Summary
The SMN complex assembles Sm cores on snRNAs, a key step in the biogenesis of snRNPs, the spliceosome's major components. Here, using SMN complex inhibitors identified by high throughput screening and a ribo-proteomic strategy on formaldehyde crosslinked RNPs, we dissected this pathway in cells. We show that protein synthesis inhibition impairs the SMN complex, revealing discrete SMN and Gemin subunits and accumulating an snRNA precursor (pre-snRNA)-Gemin5 intermediate. By high throughput sequencing of this transient intermediate's RNAs, we discovered the previously undetectable precursors of all the snRNAs and identified their Gemin5 binding sites. We demonstrate that pre-snRNA 3′-sequences function to enhance snRNP biogenesis. The SMN complex is also inhibited by oxidation and we show that it stalls an inventory-complete SMN complex containing pre-snRNAs. We propose a stepwise pathway of SMN complex formation and snRNP biogenesis, highlighting Gemin5's function in delivering pre-snRNAs as substrates for Sm core assembly and processing.
Introduction
The major components of the spliceosome, which carries out pre-mRNA splicing in eukaryotes, are small nuclear ribonucleoprotein particles (snRNPs). Each snRNP consists of a U snRNA (U1, U2, U4/U6 and U5 for the major spliceosome and U11, U12, U4atac/U6atac and U5 for the minor spliceosome), a common seven-membered ring of Sm proteins (B/B′, D1, D2, D3, E, F, and G) arranged around the snRNA's Sm site (Sm core), and several proteins that are unique to the various U snRNPs (Patel and Steitz, 2003; Will and Luhrmann, 2001). Sm core assembly is a key step in snRNP biogenesis that takes place in the cytoplasm shortly after the nuclear export of the nascent snRNA precursors (pre-snRNAs). Proper assembly of the Sm core, cap hypermethylation and 3′-end processing of the snRNAs are prerequisites for the subsequent import of snRNPs into the nucleus where they function in pre-mRNA processing (Mattaj, 1986; Patel and Bellini, 2008). The assembly of Sm cores is carried out by the SMN complex (Fischer et al., 1997; Liu et al., 1997; Meister et al., 2001a; Pellizzoni et al., 2002b). The SMN complex is comprised of SMN, Gemins 2-8 and unrip (Baccon et al., 2002; Carissimi et al., 2005; Carissimi et al., 2006; Charroux et al., 1999; Charroux et al., 2000; Grimmler et al., 2005; Gubitz et al., 2002; Liu and Dreyfuss, 1996; Pellizzoni et al., 2002a). The SMN complex binds Sm proteins and snRNAs, bringing both components together and facilitating Sm core assembly (Yong et al., 2004a; Yong et al., 2002). SMN's essential function is to confer stringent specificity toward snRNAs and prevent illicit Sm core formation (Pellizzoni et al., 2002b). The specificity for snRNAs is determined by Gemin5, which is essential for Sm core assembly (Battle et al., 2006). Binding experiments on snRNAs showed that Gemin5 recognizes a snRNP code comprised of the Sm site [A(U)5-6G] and an adjacent 3′-terminal stem-loop structure in the snRNAs, except in U1 snRNA (which has a divergent Sm site), where it consists of stem-loop 1 (Golembe et al., 2005; Yong et al., 2004a; Yong et al., 2002; Yong et al., 2004b). The snRNP code of U4atac, which lacks a stem-loop 3′-terminal to the Sm site, has not been identified. Gemin5 can bind to snRNAs directly, on its own, via its WD-repeat domain (Lau et al., 2009). In cell extracts, Gemin5 is found with SMN and also as a SMN-free subunit, but the significance of this is currently unknown (Battle et al., 2007; Paushkin et al., 2002). Despite much progress from studies in vitro, the specific steps in the assembly of Sm cores in cells have not been defined. For example, the form in which Gemin5 exists in cells, its interaction with SMN, and the step at which the SMN complex interacts with snRNAs in cells are not known.
SMN deficiency causes spinal muscular atrophy (SMA), a common motor neuron degenerative disease (Lefebvre et al., 1995; Talbot and Davies, 2001). SMA severity is directly correlated with the degree of SMN deficiency (Lefebvre et al., 1997). Furthermore, SMN deficiency results in a corresponding decrease in snRNP assembly capacity (Wan et al., 2005). Importantly, SMN deficiency alters the repertoire of snRNAs and causes widespread and cell-type specific defects in pre-mRNA splicing (Zhang et al., 2008). Thus, the SMN complex plays a major role in splicing regulation and a detailed understanding of its activity and regulation is important, both because of its fundamental function in gene expression, as well as for potential therapy for SMA. For these reasons, we developed high throughput screens (HTS) to identify modulators of the SMN complex. Using HTS based on Sm core assembly, we have recently shown that the SMN complex is readily inactivated by reactive oxygen species (ROS) both in vitro and in cells, indicating that it is a redox-sensitive assemblyosome (Wan et al., 2008). However, the step at which ROS act is not known. To obtain additional chemical tools for studying the SMN complex, we devised a high throughput microscopy screen for small molecules that affect the unique localization of SMN (Liu and Dreyfuss, 1996). Surprisingly, this screen showed that protein synthesis inhibitors cause rapid relocalization of SMN from the cytoplasm to the nucleus. Using formaldehyde-mediated protein-protein and protein-RNA crosslinking of complexes in cells, high-stringency immunopurifications, mass spectrometry and high throughput sequencing, we determined the points at which these inhibitors act. These studies identified novel intermediates of the SMN complex, suggesting a stepwise pathway for its formation and demonstrating a key role for Gemin5 as the gateway for pre-snRNAs to snRNP biogenesis. We have also discovered the hitherto unknown pre-snRNAs for all the snRNAs and showed that the 3′-end precursor sequences function to enhance snRNP biogenesis, identifying pre-snRNAs as the substrates for Sm core assembly and 3′-end processing that occur on the SMN complex.
