Abstract
Reactions of [ReOCl3(PPh3)2] with the bidentate ligands 2-(2-hydroxyphenyl) benzoxazole and (2′-hydroxyphenyl)-2-thiazoline resulted in the isolation of the complexes [ReOCl2(OC6H4-2-C=NC6H4-2-O)(PPh3)] (1) and [ReOCl2(OC6H4-2-C=NCH2CH2S)(PPh3)] (2). Reactions of 1 with the tridentate Schiff base ligands salicylaldehyde 2-hydroxyanil (H2L1b), salicylaldehyde 2-mercaptoanil (H2L2b) afnd S-benzyl-2-[(2-hydroxyphenyl)methylene] dithiocarbazate (H2L3b) yielded the ‘3+2’ rhenium(V) oxo species [ReO(OC6H4-2-C=NC6H4O) (OC6H4CH=NC6H4O)] (3), [ReO(OC6H4-2-C=NC6H4O)-(OC6H4CH=NC6H4S)] (4) and [ReO(OC6H4-2-C=NC6H4O){OC6H4CH=N–N=C(SCH2C6H5)S}] (5). Similarly, the reactions of [ReOCl2(OC6H4-2-C=NC6H4S)(PPh3)] (2) with H2L2b, H2L3b and 2-[(2-hydroxyphenyl) methylene]-N-phenyl-hydrazinecarbothioamide (H2L4b) were exploited to prepare [ReO(OC6H4-2-C=NC6H4S)(OC6H4CH=NC6H4O)] (6), [ReO(OC6H4C=NC6H4S){OC6H4CH=N–N=C (SCH2C6H5)S}] (7) and [ReO(OC6H4-2-C=NC6H4S){OC6H4CH=N–N=C(NHC6H5)}] (8), respectively.
Keywords: Oxorhenium complexes, Schiff base complexes, Crystal structures, ‘3+2’ Rhenium(V) chemistry
1. Introduction
The continued expansion of the chemistry of technetium and rhenium reflects the applications of the radionuclides of these elements to diagnostic and therapeutic nuclear medicine, respectively [1]. The Group VII congeners, technetium and rhenium, exhibit considerable chemical diversity, with complexes of oxidation states −1 to +7 having been reported. This chemical versatility is reflected in a number of robust chemical cores which have been investigated for potential applications to radiopharmaceutical development. These include the metal(V)-oxo {MO}3+ [1], the metal(V)-nitrido {MN}2+ [2–4], the metal(V)-imido {MNR}3+ [5–8], the metal(III)-hydrazino {M(NNR)}x+ [9,10] and the metal(I)-tricarbonyl {M(CO)3}+ [11–15] cores. The most extensively developed chemistry is that of the technetium and rhenium complexes of the metal(V)-oxo core [1]. While a major class of technetium and rhenium complexes are represented by the tetradentate [Nx S(4−x)] oxometal(V) compounds, another successful approach to the development of radiopharmaceuticals of technetium and rhenium has exploited Schiff base ligands to stabilize the [MO]3+ core [16–32].
In chemistry paralleling that of the {MO}3+ core with tetradentate aminothiol ligand types [Nx S(4−x)], polydentate Schiff base ligands derived from salicylaldehyde and diamines (salenes) affording [O2N2] donor sets have been extensively studied. However, the evolution of the chemistry of the {MO}3+ core by Spies and others demonstrated that mononuclear, neutral and low molecular weight technetium and rhenium complexes of the general type [MO(SXS)(SR)] were readily attainable by the reaction of tridentate dithiol ligands HSXSH (X=S, O, NR) and monodentate thiol ligand R–SH with starting materials containing the {MO}3+ [M=Tc, Re] core [33–37]. Using this approach, Tc(V) and Re(V) compounds based on the ‘3+1’ donor ligand design based on tridentate Schiff bases in combination with monothiols have also been prepared [25,28].
Not unexpectedly, these ‘3+1’ complexes are found to be relatively unstable in vitro and in vivo due to the metabolism and substitution of the labile monodentate ligand through transchelation by physiological thiols such as cysteine and glutathione [38], leading to a accumulation of the products in thiolate-rich organs such as the liver, rather than localization in the organs of interest. One approach to the preparation of more stable 18 electron structures, in contrast to the 16 electron structures of the ‘3+1’ {MO}3+ complexes, is to expand the coordination sphere at the technetium or rhenium site by exploiting ‘3+2’ ligand chemistry, which disposes a tridentate and a bidentate ligand set about the oxometal core [39–41].
We previously demonstrated that ‘3+2’ six coordinate oxorhenium complexes were readily accessible with a bidentate ligand such as 8-hydroxy-5-nitroquinoline and a variety of tridentate (ONS) and (ONO) donor ligands, including the Schiff bases HOC6H4-2-CH=NC6H4XH (X = O, S), that is salicylaldehyde-2-hydroxyanil (H2L1b) and salicylaldehyde-2-mercaptoanil (H2L2b), respectively [42]. As an extension of these studies, we have prepared a series of ‘3 + 2’ complexes of the {ReO}3+ core by reacting [ReOCl2(L1a)(PPh3)] (1) and [ReOCl2(L2a)(PPh3)] (2), where HL1a is 2-(2-hydroxyphenyl)benzoxazolate and HL2a is (2′-hydroxy-phenyl)-2-thioazoline, with the series of tridentate Schiff base ligands salicylaldehyde-2-hydroxyanil (H2L1b), salicylaldehyde-2-mercaptoanil (H2L2b), S-benzyl-2-[(2-hydroxyphenyl)methylene] dithiocarbazate (H2L3b), and 2-[(2-hydroxyphnyl)methylene]-N-phenyl-hydrazinecarbothioamide (H2L4b) (Scheme 1). The structures of [ReO(η3-OC6H4-2-CH=NC6H4-2-O)-(η2OC6H4C=NC6H4-2-O)] (3), [ReO(η3-OC6H4-2-CH=NC6H4-2-S)(η2-OC6H4C=NC6H4-2-O)]·MeOH (4-MeOH), [ReO{η3-OC6H4-2-CH=N–N=C(SCH2C6H5)-S}(η2-OC6H4C=NC6H4-2-O)]·CH2Cl2 (5-CH2Cl2) and [ReO(η3-OC6H4-2-CH=NC6H4-2-O)(η2-OC6H4C=NC6−H4S)] (6) are reported.
Scheme 1.
