upregulation of EMT-associated gene signatures
Activation of oncogenic signaling pathways has been shown to induce broad gene expression changes that can be used to generate gene expression or metagene signatures (Nevins and Potti, 2007). In order to better understand the biological basis for tumor metastasis in this model, we compared genes that were upregulated in the metastatic progression of these mouse lung tumors to known lists of transcripts associated with specific alteration in known pathways. Using gene set enrichment analyses, we identified gene signatures that were significantly enriched in our Kras/Lkb1 metastases relative to our primary Kras and Kras/Lkb1 lung tumors (Segal et al., 2004). Specifically, a collection of curated gene sets (Table S1) provided by the Molecular Signatures Database (MsigDB, broad.mit.edu/gsea/msigdb) and those collected from the literature (Ben-Porath et al., 2008; Bild et al., 2006; Cole et al., 2008; Onder et al., 2008) were used to interrogate our gene expression dataset using the Genomica analysis tool (genomica.weizmann.ac.il).
Thirty-two of the 51 curated, cancer-related, gene sets appeared to be significantly enriched and highly expressed (p<0.05, FDR <0.05) in our dataset of Kras/Lkb1 metastases relative to Kras and Kras/Lkb1 primary lung tumors (Figure 1C). This included a number of gene signatures implicated in the metastatic process such as focal adhesion alterations, EMT, increased TGF-β and β-catenin signaling, as well as embryonic stem (ES) cell expression signature and transcription factors implicated in ES cell phenotype such as TCF3 and OCT4 (Ben-Porath et al., 2008; Bild et al., 2006; Cole et al., 2008; Onder et al., 2008). A number of these findings were validated . For example, the canonical EMT markers (Kalluri and Weinberg, 2009), Vimentin, Snail, Twist1, N-cadherin, Acta2, and fibronectin seen in this signature were validated by RT-PCR (Figure S1C). Multiplex bead assay confirmed the elevated TGF-β levels in lysates from Kras/Lkb1 metastases (Figure S1D). Furthermore, immunostaining demonstrated the presence of canonical EMT markers such as vimentin and fibronectin in both Kras/Lkb1 primary and metastatic tumors. (Figure S1E). While components of these oncogenic signatures were observed in Lkb1-deficient primary tumors, their levels appeared to be higher in Lkb1-deficient metastases (Figure 1C, Figure S1C, S1D, and S1E). These results suggest that additional molecular events cooperate with Lkb1 loss to maximally activate these pathways during metastatic progression.
Phosphoscan of LKB1-deficient cells identifies activation of SRC family kinases
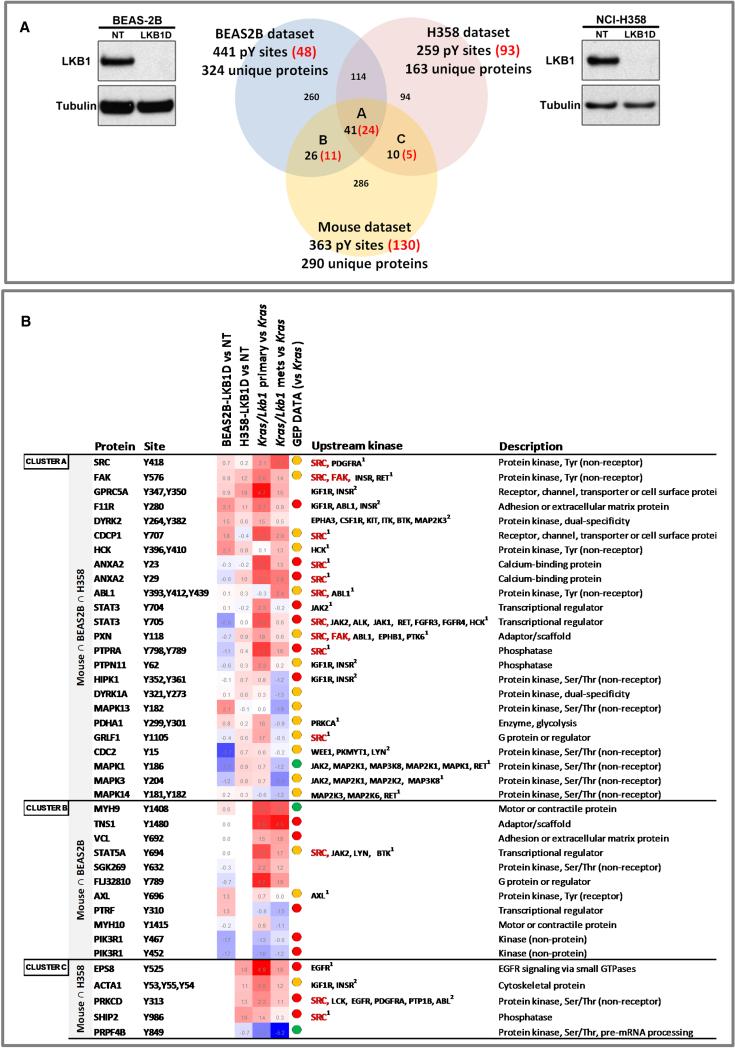
To complement our transcriptional analysis, we assessed how loss of Lkb1 impacted tyrosine kinase signaling cascades. We hypothesized that if Lkb1 loss resulted in significant activation of specific tyrosine kinases, we should detect increased phosphorylation of their respective substrates. To complement the analysis of murine tumors, we generated paired LKB1 and control knockdown cells in the BEAS-2B human bronchial epithelial cell line (Reddel et al., 1988), which is immortalized with SV40 Large T antigen, (BEAS-2B-LKB1D and BEAS-2B-NT respectively) and in the K-RAS mutant, LKB1 wild-type NSCLC cell line H358 (H358-LKB1D and H358NT), which has been previously shown not to express markers of EMT (Thomson et al., 2008) (Figure 2A). These paired human cell lines, along with our mouse lung primary and metastatic tumors, were used to generate phosphotyrosine signatures using previously described methods (Rikova et al., 2007). Specifically, whole cell extracts from cell lines or pooled tissue lysates (two pools with three tumor samples each from Kras, Kras/Lkb1 primary tumors, and Kras/Lkb1 metastases) were immunoprecipitated with a phosphotyrosine specific antibody, the immunoprecipitates were then subjected to MS analysis to identify and quantify phospho-peptides.