Results
Protein synthesis inhibition dissociates the SMN complex and impairs its activity in snRNP assembly
By immunofluorescence microscopy, the SMN complex displays a distinct cellular distribution, including staining throughout the cytoplasm and in discrete Gems bodies in the nucleus (Liu and Dreyfuss, 1996). Using SMN's unique cellular localization, we developed a high throughput high content immunofluorescence microscopy screen to detect changes in SMN sub-cellular localization. We cultured HeLa cells in 384-well plates with a different compound (at 10 μM) in each well and incubated for 4 hours. Cells were then processed for indirect immunofluorescence using the anti-SMN monoclonal antibody 2B1 (Liu and Dreyfuss, 1996). DAPI staining was simultaneously performed to define the boundary of each nucleus. Digital images were collected from six fields in each well and analyzed using imaging software for multiple parameters, including relative nuclear and cytoplasmic intensities, and number, size and signal intensity of Gems. Images of individual fields in wells flagged by the software as showing a significant change in SMN sub-cellular localization were examined directly, and active compounds were re-tested for verification. From screening of approximately 50,000 compounds, several were identified that caused a dramatic change in the cellular localization of SMN. This revealed that SMN's localization is dependent on the state of protein synthesis (Figure 1). Specifically, attenuation of protein synthesis resulted in rapid (< 4 hours) accumulation of SMN in the nucleus. The most striking nuclear accumulation of SMN was observed after treatment with the commonly used protein synthesis inhibitor, cycloheximide (CHX). The effect of cycloheximide on SMN localization was time-dependent with considerable staining of SMN in the nucleus being evident after only 2 hours of treatment (Figure S1A). Furthermore, we found that within the low micromolar concentrations tested (0.1 – 40 μM), cycloheximide exhibited a concentration-dependent accumulation of SMN in the nucleus, suggesting that an attenuation of translation rather than complete inhibition of translation is sufficient to cause this effect (Figure S1B and data not shown). To determine whether the effect of cycloheximide on localization is general to protein synthesis inhibitors, we tested several other protein synthesis inhibitors over a wide range of concentrations. Emetine, an irreversible inhibitor of translation elongation, displayed an effect on the localization of the SMN complex similar to that of cycloheximide (Figure S1C). Puromycin also showed an effect, but was more toxic and had greater cell-to-cell variability (Figure S1C). Thapsigargin, a known inducer of endoplasmic reticulum (ER) stress that attenuates protein synthesis indirectly by activating kinases that phosphorylate eIF2α, also caused SMN to accumulate in the nucleus (Figure S1C). On the other hand, cycloheximide-N-ethylethanoate (CHX-N), an inactive analog of cycloheximide, did not have any effect on SMN localization (Figure S1C). This indicates that the nuclear accumulation of SMN is dependent on ongoing protein synthesis. In addition, the nuclear accumulation is specific to the localization of SMN and not the result of general mis-localization of proteins, as the localization of many other, both nuclear and cytoplasmic proteins that we examined, including the RNA-binding proteins hnRNP A1, poly(A)-binding protein (PABP), FXR1 and snRNPs, was not significantly affected (Figure S1D). In all cases, inhibition of protein synthesis was monitored by [35S]-methionine incorporation which showed > 80% inhibition (Figure S2). To examine the effect of protein synthesis inhibition on other components of the SMN complex, HeLa cells were treated with cycloheximide, fixed and immuno-stained with antibodies to each of the Gemins. Surprisingly, inhibition of protein synthesis resulted in almost complete separation of the components of the SMN complex. SMN, Gemins 2, 6, 7 and 8 accumulated in the nucleus, whereas Gemins 3, 4, 5 and unrip remained in the cytoplasm (Figure 1A, B and data not shown). Thus, the association of the components of the SMN complex is dependent on ongoing protein synthesis. Western blots of total cell extracts showed that the proteins of the SMN complex are stable during this time (Figure 1C). As would be expected due to the separation of its components, the activity of the SMN complex after treatment with cycloheximide or another protein synthesis inhibitor, anisomycin, showed a sharp decrease (> 50%) (Figure 1D) when measured using a quantitative assay for assembly of Sm cores on U4 snRNA (Wan et al., 2005).
Figure 1. Inhibition of protein synthesis dissociates the SMN complex and impairs its activity.

(A) Immunofluorescence microscopic analysis of SMN complex components was performed before (Control) and after protein synthesis inhibition (CHX) (see also Figure S1 and S2).
(B) Immunofluorescence microscopic analysis as in (A) of the Gemin5-containing subunit.
(C) Extracts were prepared from cells treated with cycloheximide (CHX) or anisomycin (ANS) and analyzed by Western blot using antibodies specific to SMN complex proteins.
(D) Total cell extracts prepared as in (C) were used to measure the activity of the SMN complex. This activity represents the amount of Sm cores formed on biotinylated U4 snRNA. Error bars represent the mean of 2 independent experiments, each performed in triplicate.
The dissociation of the SMN complex upon protein synthesis inhibition provided an opportunity to define the composition of its subunits. For this we developed a methodology for complete ribo-proteomic analysis of in vivo captured RNPs that is illustrated in Figure 2A. This experimental approach allows proteomic as well as RNA analysis to be done in the same complex. To avoid the possibility of rearrangements of components that could occur after cell disruption, we first treated the cells with formaldehyde, which crosslinks and captures zero-distance protein-protein as well as protein-RNA interactions in intact cells (Gingras et al., 2007; Niranjanakumari et al., 2002). By an extensive series of preliminary experiments, we determined optimal conditions of mild formaldehyde crosslinking that efficiently crosslink the known SMN components but not other non-specific proteins. For these experiments, we used cell lines that stably express FLAG-tagged SMN or Gemin5 under the control of a tetracycline-inducible promoter to allow moderate expression levels of the tagged proteins. We have previously shown that the SMN complex is undisrupted and remains active under these conditions (Pellizzoni et al., 2002b). Complexes were purified from cycloheximide- or anisomycin-treated cells by immuno-affinity purification on anti-FLAG beads under highly stringent conditions, including high salt (500 mM NaCl) and detergent (1% Empigen BB). Under these conditions without prior crosslinking, each of these FLAG-tagged proteins does not retrieve any additional proteins, indicating the specificity of this procedure (Battle et al., 2006). Because of the formaldehyde crosslinking, Western blotting could not be used to determine composition. To overcome this as well as to discover potentially new components, we determined the composition of the total purified complexes by liquid chromatography tandem mass spectrometry (LC-MS/MS). Complexes purified from either FLAG-SMN or FLAG-Gemin5 cell line without treatment with inhibitors have the same protein composition and contain all of the known components (data not shown). However, treatment with protein synthesis inhibitors dramatically altered the composition of the complexes. This was measured using emPAI (exponentially modified protein abundance index) values obtained from the mass spectrometry data (Table S1). As emPAI corresponds to the absolute amount of each protein in the sample, this provided quantitation of the amount of each protein (Ishihama et al., 2005). As shown in Figure 2B, SMN immunopurified from cells treated with protein synthesis inhibitors contained almost no Gemins 4 and 5 and a much lower amount of Gemin3 (∼ 50%), indicating a separation of the SMN complex components under protein synthesis inhibition. Similarly, FLAG-Gemin5 retrieved almost no SMN or other Gemins under the same conditions (Figure 2C). These proteomics data are consistent with the imaging data and demonstrate that protein synthesis inhibition causes dissociation of the SMN complex. Furthermore, they provide evidence for a subunit comprised of SMN, Gemin2, 6, 7, 8 and unrip in cells that has not been previously identified. We note a difference in the relative amounts of Gemin8 and unrip in this subunit between the cycloheximide- and anisomycin-treated cells, but its significance is presently unknown. As unrip does not accumulate in the nucleus along with the other components of the SMN subunit upon protein synthesis inhibition (data not shown), it is possible that unrip only partially associates with it.