2. Experimental
2. 1. General considerations
1H NMR spectra were recorded on a Bruker DPX 300 (1H 300.10 MHz) spectrometer in CDCl3 (δ 7.27 ppm). IR spectra were recorded as KBr pellets with a Perkin-Elmer Paragon 1000 FTIR. Elemental analysis for carbon, hydrogen and nitrogen were carried out by Oneida Research Services, Whitesboro, NY. All chemicals were of reagent grade and were used without further purification. All reactions were preformed under a nitrogen atmosphere. [ReOCl3(PPh3)2] was prepared according to literature [43]. Bidentate ligands 2-(2-hydroxyphenyl)benzoxazolate (HL1a) [44] and (2′-hydroxyphenyl)-2-thiazoline (HL2a) [45]; tridentate Schiff base ligands salicylaldehyde 2-hydroxyanil (H2L1b) [46], salicylaldehyde 2-mercaptoanil (H2L2b) [47], S-benzyl-2-[(2-hydroxyphenyl)methylene]dithiocarbazate (H2L3b) [48], and 2-[(2-hydroxyphenyl)methylene]-N-phenyl-hydrazinecarbothioamide (H2L4b) [49] were prepared according to literature method with minor modifications.
2. 2. Synthesis
2.2.1. General procedure for the synthesis of intermediate complexes [ReOCl2(L ma)(PPh3)]
To a refluxing solution of [ReOCl3(PPh3)2] (200 mg, 0.24 mmol) in chloroform (50 cm3) was added with stirring 2-(2-hydroxyphenyl)benzoxazolate (HL1a) or (2′-hydroxyphenyl)-2-thiazoline (HL2a) (0.24 mmol) in chloroform (20 cm3). The olive green reaction mixture was refluxed for 40 min, while the color changed gradually to emerald green. The reaction mixture was cooled down and evaporated under reduced pressure to give a green solid residue, which was triturated with ethanol (30 cm3) to give the desired intermediate complex.
2.2.1.1. [ReOCl2(η2-OC6H4 -2-C = NC6H4 -2-O)(PPh3)] (1)
Yield: 136 mg, 76%. Anal. Calc. for C31H23Cl2-NO3PRe: C, 49.9; H, 3.09; N, 1.88. Found: C, 50.3; H, 3.14; N, 1.94%.
2.2.1.2. [ReOCl2(η2-OC6H4-2-C = NCH2CH2S)(PPh3)] (2)
Yield: 118. mg, 69%. Anal. Calc. for C27H23Cl2-NO2PSRe: C, 45.4; H, 3.23; N, 1.96. Found: C, 45.6; H, 3.14; N, 2.01%.
2.2.2. General procedure for the synthesis of the ‘3+2’ complexes [ReO(η2-Lma)(η3-Lmb)
To a refluxing solution of [ReOCl2(η2-Lma)(PPh3)] (0.05 mmol) in 1:1 CHCl3 + CH3OH (30 cm3) was added the appropriate tridentate ligand (0.05 mmol) with stirring. Four drops of triethylamine were then added and the solution was refluxed for additional 30 min, after which the color of the reaction mixture turned from emerald green to brownish red. The solvent was evaporated under reduced pressure and the dark red solid residue was redissolved in methylene chloride (50 cm3) and washed with water. The organic layer was separated from the mixture and dried over Na2SO4. The volume was reduced to 3 cm3 and then purified on silica gel column using gradient solvent (from 100% CHCl3 to 90:10 CHCl3: acetone).
2 2.2.1. [ReO(η3-OC6H4-2-CH=NC6H4 -2-O)(η2-OC6-H4C=NC6H4-2-O)], [ReO(η2-L1b). (η2-L1a)] (3)
Yield: 22 mg, 71%. Slow evaporation of pentane into complex 3 in minimum amount of CH2Cl2 afforded red plates suitable for X-ray analysis. FT-IR (cm−1, KBr pellet): 1597, 1530, 966. 1H NMR (CDCl3, ppm): 8.95 (s, 1H, CH=N), 7.96 (dd, 1H, J1 = 8.1 Hz, J2 = 1.5 Hz, ArH), 7.79 (d, 1H, J = 8.4 Hz, ArH), 7.70 (dd, 1H, J1 = 6,6 Hz, J2 = 1.8 Hz, ArH), 7.55–7.45 (m, 2H, ArH), 7.41 (dd, 1H, J1 = 7.8 Hz, J2 = 1.2 Hz, ArH), 7.35–7.1 (m, 3H, ArH), 7.04 (d, 1H, J = 8.1 Hz, ArH), 7.0–6.85 (m, 2H, ArH), 6.75–6.65 (m, 2H, ArH), 6.61 (d, 1H, J = 7.8 Hz, ArH). Anal. Calc. for C26H17N2O5Re: C, 50.1; H, 2.73; N, 4.49. Found: C, 49.9; H, 2.82; N, 4.38%.
2.2.2.2. [ReO(η3-OC6H4-2-CH=NC6H4-2-S)(η2-OC6 -H4C=NC6H4-2-O)] · MeOH, [ReO(η2-L2b)(η2-L1a)] · MeOH (4)
Yield: 25 mg, 76%. X-ray quality crystals were obtained by slow evaporation from a CH2Cl2–MeOH mixture. FT-IR (cm −1, KBr pellet): 1600, 1530, 965. 1H NMR (CDCl3, ppm): 8.86 (s, 1H, CH=N), 7.73 (d, 1H, J = 8.1 Hz, ArH), 7.47 (dd, 1H, J1 = 7.8 Hz, J2 = 1.8 Hz, ArH), 7.4–7.3 (m, 4H, ArH), 7.17 (t, 2H, J = 7.2 Hz, ArH), 6.9–6.8 (m, 2H, ArH), 6.75–6.65 (m, 2H, ArH), 6.50 (d, 1H, J = 8.4 Hz, ArH), 6.75–6.65 (m, 2H, ArH), 6.61 (d, 1H, J = 7.8 Hz, ArH). Anal. Calc. for C27H21N2O5SRe: C, 48.3; H, 3.13; N, 4.17. Found: C, 48.2; H, 3.08; N, 4.28%.