Figure 2. Phosphoprotein profiling of human cell lines and murine tumors.
A. Venn diagram of phosphotyrosine sites identified in Kras/Lkb1 primary versus metastatic murine tumors and isogenic paired cell lines (NCI-H358 and BEAS-2B) expressing shRNA to LKB1 (LKB1D) or a non-targeting shRNA (NT). Western blots show the effectiveness of LKB1 knockdown (LKB1D) in NCI-H358 and BEAS-2B cells. Red numbers in brackets show differentially increased or decreased phosphotyrosine sites in each comparison.
B. Table of coordinately regulated, LKB1 dependent, phosphotyrosine sites. Details of the LKB1 dependent phosphotyrosine sites found to be coordinately regulated across datasets are listed and include the phosphorylated protein, the tyrosine site, heatmaps of log2 ratios of indicated comparisons (red, positive log2 ratios; blue, negative log2 ratios), as well as the putative upstream kinase. The GEP (gene expression profiling) data column indicates the level of expression of the genes encoding these proteins between Kras/Lkb1 (primary or mets) versus Kras murine tumors comparisons (red dots = significantly overexpressed, green dots = significantly underexpressed, and orange dots = no significant change). 1 Upstream kinases obtained from PhosphoELM, HPRD and Swissprot databases; 2 Upstream kinases obtained from NetworKIN database. See also Figure S2.
We identified 441, 259, and 363 phosphotyrosine sites in BEAS-2B, H358, and mouse lung tumors, respectively, that were enriched in the absence of LKB1 (Figure 2A). Forty one of these LKB1-affected tyrosine phosphorylated peptides were noted to overlap across all 3 datasets (Figure 2A, cluster A), while an additional 26 peptides and 10 peptides were noted to overlap between the BEAS-2B / mouse tumor and H358 / mouse tumor datasets respectively (Figure 2A, clusters B and C). An examination of the putative upstream kinases known to phosphorylate these tyrosine residues suggested that a number of kinases were activated in association with LKB1 deficiency. These analyses pointed to a particularly central role for SRC activation which was not only highly phosphorylated, but is also predicted to be an upstream kinase for 13 sites of 40 differentially phosphorylated proteins among our datasets (Figure 2B). Moreover, Kras/Lkb1 primary tumors and metastases showed a significant hyperphosphorylation of a Src substrates signature, as assessed by GSEA analysis (Figure S2A).
The non-receptor tyrosine kinases, SRC and focal adhesion kinase, are activated by LKB1 loss
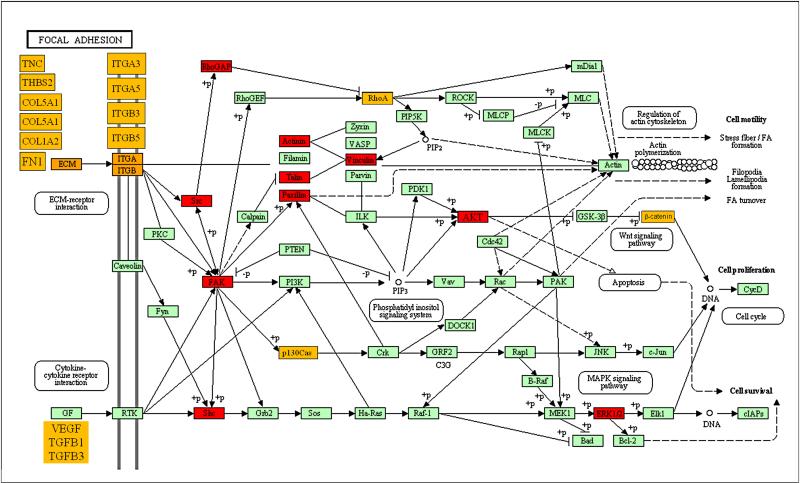
We speculated that integration of the gene expression and phosphoproteomics data sets would be a powerful approach to identify functionally significant molecular alterations in Lkb1 deficient lung cancers. We noted that gene signatures of focal adhesion activation were highly enriched in metastases from Kras/Lkb1 lung tumors (Figure 1C). Focal adhesions are dynamic subcellular structures known to mediate cell attachment to the extracellular matrix and are composed of over 50 proteins including focal adhesion kinase (FAK or PTK2), Paxillin, and SRC (Burridge et al., 1997; Playford and Schaller, 2004; Yeatman, 2004). As noted above, SRC activation was the most prominent signature emerging from our phosphoproteomic analysis and there was also strong evidence for FAK activation. To better integrate our data examining the impact of LKB1 loss on the focal adhesion pathway, components of the pathway upregulated at the gene expression or tyrosine phosphorylation level were colored yellow and red respectively and visualized on a KEGG pathway map (Figure 3). The results appear to show convergence on the activation of SRC, FAK, and their downstream pathways by Lkb1 loss and implicate alterations in the expression and activity of focal adhesion regulators in the pathogenesis of Lkb1 deficient lung cancers.
Figure 3. Focal adhesion is impaired in Kras/Lkb1 tumors and metastases.
Most relevant findings from our genomic and proteomic analysis were depicted on the focal adhesion KEGG pathway. Proteins labeled in red were hyperphosphorylated in the Phosphoscan analysis comparing Kras/Lkb1 primary tumors and metastases within the Phosphoscan analysis comparing Kras/Lkb1 primary tumors and metastases with Kras alone tumors. (Figure 2B). Proteins labeled in marked orange were found overexpressed (or their signatures were enriched) in the gene expression profiling comparing Kras/Lkb1 primary tumors and metastases with Kras alone tumors (Figures 1A and 1C).