Figure 2. Ribo-proteomic analysis of in vivo captured RNPs identifies discrete subunits of the SMN complex that accumulate upon protein synthesis inhibition.
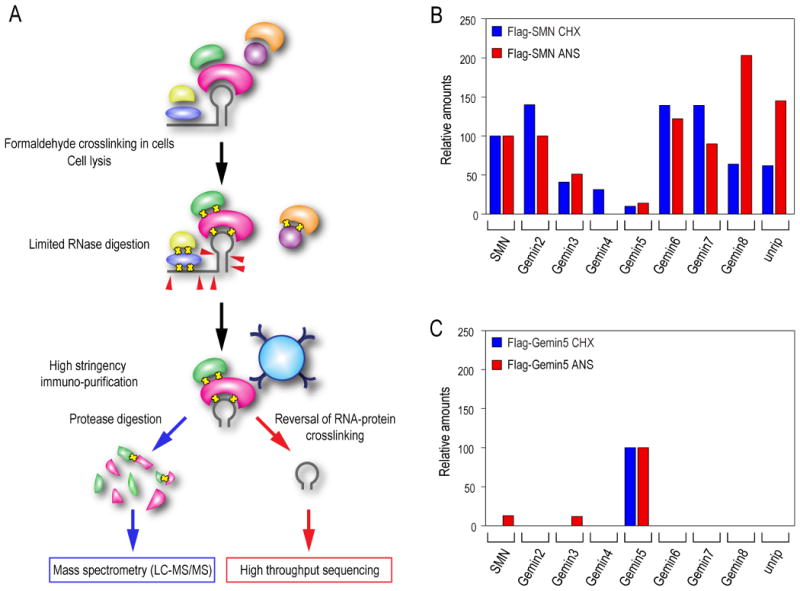
(A) An experimental strategy for in vivo capture of RNPs and their complete proteomic and RNA analysis by mass spectrometry and high throughput sequencing, respectively.
(B) Proteomic analysis of the SMN complex was performed before or after protein synthesis inhibition. The complex was purified from a FLAG-SMN expressing stable cell line and analyzed by LC-MS/MS mass spectrometry. The amount of each protein in the sample is expressed by its emPAI value and relative amounts of each component of the SMN complex associated with SMN were calculated by the ratio of their emPAI values (see also Table S1). The relative amounts are expressed as a percentage relative to its DMSO control that is normalized to 100%.
(C) Proteomic analysis of the Gemin5 subunit was performed as described in (A). The protein complex was purified from a FLAG-Gemin5 expressing stable cell line (see also Table S1).
Newly transcribed pre-snRNAs are associated with Gemin5
To determine which, if any, of the proteins of the SMN complex are associated with snRNA during protein synthesis inhibition, we used the same formaldehyde crosslinking conditions that also crosslink proteins to RNA (Niranjanakumari et al., 2002). For these experiments, we performed immunoprecipitations with antibodies to SMN (2B1) or Gemin5 (10G11) rather than immunopurify FLAG-tagged proteins as this produced lower non-specific binding to RNAs. Bound RNAs were purified and their levels quantitated by real-time RT-PCR (RT-qPCR) using primers specific for each snRNA. Strikingly, protein synthesis inhibition caused a large accumulation of every snRNA on Gemin5, particularly an up to 10-fold increase of U4atac. Both major (U2) and minor (U4atac) snRNAs were detected with Gemin5, but not U6 snRNA, which does not have an Sm site or assemble Sm cores (Figure 3A and data not shown). In contrast, there was only a marginal increase in the amount of any of the snRNAs with SMN (Figure 3B and C). To verify these results, we performed Northern blots using [γP32]ATP-labeled probes for several of the snRNAs (Figure 3B). The blots were also probed for 5S and 5.8S rRNA as a background control. Surprisingly, in the Gemin5 immunoprecipitation the snRNA probes detected almost exclusively RNAs of larger size than that of the snRNAs, which were immunoprecipitated with anti-Sm (Y12) and represent mature snRNAs. This suggested that the larger species associated with Gemin5 are likely to be precursors of the corresponding snRNAs. Under normal conditions (Figure 3B, DMSO) only very small amounts of pre-snRNAs are associated with Gemin5 or the SMN complex. Previous studies described pre-U2 snRNA that contains 11 nucleotides at the 3′-end that are removed by post-transcriptional processing (Jacobson et al., 1993; Kleinschmidt and Pederson, 1987). Indeed, both RT-qPCR and Northern blots using a probe specific for the known 3′-end pre-U2 snRNA sequence indicated that the larger U2 snRNA species that accumulated on Gemin5 are indeed pre-U2 snRNA (data not shown). Since, precursor sequences for most of the other snRNAs have not been described so far, this provides us with an opportunity to discover precursors for all the snRNAs (see later).
Figure 3. Newly synthesized precursor snRNAs accumulate on Gemin5 upon protein synthesis inhibition.
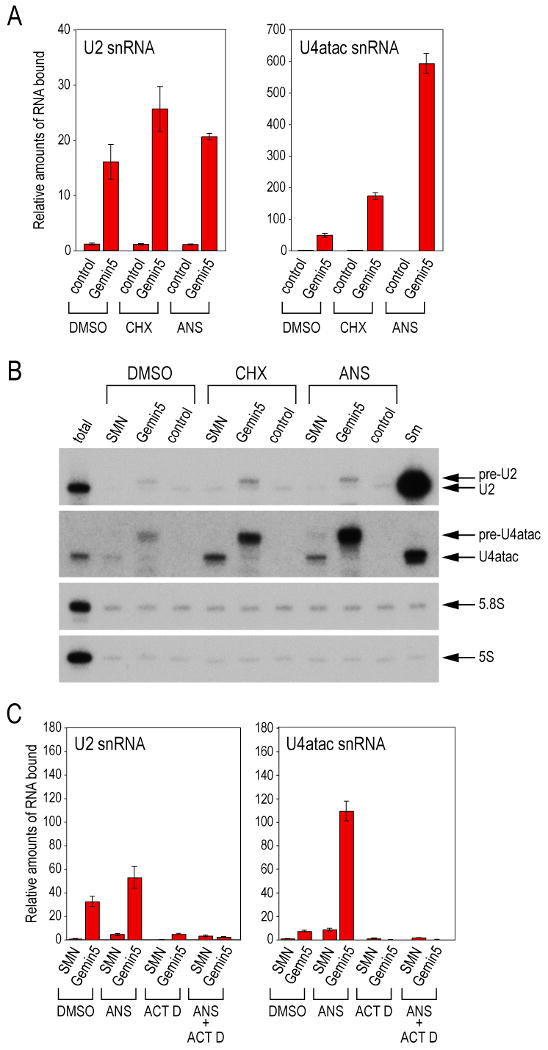
(A) Immunoprecipitations were performed using anti-Gemin5 or control antibodies to analyze snRNAs associated with Gemin5 in cells treated with cyclohexmide (CHX) or anisomycin (ANS) and crosslinked with formaldehyde. Immunoprecipitated RNA levels were quantitated by RT-qPCR using specific primers. Experiments were done at least three times; a typical experiment is shown. Error bars indicate standard deviation of three replicates.