2.2.2.3. [ReO(η3-OC6H4-2-CH=N–N=C(SCH2C6H5)-S)(η2-OC6H4C = NC6H4-2-O)] · CH2Cl2, [ReO(η2-L3b)-(η2-L1a)] · CH2Cl2 (5)
Yield: 22 mg, 71%. X-ray quality crystals were grown by slow evaporation from a CH2Cl2/MeOH mixture. FT-IR (cm−1, KBr pellet): 1579, 1540, 963. 1H NMR (CDCl3, ppm): 9.17 (s, 1H, CH3N), 8.0–7.9 (m, 2H, ArH), 7.7–7.1 (m, 10H, ArH), 7.0–6.85 (m, 2H, ArH), 7.0–6.85 (m, 2H, ArH), 6.60 (t, 1H, J = 8.4 Hz, ArH), 6.51 (d, 1H, J = 7.5 Hz, ArH), 6.42 (d, 1H, J = 7.5 Hz, ArH), 4.61 (s, 2H, CH2). Anal. Calc. for C29H22Cl2N3O4S2Re: C, 43.7; H, 2.76; N, 5.27. Found: C, 43.5; H, 2.82; N, 5.36%.
2.2.2.4. [ReO(η3-OC6H4-2-CH=NC6H4-2-O)(η2-OC6-H4C = NCH2CH2-S)], [ReO(η2-L2b)(η2-L2a)] (6)
Yield: 18 mg, 62%. Slow evaporation of ethyl either into complex 6 in minimum amount of CH2Cl2 afforded red plates suitable for X-ray analysis. FT-IR (cm−1, KBr pellet): 1607, 1542, 962. 1H NMR (CDCl3, ppm): 8.95 (s, 1H, CH=N), 7.75 (d, 1H, J = 6.9 Hz, ArH), 7.67 (dd, 1H, J1 = 7.5 Hz, J2 = 1.2 Hz, ArH), 7.32 (t, 2H, J = 6.9 Hz, ArH), 7.2–7.1 (m, 2H, ArH), 6.98 (t, 1H, J = 9 Hz, ArH), 6.82 (t, 1H, J = 7.5 Hz, ArH), 6.52, (t, 1H, J = 6.9 Hz, ArH), 6.29 (t, 2H, J = 8.7 Hz, ArH), 4.88 (m, 1H, CH2), 4.59 (m, 1H, CH2), 3.85 (m, 1H, CH2), 3.63 (m, 1H, CH2). Anal. Calc. for C22H17N2O4SRe: C, 44.7; H, 2.88; N, 4.74. Found: C, 44.8; H, 2.81;N, 4.86%.
2.2.2.5. [ReO(η3-OC6H4-2-CH=N–N=C(SCH2C6H5)-S}(η2-OC6H4C=NCH2CH2S)], [ReO(η2-L3b)(η2-L2a)] (7)
Yield: 19 mg, 64%. FT-IR (cm−1 KBr pellet): 1604, 1582, 967. 1H NMR (CDCl3, ppm): 9.14 (s, 1H, CH=N), 7.55–7.2 (m, 8H, ArH), 7.12 (t, 1H, J = 7.8 Hz, ArH), 6.7–6.6 (m, 2H, ArH), 6.41 (d, 1H, J = 8.4 Hz, ArH), 5.0–4.85 (m, 1H, CH2), 4.67 (s, 2H, CH2Ph), 4.3–4.15 (m, 1H, CH2), 3.79 (tt, 1H, J = 10.2 Hz, CH2), 3.7–3.6 (m, 1H, CH2). Anal. Calc. for C24H20-N3O3S2Re: C, 44.4; H, 3.08; N, 6.47. Found: C, 44.6; H, 2.99; N, 6.59%.
2.2.2.6. [ReO{η3-OC6H4-2-CH=N–N=C(NHC6H5)-S}(η2-OC6H4C=NCH2CH2-S)], [ReO(η2-L4b)(η2-L2a)] (8)
Yield: 17 mg, 53%. FT-IR (cm−1, KBr pellet): 1599, 1529, 968. 1H NMR (CDCl3, ppm): 9.09 (s, 1H, CH=N), 7.70 (d, 1H, J = 7.8 Hz, ArH), 7.53 (dd, 1H, J1 = 7.8 Hz, J2 = 1.5 Hz, ArH), 7.45–7.05 (m, 7H, ArH), 6.82 (t, 1H, J = 7.8 Hz, ArH), 6.67 (d, 2H, J = 7.8 Hz, ArH), 6.44 (d, 1H, J = 8.4 Hz, ArH), 5.05–4.9 (m, 1H, CH2), 4.75 – 4.6 (m, 1H, CH2), 4.35 –4.2 (m, 1H, CH2), 4.25 – 4.1 (m, 1H, CH2). Anal. Calc. for C23H19N4O3S2Re: C, 42.5; H, 2.93; N, 8.63. Found: C, 42.0; H, 2.89; N, 8.88%.
2.3. X-ray crystallography
The selected crystals of 3–6 were measured with a Bruker P4 diffractometer equipped with the SMART CCD system [50] and using graphite-monochromated Mo Kα radiation (λ= 0.71073 Å). The data collection was carried out at 89(5) K. The data were corrected for Lorentz and polarization effects, and absorption corrections were made using SADABS [51]. Neutral atom scattering factors were taken from Cromer and Waber [52]. And anomalous dispersion corrections were taken from those of Creagh and McAuley [53]. All calculations were performed using SHELXL [54]. The structures were solved by direct methods [55] and all of the non-hydrogen atoms were located from the initial solution. After locating all the initial nonhydrogen atoms in each structure, the models were refined against F2, initially using isotropic and later anisotropic thermal displacement parameters until the final value of Δ/σmax was less that 0.001. At this point the hydrogen atoms were located from the electron density difference map and a final cycle of refinements was performed, until the final value of Δ/σmax was again less that 0.001. No anomalies were encountered in the refinements. The relevant parameters for crystal data, data collection, structure solution and refinement are summarized in Table 1, and important bond lengths and angles in Tables 2–5.
Table 1.