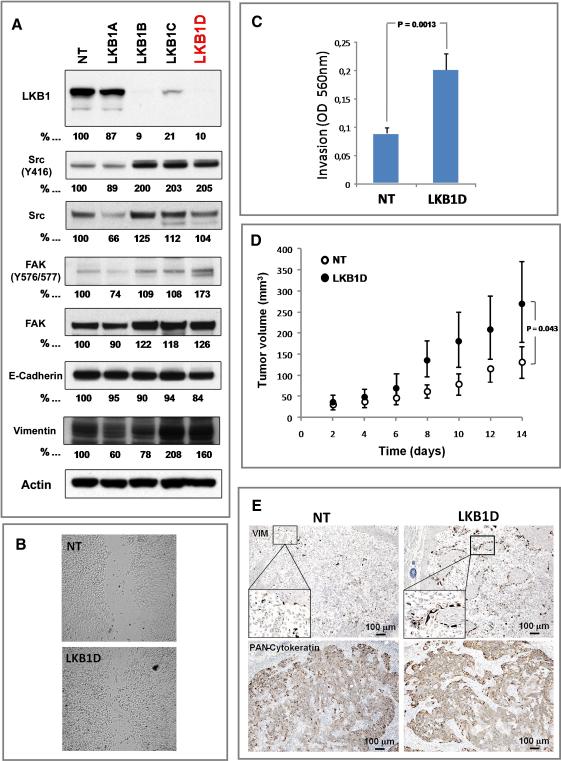
SRC and FAK activation results in focal adhesion disassembly and turnover, through downregulation of RhoA, resulting in increased cellular motility (Arthur et al., 2000; Yeatman, 2004). Given the key role of cellular motility and migration in the process of metastasis, we next determined whether LKB1 regulates these key components of focal adhesion dynamics. To this end, whole cell extracts of the H358 cells expressing control shRNA (NT) or four different shRNA sequences targeting LKB1 (A, B, C and D) were immunoblotted using antibodies against the activating tyrosine phosphorylation sites in SRC, and FAK (Y416, and Y576/577 respectively). As shown in Figure 4A, LKB1 knockdown led to activation of the focal adhesion complex, specifically SRC and FAK. Moreover, consistent with LKB1's established role as a regulator of the AMPK/TSC/mTORC1 axis (Carretero et al., 2007; Shaw et al., 2004), H358 LKB1-deficient cells showed an increase of AMPK (T172) and decrease of ACC (S79) phosphorylation, as well as an increase of S6 (S240/S244) phosphorylation (Figure S3A).
Figure 4. LKB1 knockdown activates EMT markers, mediators of focal adhesion dynamics and enhances cell motility.
A. Western blot analysis of NCI-H358 cells (LKB1 wild-type, KRAS G12C mutant) infected with lentiviruses encoding 4 different sequences of shRNAs (A-D) targeting LKB1 or non-targeting shRNA (NT). Whole cell lysates were imunoblotted with antibodies specific to LKB1 or tyrosine specific antibodies against SRC (Total and Y416), FAK (Total and Y576), paxillin (Total and Y118), E-Cadherin, and vimentin. α/β-actin (Actin) serves as a loading control.
B. Representative photographs of scratched areas of confluent monolayers of NCIH358 cells expressing a shRNA to LKB1 (LKB1D) or a non-targeting shRNA (NT) 12 hours after wounding with a pipet tip.
C. NCI-H358 cells expressing shRNA to LKB1 (LKB1D) or non-targeting shRNA (NT) were subjected to invasion assay in Boyden chambers coated with matrigel for 48 hours using 10% FBS as chemoattractant. Data is graphed as mean of 3 replicates ± SD.
D. Mean tumor volume measurements of NCI-H358 xenografts. The human lung cancer cells, NCI-H358, expressing shRNA to LKB1 (LKB1D) or a non-targeting shRNA (NT) were grown subcutaneously in athymic nude mice. After 14 days the tumors were measured and the tumor volume calculated (error bars = 1 SD; n=5, p=0.043).
E. Immunohistochemical staining of human vimentin and human cytokeratins in NCIH358 NT and LKB1D xenografts. See also Figure S3.
Since Phosphoscan and Western blot analyses detect a phosphorylation site (Y416) that is common to all SRC family kinases (SFKs) including SRC, FYN, LYN, LCK, HCK FGR and YES, we interrogated the phosphorylation status of SFKs affected by LKB1 loss in NCI-H358 and A549 cell lines using a Luminex bead assay. As shown in Figure S3B and S3C, H358 cells preferentially activate SRC and LYN, whereas A549 activates only SRC. In both cases, LKB1 loss promoted activation of SFKs. In addition, analyses of the protein lysates from H358-LKB1D and H358-NT cells with a protein array with site specific phospho-antibodies further validated SRC, FAK and other targets obtained by MS analyses of Kras/Lkb1 murine model (Figure S3D).
These changes in SRC and FAK suggest impaired adhesion and increased cellular motility (Yeatman, 2004). To assess whether loss of LKB1 affects the motility of these NSCLC cells, we performed a scratch assay. H358 cells with LKB1 knockdown (LKB1D) migrated more rapidly into the scratched space than the control cells (NT) while proliferation and viability of the two cells lines were identical in vitro (Figure 4B, Figure S3E). LKB1 loss also increased in vitro invasion of H358 cells, as assessed by Boyden chamber assay (Figure 4C). Therefore, loss of LKB1 appears to increase the kinase activity of several key proteins involved in focal adhesion dynamics and promote cell motility and invasion.
We also determined the consequences of LKB1 loss in NSCLC cells in vivo. To this end, H358-LKB1D and H358-NT cells were grown as subcutaneous xenograft tumors in nude mice, and mean tumor volume was measured two weeks after implantation. Tumors derived from H358 LKB1 deficient cells were significantly larger than those expressing a non-specific shRNA, suggesting that loss of LKB1 promotes primary tumor growth in addition to metastasis (Figure 4D). Immunohistochemical (IHC) staining with antibodies that specifically recognize human (but not mouse) antigens showed increased levels of vimentin in the H358-LKB1D xenografts than in the H358-NT tumors (Figure 4E), in agreement with the previous results suggesting that loss of LKB1 promotes EMT (Figure 3A). Elevated vimentin staining was particularly prominent at the edge of tumors suggesting that the EMT in the LKB1 deficient tumor cells may have also involved signals from the adjacent stromal cells (Figure 4E). In agreement to these observations, H358 cells without LKB1 (LKB1D) released significantly more TGF-β1/2 than H358 control (NT) cells and nuclear β-catenin accumulation was more prevalent in LKB1D than in NT cells (Figures S3F and S3G). These observations support the notion that LKB1 loss plays an important role in EMT.