(B) Northern blots were performed on RNAs analyzed in panel (A) using snRNA specific probes. RNA species larger than mature snRNAs are indicated as pre-snRNAs. RNAs from the immunoprecipitation using anti-Sm antibody served as a reference for mature snRNAs. The blot was also probed for 5S and 5.8S RNAs as controls.
(C) Experiments were performed as in (A) except cells were treated with anisomycin (ANS), actinomycin D (ACT D) or a combination of the two for 4 hrs and RNA immunoprecipitations were performed using anti-SMN and anti-Gemin5 antibodies.
To confirm that the precursor snRNAs are newly transcribed and exported to the cytoplasm, we treated cells with actinomycin D, a general transcription inhibitor. The amount of U2 and U4atac snRNAs associated with Gemin5 was then determined by RT-qPCR. As shown in Figure 3C, the accumulation of snRNAs on Gemin5, with or without anisomycin, was strongly dependent on ongoing transcription. These findings suggest that Gemin5 binds newly transcribed pre-snRNAs upon their export to the cytoplasm.
High throughput sequencing identifies numerous novel precursor snRNAs bound to Gemin5
It was evident from the Northern blots that Gemin5 bound to pre-U2 snRNA and to longer forms of snRNAs, such as U4atac, for which no information on precursors has been described (Figure 3B). We therefore wished to identify and determine the sequence of all the RNAs bound to Gemin5. To facilitate this, HeLa cells were treated with anisomycin to accumulate precursor snRNAs on Gemin5 and then exposed to formaldehyde for crosslinking. Gemin5-RNA complexes were then immunoprecipitated with anti-Gemin5 antibody (10G11). The beads were then subjected to limited RNase T1 digestion and washed extensively under highly stringent conditions (500 mM NaCl, 1% Empigen BB). The bound RNA fragments were released by mild heat treatment which reverses the formaldehyde induced RNA-protein crosslinks (Niranjanakumari et al., 2002) and their sequences were determined by high throughput sequencing. These represent Gemin5-bound RNA fragments that are relatively protected from RNase T1 digestion. More than 93,000 sequence reads, ranging in length from 15 – 30 nucleotides, were obtained and the sequence information was analyzed to map it to genomic sequences and visualized using TessLA and UCSC Genome Browser. The frequency at which a sequence is found generally reflects the relative abundance of that RNA sequence bound to Gemin5. This procedure allowed us to identify the RNAs bound to Gemin5 as well as define the specific regions of these RNAs that engage in Gemin5 binding.
As expected, the most highly represented sequence reads mapped to snRNA genes (64,091 reads). Many of the reads also contained extra 3′-end sequences (indicated by a dashed red line in Figure 4) that are not part of the mature snRNAs but are found as contiguous sequences in the corresponding genes (Table S2). These are, therefore, previously unknown precursors for all the snRNAs. Only a very limited amount of information has so far been available about snRNA precursors. Precursor sequences have been known only for pre-U2 snRNA transcribed from U2 gene clusters from chromosome 17 (Table S2), which we have also identified (Jacobson et al., 1993; Kleinschmidt and Pederson, 1987). However, in addition, we identified a pre-U2 snRNA from chromosome 11 having a different and longer 3′ extension (21 nucleotides) than previously predicted (11 nucleotides). Similarly, transcriptionally active snRNA gene loci that were not previously known to be expressed were also identified for U5 (Table S2). Previously, bands of electrophoretic mobility corresponding to precursors have been noted for U1 and U4 snRNA (approximately 25 and 7 nucleotides longer than the mature snRNAs, respectively) (Hernandez, 1985; Hernandez and Weiner, 1986; Madore et al., 1984; Yang et al., 1992). Our data identify a pre-U1 snRNA containing 49 additional nucleotides at the 3′ end. In most cases, with the exception of U4, these 3′ extensions are highly likely to form stable stem-loops based on the secondary structure predictions by Mfold (Figure 4) (Zuker, 2003).
Figure 4. High throughput sequencing identifies numerous novel precursor snRNAs bound to Gemin5.

Diagrammatic representation of the pre-snRNAs. The mature form of each snRNA is depicted as a gray line and the precursor sequences identified by high throughput sequencing are shown as a dashed red line. Nucleotide numbers for the mature and pre-snRNAs are indicated on the x-axis. Green boxes represent Sm sites. The non-canonical Sm site of U1 is shaded in light green. The previously determined snRNP code is shown as yellow bars beneath each snRNA, except for U4atac, which was identified in this study (hatched yellow bar). Regions of RNA bound to Gemin5, corresponding to high frequency sequence reads from the high throughput sequencing and protected from RNase T1 digestion, are overlayed on the snRNAs as a shaded blue region (see also Table S2).
A large number of sequence reads that overlapped with the snRNP code were obtained for each of the snRNAs (>100 - >30,000, for U11 and U1, respectively) (Figure 4). Specifically, most of the sequence reads for all the snRNAs, except pre-U4atac, mapped to the region that corresponds to 3′-terminal stem-loop of the mature snRNAs (blue area in Figure 4). This stem-loop region overlaps with the snRNP code for these snRNAs (yellow bars in Figure 4). Gemin5 binding to pre-U1 snRNA was detected in both the loop region of stem-loop 1 (SL1) that was previously shown to constitute its unique snRNP code, as well as in the SL4 region (Yong et al., 2002). It is intriguing that pre-U1 snRNAs contain, in the 3′ external sequence, a canonical Sm site sequence [A(U)5-6G] (Table S2), although it is not evolutionarily conserved. Interestingly, for pre-U4atac, most of the sequence reads mapped to the precursor sequence (Figure 4). This sequence is not part of the mature U4atac snRNA and is predicted to form a stem-loop 3′-terminal to the Sm site and thus resembles a snRNP code. For pre-U4 and pre-U4atac, sequence reads were also obtained from the 5′-most stem-loop. We note that the loop region in this stem-loop contains a consensus Cajal body box sequence (CAB) (Richard et al., 2003).