Summary of crystal data for the structures of [ReO(η3-OC6H4-2-CH=NC6H4-2-O) (η2-OC6H4C=NC6H4-2-O)] (3), [ReO(η3-OC6H4-2-CH=NC6H4-2-S)(η2-C6H4C=NC6H4-2-O)]· MeOH (4·MeOH), [ReO{η3-OC6H4-2-CH=N–N=C(SCH2C6H5)S}(η2-OC6H4C=NC6H4-2-O)] · CH2Cl2 (5·CH2Cl2) and [ReO(η3-OC6H4-2-CH=NC6H4-2-O)(η2-OC6H4C=NCH2CH2S)] (6)
| 3 | 4·MeOH | 5·CH2Cl2 | 6 | |
|---|---|---|---|---|
| Empirical formula | C26H17N2O5Re | C27H21N2O5Re | C29H22Cl2N3O4S2Re | C22H17O4N2ReS |
| Formula weight | 623.62 | 671.72 | 797.72 | 591.64 |
| Crystal size (mm3) | 0.15 × 0.13 × 0.10 | 0.40 × 0.20 × 0.05 | 0.25 × 0.20 × 0.15 | 0.20 × 0.20 × 0.15 |
| Crystal system | monoclinic | orthorhombic | triclinic | orthorhombic |
| Space group | P21/c | Pbca | P1̄ | Pbca |
| a (Å) | 9.589(1) | 12.605(2) | 8.664(1) | 7.126(1) |
| b (Å) | 25.719(4) | 16.455(2) | 12.493(2) | 22.406(3) |
| c (Å) | 9.573(1) | 22.567(3) | 13.461(2) | 24.624(4) |
| α (°) | 90 | 90 | 78.214(2) | 90 |
| β (°) | 115.012(2) | 90 | 87.953(2) | 90 |
| γ (°) | 90 | 90 | 74.632(2) | 90 |
| V (Å 3) | 2139.3(5) | 4680.9(11) | 1374.9(3) | 3931.7(10) |
| Z | 4 | 8 | 2 | 8 |
| Dcalc (g cm−3) | 1.936 | 1.906 | 1.923 | 1.999 |
| F(000) | 1208 | 2624 | 780 | 2288 |
| λ (Å) | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| μ (mm− 1) | 5.724 | 5.325 | 4.808 | 6.321 |
| θ range (°) | 1.58–28.31 | 1.80–28.42 | 1.55–28.31 | 1.65–28.30 |
| No. of unique data | 5132 | 5738 | 6363 | 4843 |
| Parameters refined | 307 | 327 | 370 | 272 |
| R1 (wR2) | 0.0574(0.1059) | 0.0610(0.1365) | 0.0532(0.1012) | 0.0574(0.0991) |
| Goodness-of-fit, S (F2) | 0.962 | 1.058 | 0.911 | 0.959 |
Table 2.
Selected bond lengths (Å) and bond angles (°) for 3
| Bond lengths | |||
| Re(1)–O(1) | 1.689(6) | Re(1)–N(2) | 2.119(8) |
| Re(1)–O(3) | 2.011(7) | N(1)–C(7) | 1.270(13) |
| Re(1)–O(4) | 2.013(6) | N(1)–C(8) | 1.428(13) |
| Re(1)–O(2) | 2.029(8) | N(2)–C(20) | 1.301(12) |
| Re(1)–N(1) | 2.058(10) | N(2)–C(21) | 1.401(12) |
| Bond angles | |||
| O(1)–Re(1)–O(3) | 102.0(3) | O(3)–Re(1)–N(2) | 91.2(3) |
| O(1)–Re(1)–O(4) | 170.3(3) | O(4)–Re(1)–N(2) | 82.1(3) |
| O(3)–Re(1)–O(4) | 84.9(3) | O(2)–Re(1)–N(2) | 92.7(3) |
| O(1)–Re(1)–O(2) | 92.0(3) | N(1)–Re(1)–N(2) | 166.3(3) |
| O(3)–Re(1)–O(2) | 165.4(3) | C(7)–N(1)–C(8) | 123.2(11) |
| O(4)–Re(1)–O(2) | 81.7(3) | C(7)–N(1)–Re(1) | 121.9(8) |
| O(1)–Re(1)–N(1) | 101 3(3) | C(8)–N(1)–Re(1) | 114.6(7) |
| O(3)–Re(1)–N(1) | 80.2(3) | C(20)–N(2)–C(21) | 106.3(8) |
| O(4)–Re(1)–N(1) | 86.6(3) | C(20)–N(2)–Re(1) | 125.7(7) |
| O(2)–Re(1)–N(1) | 93.2(3) | C(21)–N(2)–Re(1) | 128.0(6) |
| O(1)–Re(1)–N(2) | 90.9(3) | ||
Table 5.
Selected bond lengths (Å) and bond angles (°) for 6
| Bond lengths | |||
| Re(1)–O(1) | 1.696(6) | Re(1)–N(1) | 2.026(11) |
| Re(1)–O(3) | 1.991(7) | Re(1)–O(2) | 2.035(6) |
| Re(1)–O(4) | 1.998(6) | Re(1)–N(2) | 2.093(8) |
| Bond angles | |||
| O(1)–Re(1)–O(3) | 102.8(3) | O(4)–Re(1)–O(2) | 81.5(3) |
| O(1)–Re(1)–O(4) | 171.6(3) | N(1)–Re(1)–O(2) | 90.7(7) |
| O(3)–Re(1)–O(4) | 84.9(3) | O(1)–Re(1)–N(2) | 94.6(3) |
| O(1)–Re(1)–N(1) | 100.6(3) | O(3)–Re(1)–N(2) | 89.1(3) |
| O(3)–Re(1)–N(1) | 84.0(7) | O(4)–Re(1)–N(2) | 82.0(3) |
| O(4)–Re(1)–N(1) | 83.4(3) | N(1)–Re(1)–N(2) | 164.3(4) |
| O(1)–Re(1)–O(2) | 91.0(3) | O(2)–Re(1)–N(2) | 92.7(3) |
| O(3)–Re(1)–O(2) | 165.9(3) | ||
3. Results and discussion
The bidentate ligand 2-(2-hydroxyphenyl)benzoxazole (HL1a) was prepared from the condensation reaction of 2-cyanophenol and 2-aminophenol as shown in Scheme 2. The analogous bidentate ligand (2′-hydroxyphenyl)-2-thiazoline (HL2a) was isolated in a similar fashion in the reaction of 2-cyanophenol and 2-mercaptoethylalcohol. The tridentate Schiff bases salicylaldehyde-2-hydroxyanil (H2L1b) and salicylaldehyde-2-mercaptoanil (H2L2b) were synthesized in the reactions of 2-hydroxybenzyaldehyde with 2-aminophenol and 2-aminothiophenol, respectively (Scheme 3). S-benzyl-2-[(2-hydroxyphenyl) methylene]dithiocarbazate (H2L3b) was prepared by condensation of salicylaldehyde with S-benzyldithiocarbazate in ethanol solution. The tridentate Schiff base ligand 2-[(2-hydroxyphenyl)methylene]-N-phenyl-hydrazinecarbothioamide (H2L4b) was synthesized by reacting salicylaldehyde with 4-phenyl-3-thiosemicarbazide.