LKB1 loss heightens dependence on SRC and FAK signaling for cell adhesion and migration
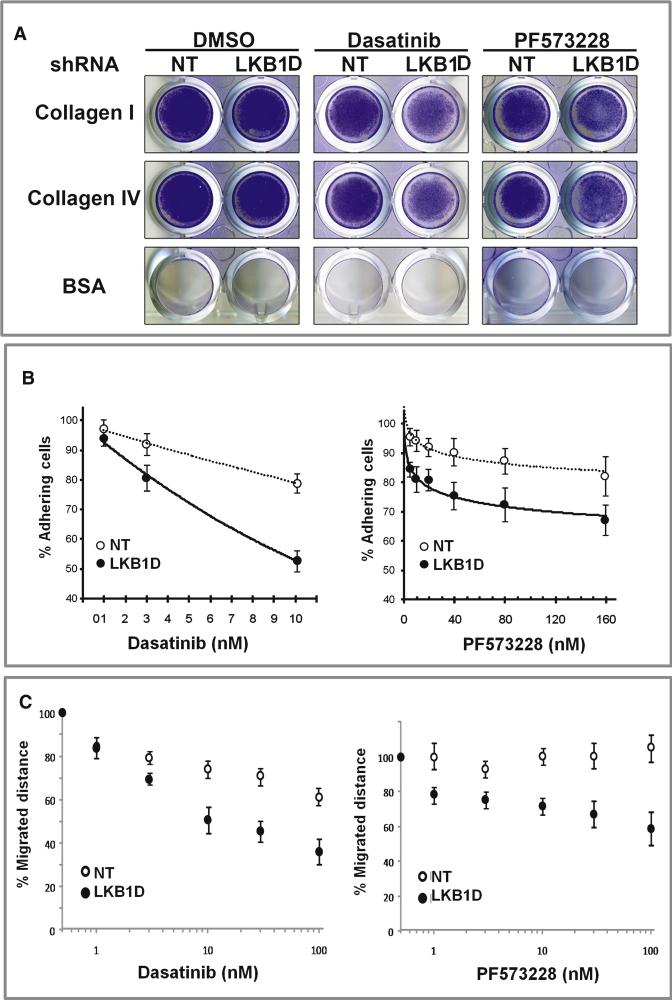
Our integrated gene expression and Phosphoscan results suggest that LKB1 loss might promote metastasis through activation of SRC and FAK signaling. Metastasis is a multi-step process involving cell interactions with extracellular matrix (ECM) components, increased cell migration and tumor invasion (Mitra and Schlaepfer, 2006). These individual processes can both modulate and be modulated by SRC and FAK activity (Huveneers and Danen, 2009; Shibue and Weinberg, 2009). We hypothesized that SRC/FAK activation induced by LKB1 loss resulted in an increased dependence upon these signaling pathways for cell adhesion to ECM. To test this hypothesis, 5×105 cells with or without LKB1 knockdown were plated on tissue culture wells coated with either collagen I, collagen IV, fibronectin, laminin I, fibrinogen or BSA in the presence of the SRC-family kinase inhibitor, Dasatinib, or the FAK inhibitor, PF573228 (Johnson et al., 2005; Slack-Davis et al., 2007). among the ECM components tested, only collagen I or IV were able to promote the adherence of H358 cells (Figure 5A and data not shown). Knockdown of LKB1 in H358 cells did not have an appreciable effect on the baseline adhesion of H358 cells to either collagen I or IV. However, treatment of H358 cells with Dasatinib or PF573228 resulted in a dose-dependent decrease in cell adhesion that was more prominent in H358 cells with LKB1 knockdown (Figure 5B). Similar results were obtained when H358 cells with or without LKB1 expression were examined for the effects of Dasatinib and PF573228 on cell migration (Figure 5C). These observations support the notion that the activation of SRC and FAK due to LKB1 loss promotes dependence upon these signaling pathways for cell migration and adhesion to extracellular matrix components as well as migration.
Figure 5. LKB1 knockdown cooperates with FAK and SRC inhibition to prevent adhesion to Collagen and migration in NCI-H358 cells.
A. Exponentially growing NCI-H358 cells expressing shRNA to LKB1 or a nonspecific shRNA (NT) were seeded at 5×105 cells in wells coated with collagen I, collagen IV, or BSA and allowed to adhere to the matrices in the presence of DMSO (control), 10nM Dasatinib (SRC inhibitor) or 100nM PF 573228 (FAK inhibitor) . After 90 minutes, loose cells were washed off and adherent cells were fixed and stained.
B. NCI-H358 cells expressing shRNA to LKB1 (LKB1D) or a non-specific shRNA (NT) were seeded at 1×105 cells/well onto collagen I coated 96 well plates containing DMSO or increasing concentrations of Dasatinib or PF573228, and incubated for 90 minutes. Data points represent the average of normalized values expressed as % of adhesion compared to DMSO-treated cells, from two independent experiments performed in triplicate; error bars = SD.
C. NCI-H358 cells expressing shRNA to LKB1 (LKB1D) or a non-specific shRNA (NT) were seeded at 1×105 cells/well onto 24well and cells were allowed to grow until forming confluent monolayers. The monolayer was scratched with a pipet tip to form a wound and the cells were grown in media containing DMSO or increasing concentrations of Dasatinib or PF573228. 12 hours after wounding of the confluent monolayers, the wound closure was measured and expressed as % closure of the original wound. Data is graphed as mean of 3 replicates ± SD.