Reactive oxygen species (ROS) arrest pre-snRNA-SMN complexes
ROS inactivate the SMN complex and cause intermolecular disulfide bond crosslinking of SMN (Wan et al., 2008). To determine the step at which ROS inhibition occurs, SMN complexes were purified from cells that were treated for 1 hr with the ROS generator β-lapachone (5 μM) or DMSO as a control. Purification was performed using FLAG-SMN and composition was further analyzed by SDS-PAGE using silver staining and Western blotting. Notably, and in contrast to protein synthesis inhibition, ROS did not dissociate the SMN complex (Figure 5A). Instead, a relative enrichment of Gemin5 was apparent, reflecting a greater degree of association between Gemin5 and SMN as well as several other components of the SMN complex, such as Gemin3. We also analyzed the snRNA and pre-snRNA content by Northern blot using specific probes for several snRNAs. This showed that ROS cause accumulation of pre-snRNAs, including pre-U2, pre-U4 and pre-U4atac, on the SMN complex (Figure 5B SMN lanes). Using precursor specific primers for RT-qPCR measurements, pre-U2 and pre-U4atac were enriched on the SMN complex upon exposure to ROS by 4- and 15-fold, respectively (Figure 5C). Pre-snRNAs were not detectable in the SMN complex in the absence of ROS exposure. Mature snRNAs were also detectable on the SMN complex in the absence of ROS (Figure 3 and 5). We also note the increased amount of precursors with Gemin5, particularly pre-U4 and pre-U4atac. These data suggest that ROS stall an SMN complex containing pre-snRNAs poised for Sm core assembly and 3′-end processing.
Figure 5. Accumulation of pre-snRNA-SMN and pre-snRNA-Gemin5 complexes.

(A) HeLa tet-ON cells expressing FLAG-SMN were treated with DMSO or β-lapachone for 1 hr. The SMN complex was purified and analyzed by SDS-PAGE and proteins were detected by silver stain or Western blot. The control lane shows background of the immunoprecipitation.
(B) HeLa cells were treated with DMSO or β-lapachone for 1 hr and then fixed with formaldehyde. RNA immunoprecipitations were carried out using antibodies against SMN or Gemin5 or control antibody. Bound RNAs were analyzed by Northern blot using specific [γP32]-labeled probes.
(C) Immunoprecipitations were performed using anti-SMN or control antibodies to analyze the snRNAs associated with SMN in cells treated with ROS and formaldehyde crosslinked. Immunoprecipitated RNA levels were quantitated by RT-qPCR using precursor specific primers. One representative experiment is shown. Standard deviations among the three replicates in a single experiment were negligible and therefore not indicated.
(D) Whole cell extracts were prepared from HeLa cells 36 hours after transfection with siRNA against SMN. Proteins were analyzed by quantitative Western blot as indicated. Cell extract from HeLa cells transfected with a control siRNA is also shown.
(E) Immunoprecipitations were performed using anti-Gemin5 or control antibody to analyze the pre-snRNAs associated with Gemin5. Cells were crosslinked with formaldehyde following SMN knockdown. Immunoprecipitated pre-snRNA levels were quantitated by RT-qPCR using primers specific to the precursor sequences. One representative experiment is shown. The standard deviations among the three replicates in a single experiment were negligible and therefore not indicated.
A prediction of this model (see Figure 7) is that the Gemin5-pre-snRNA intermediate is upstream of the ROS stalled complex. To test this, we knocked down SMN by RNAi (Figure 5D), exposed the cells to formaldehyde crosslinking and performed immunoprecipitations with antibodies to Gemin5 (10G11). The bound pre-snRNA levels were quantitated by RT-qPCR. SMN RNAi caused a large accumulation of pre-snRNAs, for both major (U2) and minor (U4atac) snRNAs, on Gemin5 (Figure 5E). This experiment, performed without the use of inhibitors, provides further evidence for the Gemin5-pre-snRNA intermediate in vivo. The results further demonstrate that Gemin5-pre-snRNA is upstream of the ROS-defined complete SMN complex (Figure 5E.)
Figure 7. Schematic representation of the discrete intermediates of the SMN complex and the stepwise assembly pathway of spliceosomal snRNPs.
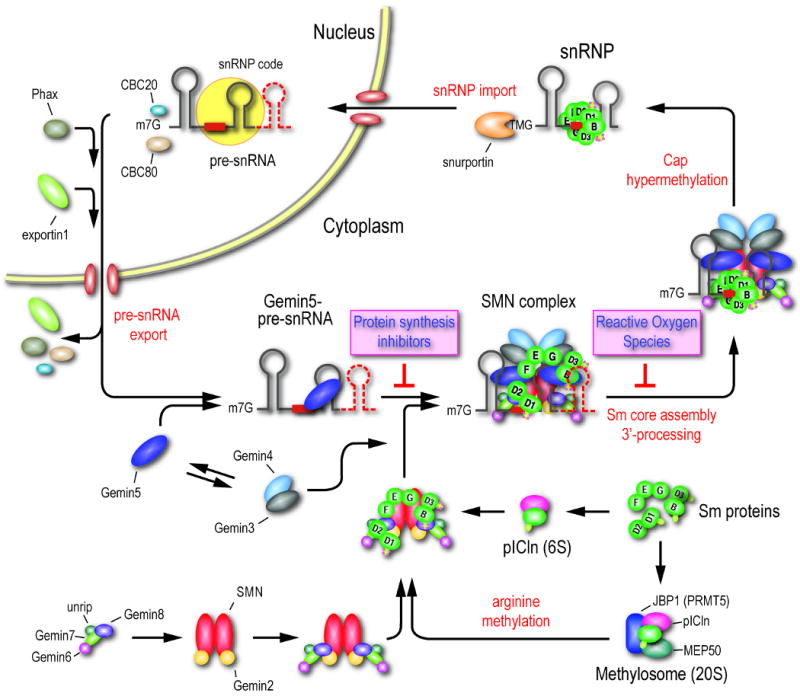
The point of inhibition of the two classes of inhibitors, protein synthesis inhibitors and ROS, identified from high throughput screens are indicated. For simplicity, the SMN complex is depicted as a dimer. A detailed description of the model is provided in the text.
The pre-snRNA 3′-extra sequences function to enhance snRNP assembly
Because the data indicated that the pre-snRNAs are the substrates for Sm core assembly, we asked if the 3′-end sequences found in the precursors have an effect on Sm core assembly. For this, we compared Sm core assembly of U1 and U4atac pre-snRNAs with their corresponding snRNAs in the in vitro snRNP assembly assay (Wan et al., 2005) (Figure 6). Both pre-U1 and pre-U4atac, at varying concentrations and reaction times, show an enhancement in Sm core assembly relative to its snRNA at every concentration and every time point tested. The results shown in Figure 6A show a strong, 1.7-fold and 1.6-fold enhancement in Sm core assembly for pre-U4atac and pre-U1, respectively. This level of enhancement is also evident as an increase in the rate of Sm core assembly for each precursor (1.7 fold) at a fixed concentration (Figure 6B), further indicating the positive effect of the precursor sequence on snRNP assembly.
Figure 6. pre-snRNAs function as enhancers of Sm core assembly.