Scheme 2.
Scheme 3.
| (1) |
The ‘3+2’ type complexes were prepared in a two step synthesis. The Re(V)-oxo precursor [ReOCl3-(PPh3)2] was reacted with H2L1a or H2L2a to give the intermediate complexes [ReOCl2(L1a)(PPh3)] (1) and [ReOCl2(L2a)(PPh3)] (2), respectively (Eq. (1)). Subsequent reactions of 1 and 2 with the appropriate tridentate Schiff base ligands yielded the ‘3+2’ complexes of the {ReO}3+ core (Eq. (2)).
| (2) |
The ‘3+2’ complexes 3–8 invariably exhibit a strong band in the infrared spectrum at 960–968 cm−1 attributed to ν(Re=O). The remainder of the spectra were dominated by ν (C=C) stretching between 1400 and 1600 cm−1 and ν (C=N) stretching ranging from 1600 to 1610 cm−1.
The 1H NMR spectra displayed resonances characteristic of aromatic protons from approximately 6.85 to 8.15 ppm. The spectra of 3–8 also featured a singlet at approximately 9.00 to 9.15 ppm associated with the imine proton of the tridentate Schiff base ligand.
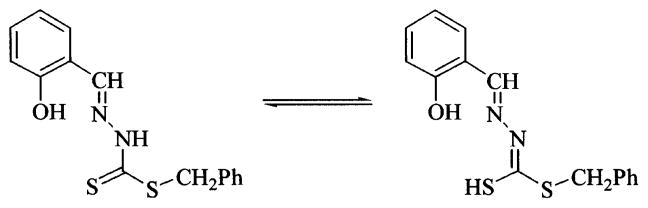
The Schiff base ligands H2L3b and H2L4b may exist in both the thione (A) and thiolo (B) forms (Scheme 4). The thione group is relatively unstable and tends to convert to the more stable C–S bond by enolization, if there is at least one proton adjacent to the thione group [56]. We have previously noted that for the case of H2L3b the thione/thiol tautomers are present in approximately 77/23 ratio based on the integration of the {CH=N} imine proton [30]. On metal complexation in 5 and 7 of {L3b}2− and in 8 of {L4b}2−, the ligands are found in the dianionic forms, involving deprotonation of the phenolic and thiolate SH groups. This is apparent from the disappearance of the ν(OH) (3110 cm−1) and ν (NH) (2970–2980 cm−1), as well as the absence of ν (SH) or ν (C=S) vibrations in the infrared spectra of 5, 7 and 8 [57]. The [ONS] mode of chelation through phenolic oxygen, azomethine nitrogen and sulfur thiol of B is confirmed from the red-shift (15–20 cm−1) of the ν (C=N) [58] and blue shift (30–40 cm−1) of the ν(C–O) vibrations of the parent ligands [59]. Furthermore, in the 1H NMR spectra of 5, 7 and 8, the phenolic proton resonance of the free ligand disappears and the azomethine proton signals are shifted downfield [60].
Scheme 4.
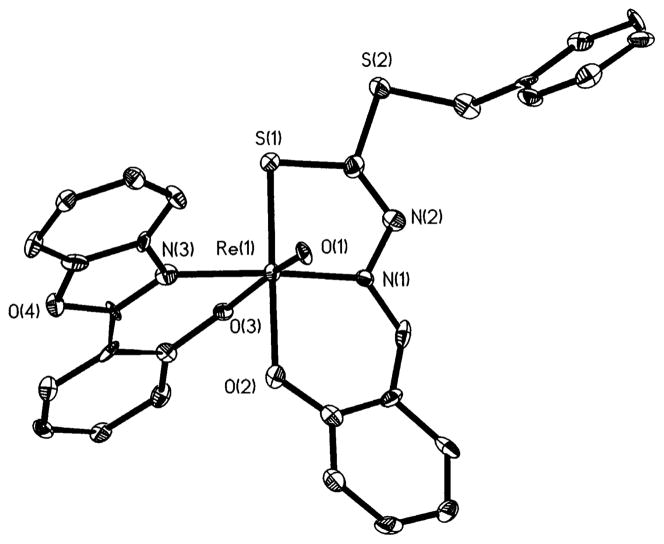
As shown in Fig. 1, the structure of 3 exhibits distorted octahedral geometry about the Re(V) center, defined by a terminal oxo-group, the oxygen and nitrogen donors of the {L1a}1− ligand, and the two oxygen and nitrogen donors of the tridentate {L1b}2− Schiff base ligand. The [ONO] donors of the tridentate Schiff base adopt a meridional disposition about the Re(V) center; consequently, the donors occupy cisoid positions relative to the oxo-group. The [ON] donors of the bidentate (L1a)1− ligand occupy the remaining positions, with the N donor in the equatorial plane with the [ONO] donors of the Schiff base. It is significant that the oxo-group of 3 does not exert an appreciable trans influence as indicated by the Re–O4 distance of 2.013(6) Å, compared to Re–O2 and Re–O3 distances of 2.029(8) and 2.011(7) Å, respectively. As shown by the data summarized in Table 6, this appears to be a characteristic feature of the structures of ‘3+2’ complexes of the {ReO}3+ core.
Fig. 1.
A view of the structure of 3, showing the atom labeling scheme.
Table 6.
Comparison of selected bond lengths (Å) for ‘3+2’ Schiff base complexes of the {ReO}3+ core
| Complex | η2-Ligand |
η3-Ligand |
Ref. | ||||
|---|---|---|---|---|---|---|---|
| Re=O | Re–O | Re–N | Re–O | Re–N | Re–S | ||
| 3 | 1.689(6) | 2.013(6) | 2.119(8), 2.011(7) | 2.029(8) | 2.058(10) | this work | |
| 4 | 1.701(6) | 2.032(6) | 2.134(7) | 2.080(6) | 2.063(7) | 2.306(2) | this work |
| 5 | 1.693(6) | 1.980(6) | 2.166(8) | 2.066(6) | 2.031(8) | 2.329(3) | this work |
| 6 | 1.696(6) | 1.998(6) | 2.093(8) | 1.991(7) | 2.026(11) | 2.035(6) | this work |
| [ReO(η3-OC6H4CH=NC6H4-2-S)-(η2-OC6H4C=NC6H4-2-S)] | 1.70(3) | 1.989(3) | 2.140(4) | 2.076(3) | 2.050(4) | 2.312(1) | [32] |
| [ReO((η2-OC6H4C=NC6H4-2-S)]3-OC6H4-2-CH=NC6H4-2-O)-((η2-OC6H4C=NC6H4-2-S)]2-8-O-5-NO2-C9H5N)] | 1.678(7) | 2.052(7) | 2.160(8) | 1.990(8) | 2.079(11) | [42] | |
The structure of 4, shown in Fig. 2, is analogous to that of 3. Once again, the [ONS] donors of the tridentate Schiff base ligand occupy meridional positions cis to the terminal oxo-group. The oxygen donor of the bidentate ligand is trans to the oxo-group, and the nitrogen donor occupies the fourth positions of the equatorial plane. In this case also, there is no evident trans influence of the terminal oxo-group.