Combined inhibition of PI3K-mTOR, MEK, and SRC family kinases results in synergistic in vivo tumor response in LKB1 defective lung tumors
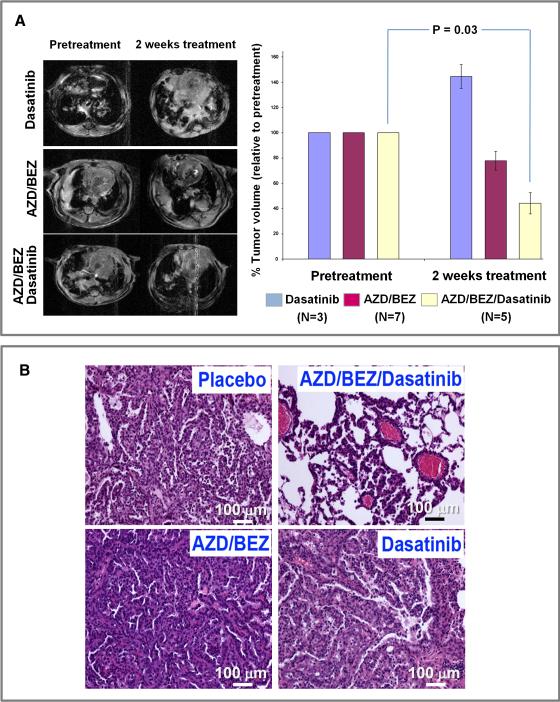
We had previously observed that dual inhibition of PI3K-mTOR and MEK using BEZ235 (a PI3K-mTOR inhibitor) and AZD2644 (a MEK1/2 inhibitor) resulted in an 80% reduction of tumor volume of Kras dependent murine lung tumor model (Engelman et al., 2008). Surprisingly, treatment of Kras/Lkb1 mice with the same combination showed that these lung tumors were largely unresponsive (Figure 6A). Since our integrated gene expression and phosphotyrosine profiling as well as in vitro cell culture studies demonstrated that SRC is activated by LKB1 loss (Figure 1C, Figure 2B, Figures S2 S3A, S3B and S3C), we hypothesized that inhibition of SRC by Dasatinib might restore sensitivity of Kras/Lkb1 lung tumors to PI3K and MEK inhibition. To examine the therapeutic impact of SRC inhibition in Kras/Lkb1 lung cancers, we treated established Kras/Lkb1 lung cancers with Dasatinib alone, the combination of BEZ235 and AZD2644 (AZD/BEZ), or the combination of all three agents (Dasatinib/AZD/BEZ). While Kras/Lkb1 lung tumors did not respond to Dasatinib or BEZ/AZD, the triple combination Dasatinib/BEZ/AZD resulted in significant tumor regression as detected by serial MRI imaging (Figure 6A) and histology (Figure 6B). This in vivo data suggests that Lkb1 loss results in reduced sensitivity to the combined inhibition of PI3K and MEK, at least in part, through activation of SRC. Moreover, although Dasatinib alone in Kras/Lkb1 lung cancer model did not provide overall survival benefit over vehicle in this Kras/Lkb1 lung cancer model, it blocked metastasis (Table S2).
Figure 6. Combination therapy with PI3K, MEK, and SRC inhibitors results in synergistic in vivo response of Lkb1 deficient murine lung tumors.
A. Representative MRI images before and after treatment for each group. Mice in each group were MRI imaged then treated daily for two weeks with the following doses: 45 mg/kg of BEZ235, 25mg/kg of AZD6244 and 15 mg/kg of Dasatinib and imaged again. Tumor volumes were normalized to pretreatment tumor volumes. The average tumor volumes of 3-7 mice in each treatment group are shown. Error bars = 1 SD (p < 0.05 BEZ/AZD/Dasatinib-treated versus pretreatment).
B. H&E staining. H&E sections of representative Kras/Lkb1 tumors from animals treated with the indicated drugs. See also Tables S2 and S3.
Gene expression profiling of short term treated tumors showed that all treatments were able to revert Lkb1 and Mets signatures (Table S2). Interestingly, Dasatinib alone induced downregulation of SRC, TGF-β and MEK signatures, but not AKT, EMT and ES signatures. Triple combination downregulated SRC, TGF-β, MEK as well as AKT, EMT and ES signatures. This analysis supports the observation that Dasatinib synergizes with BEZ/AZD in inhibiting signal transduction pathways that are important in the proliferation and survival of Kras/Lkb1 driven cancer cells.
Discussion
Cancer genomic studies have established a number of oncogene and tumor suppressor pathways as important for the initiation and maintenance of neoplastic lesions in NSCLC. However, the molecular alterations necessary for invasion and metastasis of NSCLC are less well-defined. We have previously reported that deletion of the Lkb1 tumor suppressor gene in the context of Kras-driven murine lung tumors promotes invasion and metastasis (Ji et al, 2007). Here we extend these findings to show that Kras/Lkb1 primary and metastatic tumors have upregulated expression of markers and inducers of EMT. Furthermore, through an integrated genomic and phosphoproteomic analysis of mouse lung primary and metastatic tumors, we have determined that two key modulators of focal adhesion dynamics, SRC and FAK, are upregulated by Lkb1 loss during NSCLC progression. Similarly, LKB1 loss in vitro also resulted in SRC activation, increased motility and SRC-dependent adhesion. In fact, migration was selectively abrogated by SRC and FAK inhibition in LKB1 deficient cells. Finally, while Kras mutant lung tumors are sensitive to the combined inhibition of the PI3K and MEK pathways, we have found that Kras/Lkb1 tumors are resistant to these inhibitors, and that sensitivity can be restored by additional targeting of SRC. It also important to note that addition of Dasatinib to combined PI3K/MEK inhibition induced tumor shrinkage in the Lkb1 deficient tumors. This reveals an important role for SFKs in tumor growth and promoting resistance to combined PI3K/MEK inhibition. Indeed, in addition to their prominent roles in tumor migration and adhesion, SFKs are also essential for transducing signals from RTKs and integrins to promote cell survival and proliferation (Yeatman, 2004). It was somewhat surprising that single-agent Dasatinib led to increased volume of Kras/Lkb1 tumors (Figure 6) and persisting Akt and EMT signatures (Table S3). Although we cannot fully explain this observation, it is tempting to speculate that SRC inhibition led to loss of a feedback mechanism resulting in increased tumor growth or to a change in the interactions between tumor cells and stroma or immune cells resulting in tumor growth. Collectively, these results point towards a mechanism underlying the increased propensity for metastases seen in Lkb1 deficient lung tumors and identify SRC as a molecularly targetable pathway for the treatment of LKB1-deficient NSCLC in humans.