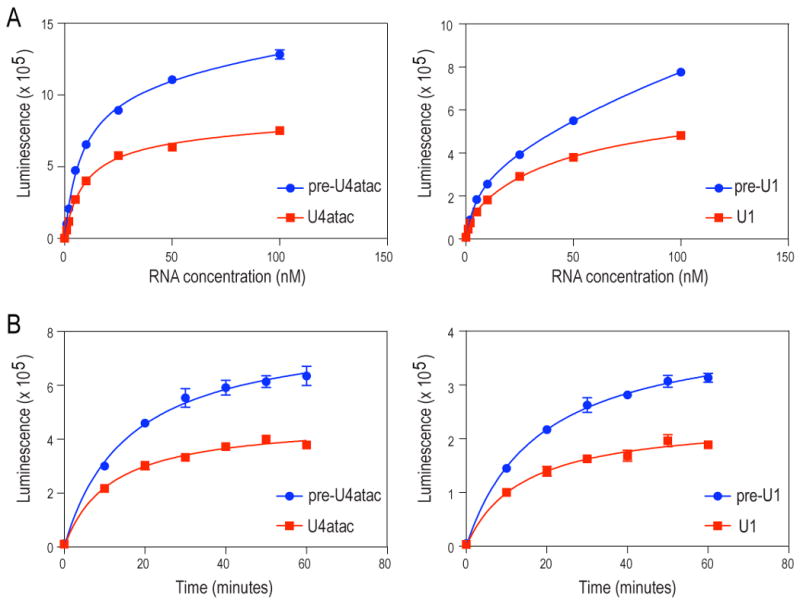
Quantitative in vitro snRNP assembly reactions were performed as previously described (Wan et al., 2005) except pre-snRNA and snRNA concentrations were varied (A) or reaction times were varied (B). Error bars represent the standard deviation of three replicates in a single experiment. One representative experiment is shown.
Discussion
snRNP biogenesis is critical for gene expression in eukaryotes. The SMN complex plays an essential role in this process and a considerable amount of information is available about its components, interactions and activity in snRNP assembly in vitro (Chari et al., 2008; Fischer et al., 1997; Meister et al., 2001a; Narayanan et al., 2004; Pellizzoni et al., 2002b). In contrast, little is known about the activity and regulation of the SMN complex in cells. Here, we used two classes of SMN complex inhibitors discovered by high throughput screening, protein synthesis inhibitors and ROS, to dissect this pathway in cells. By formaldehyde-mediated protein-protein and protein-RNA crosslinking, mass spectrometry and high throughput sequencing, we defined the points of inhibition for each of these inhibitors, providing important insights into snRNP biogenesis. One of the surprising observations we describe is that protein synthesis inhibition, by a variety of mechanisms, results in accumulation of separate subunits of the SMN complex. One including SMN then translocates from the cytoplasm to the nucleus. This is an unexpected biological effect of these widely used reagents whose effects are frequently interpreted as resulting from inhibition of protein synthesis alone. The mechanism by which protein synthesis inhibition causes this is not presently known. It could be the result of a decrease in the level of a rapidly turning over protein(s), including the supply of newly synthesized Sm proteins. The components of the SMN complex themselves are stable under these conditions. It could also, however, be the result of a stress signal generated as a result of attenuation of the translation machinery. Nevertheless, protein synthesis inhibitors provided invaluable information on the subunits and intermediates of the SMN complex.
A summary of our major findings and view of the pathway is presented as a model in Figure 7. Nascent pre-snRNA transcripts produced by RNA polymerase II become associated with the cap binding complex (CBC) proteins, CBC20 and CBC80, that recruit an adaptor protein, Phax, which then recruits the nuclear export receptor exportin 1 (CRM1) (Ohno et al., 2000). The fate of the nascent pre-snRNAs immediately after their export to the cytoplasm was not previously known and our data demonstrate that they associate with Gemin5. Because Gemin5 is almost entirely cytoplasmic, this association likely occurs after the export of the pre-snRNAs from the nucleus, but the possibility that Gemin5 can shuttle in and out of the nucleus and associate with pre-snRNAs in the nucleus prior to export cannot be ruled out. This suggests that Gemin5 directs pre-snRNAs to the SMN complex or subunit. We further identified a subunit containing SMN together with Gemins 2/6/7/8 and unrip. The association of this subunit with the Gemin5-pre-snRNA subunit is dependent on protein synthesis. Smaller, as well as, additional subunits may exist. For example, SMN/Gemin2, Gemin6/7/8/unrip and Gemin3/4/5 have been described (Battle et al., 2007; Carissimi et al., 2005; Carissimi et al., 2006). As we have not detected a Gemin3/4/5 complex, but because Gemin3 and Gemin4 are in the SMN complex (Figure 5), we depict Gemin3/4 as a subunit that joins the SMN complex separately. Proteomic analyses showed that the SMN subunit also contains the Sm proteins, however, it has not been possible to quantitate them due to the small size, paucity of trypsin cleavage sites and low concentration of these proteins in this subunit. Previous studies have shown that the Sm proteins are found in complexes with pICln (6S complex) and methylosome/PRMT5 (Friesen et al., 2001; Friesen et al., 2002; Meister et al., 2001b; Pu et al., 1999), which makes symmetric dimethyl-arginine (sDMA) modification on some of the Sm proteins and thereby enhances their association with SMN.
In addition to protein synthesis inhibitors, formaldehyde crosslinking of complexes in living cells, which we optimized to capture the protein-protein and protein-RNA interactions that occur in vivo, was key to discovering intermediates. This ribo-proteomic strategy provides a powerful tool and therefore should be widely applicable for studies of all aspects of RNA metabolism. Formaldehyde crosslinking of protein-RNA complexes in cells is very efficient compared to UV crosslinking (Choi and Dreyfuss, 1984a, b; Dreyfuss et al., 1984; Niranjanakumari et al., 2002). Unlike UV crosslinked protein-RNA complexes, these can be readily reversed and we show that the recovered RNAs are amenable to further analysis. In addition, the protein composition of the same complexes can be determined using mass spectrometry, whereas UV does not generate protein-protein crosslinks. These advantages of formaldehyde crosslinking, together with the ability of protein synthesis inhibitors to affect a large accumulation of pre-snRNAs on Gemin5, allowed us to trap the transient Gemin5-pre-snRNA intermediate and determine the RNA sequences.
The wealth of sequence information led to the discovery of precursors for all the major and minor snRNAs. To date, only limited information on precursor sequences was available for snRNAs (Hernandez, 1985; Hernandez and Weiner, 1986; Kleinschmidt and Pederson, 1987; Neuman de Vegvar and Dahlberg, 1989), however, our studies revealed multiple precursor species for each snRNA. Since there are multiple gene copies for most of the major snRNAs with only slight sequence variation among them, distinguishing between transcriptionally active snRNA genes from pseudogenes has been difficult and is not easily achieved. Our data provide information on the loci that are transcribed and identify those with previously uncharacterized transcriptional activity. It is of importance to note that the majority of the sequence reads in most of the snRNAs did not map to the precursor sequences but rather to the regions that corresponded to the snRNP code existing in the mature snRNAs (Figure 4). These in vivo mapping data from high throughput sequencing are consistent with previous mapping results performed in vitro (Yong et al., 2004a). RNase T1 digestion readily removed the Sm site suggesting that it is exposed in its Gemin5-bound state.