Fig. 2.
A view of the structure of 4.
The structure of 5, shown in Fig. 3, again features the [ONS] donors of the tridentate Schiff base ligand occupying meridional positions cis to the terminal oxo-group. The oxygen donor of the bidentate ligand is trans to the oxo-group, while the nitrogen donor completes the bonding in the equatorial plane. There is no apparent trans influence associated with the terminal oxo-group.
Fig. 3.
A view of the structure of 5.
As shown in Fig. 4, the structure of 6 is analogous to that of 3 with the ligand {L2a}1− replacing the bidentate {L1a}1− of 3. The metrical parameters are unexceptional.
Fig. 4.
A view of the structure of 6.
In all cases, the N1–C7 distance (1.27(1)–1.31(1) Å) and the valence angles at N1 (114.6(7)–123.4(8)°) confirm that the N1 site is sp2 hybridized and the ligand is in the Schiff base form. Similarly, the N2–C2 bond lengths (1.28(2)–1.32(1) Å) and the N2 valence angles (106.3(8)–128.3(4)°) in 3, 4, and 6 and the N3–C27 bond length (1.32(1) Å) and the N3 valence angles (107.6(5)–126.3(6)°) in 5 are consistent with C–N double bond character and sp2 hybridization at the N2 site in 3, 4 and 6 and at the N3 site in 5. This observation confirms that the bidentate ligands in 3, 4 and 6 adopt the oxidized 2-(2-hydroxyphenyl)benzoxazolate form, while that in 5 is in the (2-hydroxyphenyl)-2-thiazolate form.
4. Conclusions
The ‘3+2’ chemistry of the {ReO}3+ core is readily accessed by exploiting tridentate Schiff base ligands. Thus, a variety of Schiff base complexes with [ONO] or [ONS] donor sets react smoothly with [ReOCl(η 2-Lma)(PPh3)] complexes 1 and 2 to provide a series of ‘3+2’ compounds of the general type [ReO(η 2-Lma)(η 3-Lmb)] (4–8). These results confirm that the Schiff base chemistry of the {ReO}3+ core may be exploited in the elaboration of materials whose size, shape, molecular weight, charge and lipophilicity may be systematically varied.
Table 3.
Selected bond lengths (Å) and bond angles (°) for 4·MeOH
| Bond lengths | |||
| Re(1)–O(1) | 1.701(6) | Re(1)–S(1) | 2.306(2) |
| Re(1)–O(3) | 2.032(6) | N(1)–C(7) | 1.312(11) |
| Re(1)–N(1) | 2.063(7) | N(1)–C(8) | 1.442(10) |
| Re(1)–O(2) | 2.080(6) | N(2)–C(20) | 1.322(11) |
| Re(1)–N(2) | 2.134(7) | N(2)–C(21) | 1.420(11) |
| Bond angles | |||
| O(1)–Re(1)–O(3) | 167.4(3) | O(3)–Re(1)–S(1) | 88.6(2) |
| O(1)–Re(1)–N(1) | 99.5(3) | N(1)–Re(1)–S(1) | 84.1(2) |
| O(3)–Re(1)–N(1) | 87.5(3) | O(2)–Re(1)–S(1) | 167.2(2) |
| O(1)–Re(1)–O(2) | 90.4(3) | N(2)–Re(1)–S(1) | 95.71(19) |
| O(3)–Re(1)–O(2) | 78.8(2) | C(7)–N(1)–C(8) | 121.0(7) |
| N(1)–Re(1)–O(2) | 92.9(3) | C(7)–N(1)–Re(1) | 121.4(6) |
| O(1)–Re(1)–N(2) | 91.3(3) | C(8)–N(1)–Re(1) | 117.4(6) |
| O(3)–Re(1)–N(2) | 81.4(3) | C(20)–N(2)–C(21) | 106.4(7) |
| N(1)–Re(1)–N(2) | 169.0(3) | C(20)–N(2)–Re(1) | 125.1(6) |
| O(2)–Re(1)–N(2) | 84.8(2) | C(21)–N(2)–Re(1) | 128.3(5) |
| O(1)–Re(1)–S(1) | 102.4(2) | ||
Table 4.