Tumor metastasis is a complex, multistage process that involves both initiating functions, such as invasion and angiogenesis, as well as metastasis progression functions such as extravasation and survival (Nguyen and Massague, 2007). EMT has been implicated in metastasis initiation and involves loss of cell-cell junction proteins as well as a conversion of stationary cells to motile cells capable of invading through the ECM (Yang and Weinberg, 2008). Our gene expression analysis suggests that LKB1 loss results in the aberrant expression of a number of oncogenic pathways and transcription factors known to be inducers of EMT such as SNAI1, TWIST1, and the TGF-β and β-catenin pathways (Kalluri and Weinberg, 2009; Nguyen and Massague, 2007; Yang and Weinberg, 2008). While the mechanism by which LKB1 loss promotes EMT remains to be determined, we observe that NSCLC cells lacking LKB1 exhibit higher nuclear localization of β-catenin and increased release of TGF-β (Figures S3F and S3G) suggesting that LKB1 loss may have a direct cell autonomous role in promoting the EMT program. The expression signatures derived from Kras/Lkb1 metastasis enriched for EMT correlate with NSCLC patients with metastatic potential and worsened survival (Figure 1B) while expression signatures derived from Kras primary tumors or Kras/Lkb1 tumors did not correlate with survival thus have no prognostic values. Importantly, the presence of Mets signature in primary tumor samples, including a subset of double KRAS and LKB1 mutants, suggests that a subset of primary tumors harbor cells destined for metastasis.
Part of the roadblock in defining the pathways involved in invasion and metastases in human NSCLC has been the difficulty in obtaining tissue from metastatic sites. Through the use of a Kras-driven, Lkb1-deficient genetically engineered mouse model of lung cancer, we have been able to collect matched primary and metastatic tumors and apply integrated gene expression and phosphoproteomic analysis to define altered signal transduction pathways. The utilization of complex murine cancer models to identify signal transduction pathways altered in cancer progression and metastases might serve as a paradigm for other solid tumors that suffer from similar barriers to tissue acquisition.
Previous work from our lab has shown that dual inhibition of the PI3K and MEK pathways in Kras driven murine NSCLC results in synergistic tumor responses (Engelman et al., 2008). Strikingly, Lkb1 loss renders these tumors substantially more resistant to this therapeutic regimen; therefore Lkb1 loss could serve as a negative predictor of the responses to the therapy. Despite the broad gene expression changes seen in the setting of Lkb1 loss and progression to metastases, it is notable that we were able to define a candidate mediator of Lkb1-deficient tumor growth, SRC, in which additional inhibition — with Dasatinib— was able to restore sensitivity to inhibition of PI3K and MEK in vivo. A limitation of this analysis is that Dasatinib is a potent inhibitor of other kinases, e.g. ABL, which was also activated to a lesser degree in Lkb1-deficient tumors. Future studies will determine the relative importance of SFKs vs. other Dasatinib targets in mediating the response to this agent. Lastly, Kras/Lkb1 mice treated with Dasatinib alone did not show any evidence of metastasis (Table S2). This further reinforces our data that the Src family members are important mediators of metastatic progression in Kras/Lkb1 driven lung cancer. In conclusion, our results imply that despite the complex transcriptional and signaling changes that occur in the setting of LKB1 loss and progression of NSCLC, these tumors may still be addicted to isolated oncogenic events that can be successfully therapeutically targeted.
Experimental Procedures
Mouse colony and tumor collection
All mice were housed and treated in accordance with protocols approved by the institutional care and use committees for animal research at the Dana-Farber Cancer Institute. Kras and Kras/Lkb1 mice were treated with 5×106 p.f.u. adeno-Cre (University of Iowa viral vector core) intranasally as previously described (Ji et al., 2007). Primary lung adenocarcinomas and metastases were macrodissected and formalin fixed or frozen in liquid nitrogen and stored at −80° until use.
Histology and Immunohistochemical analysis
Formalin fixed tissues were paraffin embedded, sectioned at 5 μm, and haematoxylin and eosin stained (Department of Pathology in Brigham and Women's Hospital). Unstained slides were deparaffinized in xylene and rehydrated sequentially in ethanol. Antigen retrieval was performed by boiling slides in 10mM sodium citrate (pH 6.0) for 30 minutes. Primary antibodies were diluted in TBST with 5% goat serum and incubated overnight at 4ºC (see Supplemental experimental procedures for detailed list of primary antibodies). Detection was performed using the avidin-biotin horseradish peroxidase technique in which 3,3”-diaminobenzidine was the chromogenic substrate.
Analysis of microarray gene expression data
Gene expression profiling methods (Total RNA isolation and microarray processing) are available in Supplemental Methods. Clustering and differential expression analyses were performed using the Gepas Analysis suite (Tarraga et al., 2008) and GenePattern (Reich et al., 2006) with procedures described in Supplemental methods. Genomica software (http://genomica.weizmann.ac.il/) was used to identify enrichment patterns of a custom collection of experimental signatures associated with different phenotypes and cancer-related events (Ben-Porath et al., 2008; Segal et al., 2004) (see Table S1 for detailed description). Briefly, the data was log2 transformed and mean-centered. Genes whose expression was 2 fold or greater than the mean expression level were scored. Enrichment of overexpressed or underexpressed genes that belong to each tested gene signature was calculated using a hypergeometric test and a false discovery rate (FDR) calculation to account for multiple hypothesis testing. (P < 0.05, FDR<0.05). Finally, the fraction of tumors showing significant enrichment for a particular gene signature in each class (Kras tumors, Kras/Lkb1 primary tumors and metastasis) was calculated and assigned a P value according to the hypergeometric distribution (P < 0.01, FDR<0.05).
Gene Set Enrichment analysis of short term treated Kras/Lkb1 tumors was performed using GSEA (broad.mit.edu/gsea) across the complete list of genes ranked by signal-to-noise ratio.
UNC human lung cancer microarrays (Agilent custom 44,000 probe) annotated with KRAS and LKB1 genetic status were mapped to mouse genes using NCBI Homologene, retaining only one-to-one orthologs. Microarray data were sample and gene standardized, and Lkb1 and Mets signature scores were calculated as the median of the genes across the gene set. Statistical significance was calculated using the Wilcoxon rank sum test.