One of the most striking findings relates to the precursor sequence of U4atac. In contrast to the other snRNAs, most of the Gemin5 bound fragments included the 3′-end of pre-U4atac, a sequence predicted to form a stem-loop that is later removed as the mature U4atac forms (Figure 4). Notably, this pre-U4atac sequence, in contrast to precursor sequences of the other snRNAs, is evolutionarily conserved. It was difficult to understand how an Sm core could assemble on mature U4atac, as unlike other snRNAs, it does not have a canonical snRNP code as it lacks a stem loop 3′-terminal to the Sm site. These data strongly suggest that the snRNP code for U4atac is contained in its precursor sequence. Thus, Gemin5 binds specifically to pre-snRNAs and these, but not mature snRNAs, are the substrates for the SMN complex. Importantly, the extra 3′ sequences found in the pre-snRNAs are not merely neutral appendages that are later trimmed, but rather have an important function(s) in Sm core assembly. Our measurements for pre-U1 and pre-U4atac snRNAs demonstrate that they significantly enhance a critical step of snRNP biogenesis. We suggest that these precursor sequences are subsequently removed by 3′-end processing shortly after the step in the assembly that they facilitate because they are not needed, and may potentially interfere with the function of the mature snRNA.
ROS inhibit the activity of the SMN complex but do not dissociate it. In fact, it contains all the known protein components of the SMN complex, as well as pre-snRNAs and Sm proteins (data not shown). The mechanism by which ROS inactivate the SMN complex and inhibit snRNP assembly is not known. However, ROS cause SMN-SMN intermolecular disulfide bridging and it is possible that this is, at least in part, the basis of inhibition (Wan et al., 2008). The SMN-SMN oxidative disulfide crosslinking also provides evidence that SMN in cells is oligomeric. We depict SMN as a dimer (Figure 7) for simplicity and because the number of subunits of SMN in oligomers is not known. The hyper-stoichiometric amount of Gemin5 and the presence of pre-snRNA in the SMN complex support the idea that Gemin5 delivers pre-snRNAs to this complex and that this function is not inhibited by ROS. The RNAi experiments suggest that the competition of the various pre-snRNA-Gemin5 complexes for a limiting amount of SMN subunit could contribute to the altered stoichiometry of snRNPs and the splicing abnormalities that likely result from it, as seen in the SMN deficient SMA mouse model (Zhang et al., 2008). The presence of pre-snRNAs and low levels of mature snRNAs in the ROS stalled SMN complex suggests that 3′-end processing occurs on the SMN complex (Figure 5B). Previous studies have shown that 5′-end modification of the monomethyl-guanosine (m7G) to trimethyl-guanosine (TMG) cap also occurs on the SMN complex by Tgs1 (Mouaikel et al., 2003). We suggest that the Gemin5-pre-snRNA and the SMN subunits associate when the SMN complex is loaded with Sm proteins. The structure of Gemin6 and Gemin7, both of which contain Sm-like folds and interact with each other in a similar manner to Sm-Sm (Ma et al., 2005), together with the role of Gemin6/7/8/unrip in assembly (Carissimi et al., 2006) suggests a role for these proteins in forming the Sm core.
These studies using ribo-proteomic analysis of in vivo captured RNPs provide a new view of the biology of the SMN complex. They reveal previously unknown subunits of the SMN complex, identify activities required for their interactions, suggest a specific order of steps in snRNP biogenesis, and led to the discovery of precursors for all the snRNAs. The fate of nascent pre-snRNAs immediately after their export to the cytoplasm was not known. Our data show that Gemin5 is the link between newly transcribed and exported pre-snRNAs and their subsequent assembly on the SMN complex. We further show that the snRNA precursor's 3′ sequences function to enhance snRNP biogenesis. The ROS stalled SMN complex represents the active transient intermediate poised for Sm core assembly and pre-snRNA processing. We suggest that the pre-snRNAs are the substrates for the SMN complex. Almost without exception, previous studies of the SMN complex and Sm core assembly used snRNAs because the pre-snRNAs were not considered to be the substrates and their sequences were not known. Previous experiments can now be revisited and future studies can now be designed using pre-snRNAs as substrates, from which additional aspects of the complexity and regulation of snRNP biogenesis will likely emerge.
Experimental Procedures
Cell culture, drug treatment and formaldehyde crosslinking
HeLa PV cells were grown in DMEM supplemented with 10% FBS. Where indicated, cells were treated with cycloheximide (20 μM), anisomycin (1 μg/ml), actinomycin D (5 μg/ml) or β-lapachone (5 μM). Protein synthesis inhibitors were treated for 6 hrs and β-lapachone for 1 hr. At the end of the incubation, cells were harvested, washed twice with ice-cold 1X PBS and fixed with 0.2% formaldehyde for 10 min while rotating at room temperature. The fixation was quenched in 0.15 M glycine/pH 7.0 for 10 min while rotating at room temperature followed by washing with 1X PBS. Cells were lysed in 20 mM Tris-HCl pH 7.5 buffer containing 2.5 mM MgCl2, 500 mM NaCl and 1% Empigen BB.
Immunoprecipitation, immunofluorescence, Western blotting and antibodies
Antibodies used in this study: α-SMN (2B1 or 62E7), α-Gemin2 (2E17), α-Gemin3 (12H12), α-Gemin4 (64I1), α-Gemin5 (10G11), α-Gemin6 (6H5 or rabbit polyclonal), α-Gemin7 (7E2), α-Gemin8 (13C3), α-Sm (Y12) and α-unrip (BD transduction). Western blots and immunofluorescence were performed as described previously (Liu and Dreyfuss, 1996). Immunoprecipitation of RNA-protein complexes was carried out as described (Choi and Dreyfuss, 1984a, b; Dreyfuss et al., 1984).
Reverse transcription and RT-qPCR measurement of snRNAs
Specific primers for each snRNA were used to generate cDNA using Advantage RT-for-PCR kit (Clontech) on immunopurified RNAs. Half of the RNA was used as a template in a 20 μl reaction. 1.5 and 2.5% percent of the cDNA was used for each RT-qPCR reaction for the major and the minor snRNAs, respectively. Quantification was performed relative to DMSO. Primer sequences for pre-U2 are 5′-GTTCTCTCCCCGAAGGGAGA-3′ and 5′-ATCCCCGGAGGGGGTGC-3′ and for pre-U4atac is 5′-GATAAGTCTATGAAAACAGG-3′. Further experimental details and all other primers and probes used have been described elsewhere (Zhang et al., 2008).
Assay for in vitro assembly of snRNPs
An automated and quantitative assay for in vitro snRNP assembly was performed as previously described (Wan et al., 2005).