Selected bond lengths (Å) and bond angles (°) for 5·CH2Cl2
| Bond lengths | |||
| Re(1)–O(1) | 1.693(6) | N(1)–C(7) | 1.313(12) |
| Re(1)–O(3) | 1.980(6) | N91)–N(2) | 1.421(10) |
| Re(1)–N(1) | 2.031(8) | N(2)–C(8) | 1.295(12) |
| Re(1)–O(2) | 2.066(6) | N(3)–C(22) | 1.315(12) |
| Re(1)–N(3) | 2.166(8) | N(3)–C(23) | 1.428(11) |
| Re(1)–S(1) | 2.329(3) | ||
| Bond angles | |||
| O(1)–Re(1)–O(3) | 166.8(3) | O(3)–Re(1)–S(1) | 91.6(2) |
| O(1)–Re(1)–N(1) | 98.4(3) | N(1)–Re(1)–S(1) | 82.2(2) |
| O(3)–Re(1)–N(1) | 90.0(3) | O(2)–Re(1)–S(1) | 170.2(2) |
| (O(1)–Re(1)–O(2) | 89.4(3) | N(3)–Re(1)–S(1) | 98.6(2) |
| O(3)–Re(1)–O(2) | 80.1(3) | C(7)–N(1)–N(2) | 113.4(8) |
| N(1)–Re(1)–O(2) | 92.4(3) | C(7)–N(1)–Re(1) | 124.0(7) |
| O(1)–Re(1)–N(3) | 90.2(3) | N(2)–N(1)–Re(1) | 122.6(6) |
| O(3)–Re(1)–N(3) | 81.0(3) | C(8)–N(2)–N(1) | 112.8(8) |
| N(1)–Re(1)–N(3) | 171.0(3) | C(22)–N(3)–C(23) | 107.6(8) |
| O(2)–Re(1)–N(3) | 85.4(3) | C(22)–N(3)–Re(1) | 126.3(6) |
| O(1)–Re(1)–S(1) | 99.5(2) | C(23)–N(3)–Re(1) | 125.9(6) |
References
- 1.(a) Jurisson SS, Lydon JD. Chem Rev. 1999;99:2205. doi: 10.1021/cr980435t. [DOI] [PubMed] [Google Scholar]; (b) Liu S, Edwards DS. Chem Rev. 1999;99:2235. doi: 10.1021/cr980436l. [DOI] [PubMed] [Google Scholar]; (c) Prats E, Aisa F, Abos MD, Villavieja L, Garcia-Lopez F, Asenjo MJ, Razola P, Banzo J. J Nucl Med. 1999;40:296. [PubMed] [Google Scholar]; (d) Dilworth JR, Parrot SJ. Chem Soc Rev. 1998;27:43. [Google Scholar]; (e) Hom RK, Katzenllenbogen JA. Nucl Med Biol. 1997;24:485. doi: 10.1016/s0969-8051(97)00066-8. [DOI] [PubMed] [Google Scholar]; (f) Liu S, Edwards DS. Bioconjugate Chem. 1997;8:621. doi: 10.1021/bc970058b. [DOI] [PubMed] [Google Scholar]; (g) Schwochan K. Angew Chem, Int Ed Engl. 1994;33:2258. [Google Scholar]; (h) Jurisson S, Berning D, Jia W, Ma D. Chem Rev. 19993;93:1137. [Google Scholar]; (i) Volkert W, Deutsch EA. Adv Met Med. 1993;1:115. [Google Scholar]; (j) Steigman I, Eckelman WC. The Chemistry of Technetium in Medicine. National Academy Press; Washington, DC: 1992. [Google Scholar]; (k) Verbruggen A. Eur J Nucl Med. 1990;17:346. doi: 10.1007/BF01268027. [DOI] [PubMed] [Google Scholar]
- 2.Abram U, Braun M, Abram S, Kirmse R, Voight A. J Chem Soc, Dalton Trans. 1998:231. [Google Scholar]
- 3.Gerber TIA, Bruwer J, Bandoli G, Perils J, du Preez JGH. J Chem Soc, Dalton Trans. 1995:2189. [Google Scholar]
- 4.Bolzati C, Boschi A, Uccelli L, Malago E, Bandoli G, Tisato F, Refosco F, Pasqualini R, Duatti A. Inorg Chem. 1999;38:4479. doi: 10.1021/ic9903477. [DOI] [PubMed] [Google Scholar]
- 5.Arterburn JA, Fogarty IM, Hall KA, Ott KC, Bryan JC. Angew Chem, Int Ed Engl. 1996;35:2877. [Google Scholar]
- 6.Rochon FD, Melanson R, Kong PC. Inorg Chem. 1995;34:2273. doi: 10.1021/ic970573l. [DOI] [PubMed] [Google Scholar]
- 7.Bandoli G, Gerber TIA, Perils J, duPreez JGH. Inorg Chim Acta. 1998;278:96. [Google Scholar]
- 8.Bakir M, Sullivan BP. J Chem Soc, Dalton Trans. 1995:1733. [Google Scholar]
- 9.Liu S, Edwards S, Harris AR, Hemingway SJ, Barrett JA. Inorg Chem. 1999;38:1326. doi: 10.1021/ic980973o. [DOI] [PubMed] [Google Scholar]
- 10.Rose DJ, Maresca KP, Nicholson T, Davison A, Jones AG, Babich J, Fischman A, Graham W, DeBord JRD, Zubieta J. Inorg Chem. 1998;37:2701. doi: 10.1021/ic970352f. [DOI] [PubMed] [Google Scholar]
- 11.Alberto R, Schibli R, Angst D, Schubiger PA, Abram U, Abram S, Kaden TkA. Transition Met Chem. 1997;22:597. [Google Scholar]
- 12.Alberto R, Schibli R, Egli A, Schubiger AP. J Am Chem Soc. 1998;120:7987. doi: 10.1021/ja003932b. [DOI] [PubMed] [Google Scholar]
- 13.Alberto R, Schibli R, Schubiger AP. J Am Chem Soc. 1999;121:6076. doi: 10.1021/ja003932b. [DOI] [PubMed] [Google Scholar]
- 14.Waibel R, Alberto R, Willude J, Finnern R, Schibli R, Stichelberger A, Egli A, Abram U, Mack JP, Pluchthun A, Schubiger PA. Nature Biotechnology. 1999;17:897. doi: 10.1038/12890. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Femia FJ, Babich JW. J Zubieta, Inorg Chem. 2001 doi: 10.1021/ic000446g. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middleton AR, Masters AF, Wilkinson G. J Chem Soc, Dalton Trans. 1979:542. [Google Scholar]
- 17.Bandoli G, Nicolini M, Mazzi U, Refosco F. J Chem Soc, Dalton Trans. 1984:2505. [Google Scholar]
- 18.Jurisson S, Lindoy LF, Dancey KP, McPartlin M, Tasker PA, Uppal DK, Deutsch E. Inorg Chem. 1984;23:227. [Google Scholar]
- 19.Marchi A, Duatti A, Rossi R, Magon L, Mazzi U, Pasquetto A. Inorg Chim Acta. 1984;81:15. [Google Scholar]
- 20.Tisato F, Refosco F, Mazzi U, Bandoli G, Dolmella A. Inorg Chim Acta. 1989;164:127. [Google Scholar]
- 21.Tisato F, Refosco F, Mazzi U, Bandoli G, Nicolini M. Inorg Chim Acta. 1991;189:97. [Google Scholar]
- 22.van Bommel KJC, Verboom W, Kooijman H, Spek AL, Reinhoudt DN. Inorg Chem. 1998;37:4197. doi: 10.1021/ic980125t. [DOI] [PubMed] [Google Scholar]