Survival analysis
The Director's Challenge Consortium human primary lung adenocarcinoma dataset was downloaded from caarraydb.nci.nih.gov (experiment ID 1015945236141280:1) (Shedden et al., 2008). MSKK dataset 2 was downloaded from cbio.mskcc.org/Public/lung_array_data (Nguyen el al., 2009). Datasets were first log2 transformed, mean-centered dataset and loaded into Genomica software. Enrichment of genes that belong to our murine derived Mets signature was calculated as described above and each patient was assigned an enrichment score. Positive and negative enrichment scores for each individual were matched with survival data (time to last contact or death) or recurrence of metastasis (metastasis free survival). Kaplan-Meier plots were created with Graphpad software. Statistical significance was calculated using the log rank test.
Phosphopeptide immunoprecipitation, LC-MS/MS and phosphopeptide analysis
We performed phosphotyrosine peptide identification and quantification as described previously (Rikova et al., 2007). See Supplemental Methods for sample preparation and handling. For differential phosphorylation analysis, duplicated intensities were averaged, and fold-changes between different experimental conditions were calculated. For KEGG classification of the phosphoproteins, we used FatiGO software (www.babelomics.org) (Al-Shahrour et al., 2004). A modification of the Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) using phosphoprotein sets instead gene sets (Figure S2A) was used to interrogate the PhosphoPoint database of tyrosine kinase substrates (kinase.bioinformatics.tw) (Yang et al., 2008).
Quantitative RT-PCR
50 ng RNA of total RNA was reverse-transcribed with the Superscript cDNA Synthesis Kit (Invitrogen). Real time PCR reactions were prepared in triplicate for each sample using primers and TaqMan probes purchased from Applied Biosystems (see also Supplemental Experimental Procedures). Reactions were run on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). GAPDH was used as a reference for all reactions. Relative levels of a gene was determined by ΔΔCt method.
Multiplex bead analysis
Quantification of TGF-β isoforms and Src family kinase activities were assayed using Human TGF-β 1,2,3 Assay and SRC Family Kinase 8-Plex (Millipore), respectively. Assays were performed according to the manufacturer's specifications and analyzed with a Luminex 100 platform. Fluorescence intensity from duplicate samples was averaged and transformed into protein concentration using a calibration curve obtained with TGFβ1, 2 and 3 standards. For Src assay, activity was expressed in mean fluorescence intensity (MFI) with background correction from duplicate samples. Statistical significance was calculated using a two-sided Student's exact t-test.
Western blot and kinase array analysis
Parameters for Western blot and antibodies are available in Supplemental Experimental Procedures.. Human phospho-kinase arrays (R&D Systems) were performed according to the manufacturer's instructions. The percent control activation/deactivation was determined by densitometric analysis using ImageJ Software (NIH).
Short hairpin RNA constructs, lentiviral infection, and small interfering RNA infection
Short hairpin RNA (shRNA) constructs cloned in pLKO.1 puro vector were designed by the RNAi consortium and distributed by Open Biosystems. shRNA sequences are provided with supplemental experimental procedures. Stable polyclonal cell lines are established as described previously (Shimamura et al., 2008).
Cell migration, invasion and adhesion assays
Cell migration in cultures was measured using a two-dimensional in vitro scratch motility assay. 1mm wide scratch was made on confluent monolayer in tissue culture slides and allowed to grow under standard conditions for 12 h. Repopulation of the cleared field was recorded using a Nikon inverted TE2000 and migrated distance was quantified using Image J software. Invasion and adhesion assays kits were obtained from Cell Biolabs and assays were performed according to manufacturer's specifications. Details are available in Supplemental Methods.
Immunofluorescence analysis
Cells were fixed on four-well chamber glass slides in methanol/acetone (1:1) at –20°C for 10 min and air-dried. Primary block was done with 10% goat serum (Sigma) and 0.05% Tween 20 in PBS buffer for 1 hour at room temperature. A mouse monoclonal antibody for activated β-catenin (Millipore) was diluted 1:50 in blocking buffer and incubated overnight at 4°C. Rabbit anti-mouse secondary antibody conjugated with Alexa488 dye (Invitrogen) was diluted 1:100 in blocking buffer and incubated for 1 h at room temperature. Images were acquired using a Nikon inverted TE2000 microscope equipped a Hamamatsu Orca ER digital CCD camera.
In Vivo Xenograft Experiments
Five-to 6-week-old female nu/nu mice (Charles River) were maintained under pathogen-free conditions. Tumors were generated by injecting s.c. 5×106 cells mixed with reconstituted basement membrane Matrigel (BD Biosciences) with 1:1 ratio in PBS. Animals were sacrificed 14 days after inoculation, and tumors were excised and fixed in formalin. Tumor measurements were obtained using electronic calipers. Tumor volume was calculated using: Tumor volume = (length × width2)/2. We compared tumor volumes of mice using a two-sided Student's exact t-test.
Cancer therapy using inhibitors
Dual PI3K-mTOR inhibitor, NVP-BEZ235-AN (Novartis Institutes for Biomedical Research) was reconstituted in one volume of N-methyl-2-pyrrolidone (69118, Fluka) and nine volumes of PEG300 (81160, Fluka) and administered by oral gavage. MEK inhibitor ARRY-142886 (AZD6244, Otava Chemicals) and Dasatinib (LC Labs) were reconstituted in 0.5% methyl cellulose (Fluka) and 0.4% polysorbate (Tween 80; Fluka) and administered by oral gavage. Dosages are indicated in the figure legends.
Magnetic resonance imaging and tumor volume measurement
We performed MRI measurements as described previously (Engelman et al., 2008). Brief descriptions are available in supplemental methods.