RNA interference
Transfection of control siRNA and siRNA targeted against SMN (Feng et al., 2005) into HeLa cells was performed using Lipofectamine RNAiMAX (Invitrogen) as specified by the manufacturer. Transfected cells were analyzed 36-40 hours post transfection. All siRNAs were purchased from Dharmacon.
Liquid chromatography tandem mass spectrometry (LC-MS/MS)
FLAG-SMN or FLAG-Gemin5 complexes were immuno-affinity purified from cells fixed with 0.2% formaldehyde under highly stringent conditions (20 mM Tris-HCl pH 7.5, 2.5 mM MgCl2, 500 mM NaCl and 1% Empigen BB). Protein complexes were trypsinized and analyzed with a nanoLC/nanospray/LTQ mass spectrometer by the Proteomics Core facility at the University of Pennsylvania School of Medicine.
High throughput sequencing
Immunoprecipitation using anti-Gemin5 antibody (10G11) was performed in cell extracts prepared from 0.2% formaldehyde fixed HeLa cells treated with anisomycin. Gemin5-bound RNAs were treated with limited amounts of RNase T1 on the beads and reverse-crosslinking was done as described (Niranjanakumari et al., 2002). Construction of a cDNA library was performed according to the manufacturer's protocol (Illumina). High throughput sequencing was performed with an Illumina Genome Analyzer and data analysis was carried out at the Functional Genomics Core at the University of Pennsylvania School of Medicine. The sequence data are available at the Gene Expression Omnibus under the accession number GSE20751.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratory for helpful discussions and comments on this manuscript. We thank Dr. Chao-Xing Yuan of the Proteomics Core facility for expert help with mass spectrometry experiments. The facility is supported by grants P30CA016520 (Abramson Cancer Center) and ES013508-04 (CEET). We thank Dr. Jonathan Schug of the Functional Genomics Core at the University of Pennsylvania School of Medicine for excellent help with high throughput sequencing and data analysis. We also thank Dr. Larry N. Singh for help with processing and depositing the data. This work was supported by the Association Française Contre les Myopathies (AFM). G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baccon J, Pellizzoni L, Rappsilber J, Mann M, Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J Biol Chem. 2002;277:31957–31962. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Wang J, Dreyfuss G. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J Biol Chem. 2007;282:27953–27959. doi: 10.1074/jbc.M702317200. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell. 2006;23:273–279. doi: 10.1016/j.molcel.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Carissimi C, Baccon J, Straccia M, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L. Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett. 2005;579:2348–2354. doi: 10.1016/j.febslet.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Carissimi C, Saieva L, Baccon J, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L. Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. J Biol Chem. 2006;281:8126–8134. doi: 10.1074/jbc.M512243200. [DOI] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YD, Dreyfuss G. Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc Natl Acad Sci U S A. 1984a;81:7471–7475. doi: 10.1073/pnas.81.23.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YD, Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol. 1984b;99:1997–1204. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Choi YD, Adam SA. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol Cell Biol. 1984;4:1104–1114. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Gubitz AK, Wan L, Battle DJ, Dostie J, Golembe TJ, Dreyfuss G. Gemins modulate the expression and activity of the SMN complex. Hum Mol Genet. 2005;14:1605–1611. doi: 10.1093/hmg/ddi168. [DOI] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Wyce A, Paushkin S, Abel L, Rappsilber J, Mann M, Dreyfuss G. A novel WD repeat protein component of the methylosome binds Sm proteins. J Biol Chem. 2002;277:8243–8247. doi: 10.1074/jbc.M109984200. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Dreyfuss G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol Cell Biol. 2005;25:10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler M, Otter S, Peter C, Muller F, Chari A, Fischer U. Unrip, a factor implicated in cap-independent translation, associates with the cytosolic SMN complex and influences its intracellular localization. Hum Mol Genet. 2005;14:3099–3111. doi: 10.1093/hmg/ddi343. [DOI] [PubMed] [Google Scholar]
- Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J Biol Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985;4:1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N, Weiner AM. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Rhoadhouse M, Pederson T. U2 small nuclear RNA 3′ end formation is directed by a critical internal structure distinct from the processing site. Mol Cell Biol. 1993;13:1119–1129. doi: 10.1128/mcb.13.2.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt AM, Pederson T. Accurate and efficient 3′ processing of U2 small nuclear RNA precursor in a fractionated cytoplasmic extract. Mol Cell Biol. 1987;7:3131–3137. doi: 10.1128/mcb.7.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Bachorik JL, Dreyfuss G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat Struct Mol Biol. 2009;16:486–491. doi: 10.1038/nsmb.1584. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Ma Y, Dostie J, Dreyfuss G, Van Duyne GD. The Gemin6-Gemin7 heterodimer from the survival of motor neurons complex has an Sm protein-like structure. Structure. 2005;13:883–892. doi: 10.1016/j.str.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Madore SJ, Wieben ED, Kunkel GR, Pederson T. Precursors of U4 small nuclear RNA. J Cell Biol. 1984;99:1140–1144. doi: 10.1083/jcb.99.3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001a;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001b;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- Mouaikel J, Bujnicki JM, Tazi J, Bordonne R. Sequence-structure-function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res. 2003;31:4899–4909. doi: 10.1093/nar/gkg656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Achsel T, Luhrmann R, Matera AG. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol Cell. 2004;16:223–234. doi: 10.1016/j.molcel.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Neuman de Vegvar HE, Dahlberg JE. Initiation and termination of human U1 RNA transcription requires the concerted action of multiple flanking elements. Nucleic Acids Res. 1989;17:9305–9318. doi: 10.1093/nar/17.22.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Patel SB, Bellini M. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 2008;36:6482–6493. doi: 10.1093/nar/gkn658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J Biol Chem. 2002a;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002b;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Pu WT, Krapivinsky GB, Krapivinsky L, Clapham DE. pICln inhibits snRNP biogenesis by binding core spliceosomal proteins. Mol Cell Biol. 1999;19:4113–4120. doi: 10.1128/mcb.19.6.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Davies KE. Spinal muscular atrophy. Semin Neurol. 2001;21:189–197. doi: 10.1055/s-2001-15264. [DOI] [PubMed] [Google Scholar]
- Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Ottinger E, Cho S, Dreyfuss G. Inactivation of the SMN complex by oxidative stress. Mol Cell. 2008;31:244–254. doi: 10.1016/j.molcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Yang H, Moss ML, Lund E, Dahlberg JE. Nuclear processing of the 3′-terminal nucleotides of pre-U1 RNA in Xenopus laevis oocytes. Mol Cell Biol. 1992;12:1553–1560. doi: 10.1128/mcb.12.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol Cell Biol. 2004a;24:2747–2756. doi: 10.1128/MCB.24.7.2747-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Pellizzoni L, Dreyfuss G. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 2002;21:1188–1196. doi: 10.1093/emboj/21.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Wan L, Dreyfuss G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 2004b;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN Deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