- 23.Tisato F, Refosco F, Mazzi U, Bandoli G, Nicolini M. J Chem Soc, Dalton Trans. 1987:1693. [Google Scholar]
- 24.Duatti A, Marchi A, Rossi R, Magon L, Deutsch E, Bertolasi V, Bellucci F. Inorg Chem. 1988;27:4208. [Google Scholar]
- 25.Bandoli G, Mazzi U, Pietzsch HJ, Spies H. Acta Crystallogr, Sect C. 1992;48:1422. [Google Scholar]
- 26.Bandoli G, Mazzi U, Wilcox BE, Jurisson S, Deutsch E. Inorg Chim Acta. 1984;95:217. [Google Scholar]
- 27.Mazzi U, Refosco F, Tisato F, Bandoli G, Nicolini M. J Chem Soc, Dalton Trans. 1986:1623. [Google Scholar]
- 28.Pietzsch HJ, Spies H, Hoffmann S, Scheller D. Appl Radiat Isot. 1990;41:185. [Google Scholar]
- 29.Bandoli G, Mazzi U, Clemente DA, Roncari E. J Chem Soc, Dalton Trans. 1982:2455. [Google Scholar]
- 30.Chen X, Femia FJ, Babich JW, Zubieta J. Inorg Chim Acta. 2000;307:154. doi: 10.1016/S0020-1693(00)00193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Femia FJ, Chen X, Babich JW, Zubieta J. Inorg Chim Acta. 2000;300–302:517. doi: 10.1016/S0020-1693(99)00586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Femia FJ, Babich JW, Zubieta J. Inorg Chim Acta. 2000;307:149. doi: 10.1016/s0020-1693(00)00191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietzsch HJ, Spies H, Hoffmann S. Inorg Chim Acta. 1989;165:163. [Google Scholar]
- 34.Spies H, Fietz T, Pietzsch HJ, Johannsen B, Leibnitz P, Reck G, Scheller D, Klostermann K. J Chem Soc, Dalton Trans. 1995:2277. [Google Scholar]
- 35.Maresca KP, Femia FJ, Babich JW, Zubieta J. Inorg Chem Commun. 1998;1:209. [Google Scholar]
- 36.Maresca KP, Bonavia GH, Babich JW, Zubieta J. Inorg Chim Acta. 1999;284:252. [Google Scholar]
- 37.Maresca KP, Femia FJ, Bonavia GH, Babich JW, Zubieta J. Inorg Chim Acta. 2000;297:98. [Google Scholar]
- 38.(a) Shyer R, Seifert S, Spies H, Gupta A, Johannsen B. Eur J Nucl Med. 1998;25:793. doi: 10.1007/s002590050284. [DOI] [PubMed] [Google Scholar]; (b) Nock B, Maina T, Yannoukakos D, Pirmettis IC, Papadopoulos MS, Chiotellis E. J Med Chem. 1999;42:1066. doi: 10.1021/jm980174f. [DOI] [PubMed] [Google Scholar]; (c) Nock B, Tsoukalas C, Maina T, Pirmettis I, Papadopoulos M, Johannsen B, Chiotellis E.Eur J Nucl Med 2419971799021115 [Google Scholar]; (d) Pelecanou M, Pirmettis IC, Nock BA, Papadopoulos M, Spies H, Johannsen B, Stassinopoulou CI. Inorg Chim Acta. 1988;281:148. [Google Scholar]
- 39.Nock B, Maina T, Tisato F, Papadopoulos MS, Raptopoulou CP, Terzis A, Chiotellis E. Inorg Chem. 1999;38:4197. doi: 10.1021/ic991341k. [DOI] [PubMed] [Google Scholar]
- 40.Nock B, Maina T, Tisato F, Papadopoulos M, Raptopoulou CP, Terzis A, Chiotellis E. Inorg Chem. 2000;39:2178. doi: 10.1021/ic991341k. [DOI] [PubMed] [Google Scholar]
- 41.Femia FJ, Babich JW, Zubieta J. Inorg Chim Acta. 2000;300–302:462. doi: 10.1016/S0020-1693(99)00586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Femia FJ, Babich JW, Zubieta J. Inorg Chim, Acta. 2000;308:80. doi: 10.1016/S0020-1693(00)00218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatt J, Rowe GA. J Chem Soc. 1962:4019. [Google Scholar]
- 44.Orlando CM, Jr, Wirth JG, Heath DR. J Org Chem. 1970;35:3147. [Google Scholar]
- 45.Shutter E, Hoveyda HR, Karunaratne V, Rettig SJ, Orvig C. Inorg Chem. 1996;35:368. doi: 10.1021/ic9507528. [DOI] [PubMed] [Google Scholar]
- 46.Morisige K. Anal Chim Acta. 1974;73:245. [Google Scholar]
- 47.Mookerjee MN, Singh RV, Tandon JP. J Prakt Chem. 1983;325:319. [Google Scholar]
- 48.Ali MA, Bose R. J Inorg Nucl Chem. 1977;39:265. [Google Scholar]
- 49.Purohit S, Koley AP, Prasad LS, Manoharan PT, Ghosh S. Inorg Chem. 1989;28:3735. [Google Scholar]
- 50.Siemens. SMART Software Reference Manual. Siemens Analytical X-ray Instruments Inc; Madison, WI: 1994. [Google Scholar]
- 51.Sheldrick GM. SADABS: program for empirical absorption corrections. University of Göttingen; Germany: 1996. [Google Scholar]
- 52.Cromer DT, Waber JT. International Tables for X-ray Crystallography. IV. Kynoch; Birmingham, UK: 1974. [Google Scholar]
- 53.Creagh DC, McAuley JWJ. International Tables for X-ray Crystallography, Table 4. C. Kluwer Academic; Boston, MA: 1992. [Google Scholar]
- 54.SHELXL PC™. Siemens Analytical X-ray. Instruments Inc; Madison, WI: 1990. [Google Scholar]
- 55.TEXAN. Texray structural analysis package (revised) Molecular Structure Corporation; The Woodlands, TX: 1992. [Google Scholar]
- 56.Ali MA, Bose R. J Inorg Nucl Chem. 1977;39:265. [Google Scholar]
- 57.Ali MA, Chowdhury DA, Nazimuddin IM. Polyhedron. 1984;3:595. [Google Scholar]
- 58.Purohit S, Koley AP, Prasad LS, Manoharan PT, Ghosh S. Inorg Chem. 1989;28:3735. [Google Scholar]
- 59.Kovacic JE. Spectrochim Acta A. 1967;23:183. [Google Scholar]
- 60.Saxena A, Tandon JP, Molloy KC, Zucherman JJ. Inorg Chim Acta. 1982;63:71. [Google Scholar]