Supplementary Material
Acknowledgements
J.C. was a fellow of Spanish Ministry of Science and Innovation (MICINN) and Spanish Association against Cancer (AECC). T.S. was supported by the Claudia Adams Barr Program in Innovative Basic Cancer Research. K.K Wong and J.A. Engelman are founders of Gatekeeper Therapeutics. This work was supported by Dana-Farber–Harvard Cancer Center Lung Cancer Specialized Program of Research Excellence (SPORE) grant P50 CA090578 (J.A. Engelman and K-K. Wong); R01 AG2400401 (K-K. Wong), R01 CA122794 (K-K. Wong), R01 CA140594 (J.A. Engelman and K-K. Wong), K08 grant CA120060 (J.A. Engelman), R01CA137008 (J.A. Engelman), DF/HCC Gastrointestinal Cancer SPORE P50 CA127003 (J.A. Engelman); American Association for Cancer Research (J.A. Engelman); V Foundation (J.A. Engelman); American Cancer Society RSG-06-102-01-CCE (J.A. Engelman); and Ellison Foundation Scholar (J.A. Engelman). K. Rikova, K. Crosby, J. C. Silva and T-L Gu are employees of Cell Signaling Technology. C. Garcia-Echeverria is an employee of Novartis Institutes for Biochemical Research. We thank H. Voelker for help in preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Significance
While large-scale genomic analyses of non-small cell lung cancers (NSCLC) have yielded a better understanding of lung cancer genetic alterations, studies defining the pathways deregulated in tumor progression and metastases are limited. LKB1 inactivation is found in up to 30% of human NSCLC and Lkb1 deficient lung tumors in mice have a high penetrance of lymph node and distant metastases. Our analyses of primary and metastatic Lkb1-deficient mouse lung tumors have shown that progression to metastatic lung cancer is associated with unique gene and phosphoprotein signatures. In this work, we employ these signatures to design an effective, rational therapeutic strategy to treat Lkb1-deficient lung cancers.
Microarray Data
Microarray data have been deposited at GEO with the accession number GSE21581.
References
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- Arthur WT, Petch LA, Burridge K. Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr Biol. 2000;10:719–722. doi: 10.1016/s0960-9822(00)00537-6. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Burridge K, Chrzanowska-Wodnicka M, Zhong C. Focal adhesion assembly. Trends Cell Biol. 1997;7:342–347. doi: 10.1016/S0962-8924(97)01127-6. [DOI] [PubMed] [Google Scholar]
- Carretero J, Medina PP, Blanco R, Smit L, Tang M, Roncador G, Maestre L, Conde E, Lopez-Rios F, Clevers HC, Sanchez-Cespedes M. Dysfunctional AMPK activity, signalling through mTOR and survival in response to energetic stress in LKB1-deficient lung cancer. Oncogene. 2007;26:1616–1625. doi: 10.1038/sj.onc.1209951. [DOI] [PubMed] [Google Scholar]
- Carretero J, Medina PP, Pio R, Montuenga LM, Sanchez-Cespedes M. Novel and natural knockout lung cancer cell lines for the LKB1/STK11 tumor suppressor gene. Oncogene. 2004;23:4037–4040. doi: 10.1038/sj.onc.1207502. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearle N, Schumacher V, Menko FH, Olschwang S, Boardman LA, Gille JJ, Keller JJ, Westerman AM, Scott RJ, Lim W, et al. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res. 2006;12:3209–3215. doi: 10.1158/1078-0432.CCR-06-0083. [DOI] [PubMed] [Google Scholar]
- Huveneers S, Danen EH. Adhesion signaling -crosstalk between integrins, Src and Rho. J Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- Iwanaga K, Yang Y, Raso MG, Ma L, Hanna AE, Thilaganathan N, Moghaddam S, Evans CM, Li H, Cai WW, et al. Pten inactivation accelerates oncogenic K-ras-initiated tumorigenesis in a mouse model of lung cancer. Cancer Res. 2008;68:1119–1127. doi: 10.1158/0008-5472.CAN-07-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Li D, Chen L, Shimamura T, Kobayashi S, McNamara K, Mahmood U, Mitchell A, Sun Y, Al-Hashem R, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006a;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, Jung BL, McNamara K, Xia H, Glatt KA, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2006b;103:7817–7822. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FM, Saigal B, Talpaz M, Donato NJ. Dasatinib (BMS-354825) tyrosine kinase inhibitor suppresses invasion and induces cell cycle arrest and apoptosis of head and neck squamous cell carcinoma and non-small cell lung cancer cells. Clin Cancer Res. 2005;11:6924–6932. doi: 10.1158/1078-0432.CCR-05-0757. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Perera S, Zhou B, Carretero J, Yeh J, Heathcote S, Jackson A, Nikolinakos P, Ospina B, Naumov G, et al. HIF2α cooperates with RAS to promote lung tumorigenesis in mice. Journal of Clinical Investigation. 2009;119:2160–2170. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen JP, Kim J, Lee J, Rogers AM, Park JO, Zhao X, Naoki K, Okamoto I, Nakagawa K, Yeap BY, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008;99:245–252. doi: 10.1038/sj.bjc.6604469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Iwakawa R, Takahashi K, Kohno T, Nakanishi Y, Matsuno Y, Suzuki K, Nakamoto M, Shimizu E, Minna JD, Yokota J. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26:5911–5918. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Current opinion in cell biology. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601–609. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene. 2004;23:7928–7946. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- Reddel RR, Ke Y, Kaighn ME, Malan-Shibley L, Lechner JF, Rhim JS, Harris CC. Human bronchial epithelial cells neoplastically transformed by v-Ki-ras: altered response to inducers of terminal squamous differentiation. Oncogene Res. 1988;3:401–408. [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–3662. [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T, Weinberg RA. Integrin beta1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc Natl Acad Sci U SA. 2009;106:10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura T, Li D, Ji H, Haringsma HJ, Liniker E, Borgman CL, Lowell AM, Minami Y, McNamara K, Perera SA, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res. 2008;68:5827–5838. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, Luzzio MJ, Cooper B, Kath JC, Roberts WG, Parsons JT. Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem. 2007;282:14845–14852. doi: 10.1074/jbc.M606695200. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarraga J, Medina I, Carbonell J, Huerta-Cepas J, Minguez P, Alloza E, Al-Shahrour F, Vegas-Azcarate S, Goetz S, Escobar P, et al. GEPAS, a web-based tool for microarray data analysis and interpretation. Nucleic Acids Res. 2008;36:W308 314. doi: 10.1093/nar/gkn303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clin Exp Metastasis. 2008;25:843–854. doi: 10.1007/s10585-008-9200-4. [DOI] [PubMed] [Google Scholar]
- Yang CY, Chang CH, Yu YL, Lin TC, Lee SA, Yen CC, Yang JM, Lai JM, Hong YR, Tseng TL, et al. PhosphoPOINT: a comprehensive human kinase interactome and phospho-protein database. Bioinformatics. 2008;24:i14–20. doi: 10.1093/bioinformatics/btn297. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Developmental Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Yeatman TJ. A renaissance for SRC. Nature Rev Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.