Abstract
Mechanical forces are central to developmental, physiological and pathological processes1. However, limited understanding of force transmission within sub-cellular structures is a major obstacle to unravelling molecular mechanisms. Here we describe the development of a calibrated biosensor that measures forces across specific proteins in cells with pico-Newton (pN) sensitivity, as demonstrated by single molecule fluorescence force spectroscopy2. The method is applied to vinculin, a protein that connects integrins to actin filaments and whose recruitment to focal adhesions (FAs) is force-dependent3. We show that tension across vinculin in stable FAs is ~2.5 pN and that vinculin recruitment to FAs and force transmission across vinculin are regulated separately. Highest tension across vinculin is associated with adhesion assembly and enlargement. Conversely, vinculin is under low force in disassembling or sliding FAs at the trailing edge of migrating cells. Furthermore, vinculin is required for stabilizing adhesions under force. Together, these data reveal that FA stabilization under force requires both vinculin recruitment and force transmission, and that, surprisingly, these processes can be controlled independently.
Focal adhesions (FAs) are complex intracellular linkages between integrins and the F-actin cytoskeleton that both transmit and respond to mechanical forces. FAs show complex mechanosensitivity such that they form or enlarge when force increases, and shrink or disassemble when force decreases3. However, mechanical forces can also induce FA disassembly, including sliding, a form of controlled disassembly4. In the absence of a method to measure forces across proteins in cells, these distinct processes have been difficult to elucidate and are poorly understood.
Vinculin is an intracellular FA protein comprised of a head domain (Vh) and a tail domain (Vt) separated by a flexible linker5. Binding of Vh to talin recruits vinculin to FAs, whereas Vt binds to F-actin and paxillin6. Interestingly, vinculin seems intimately linked to FA mechanosensitivity. Its recruitment to FAs is regulated by externally-or internally-generated mechanical forces7,8; vinculin-deficient cells display impaired cell spreading and cell migration, are less stiff than normal cells and exert lower traction forces9–11. Furthermore, vinculin seems to be a key element in the molecular “clutch” that links the actin cytoskeleton and extracellular matrix12 and co-localizes with areas of high force during leading edge protrusion13. These and other data have led to the concept of adhesion strengthening, in which adhesions under force recruit additional vinculin and enlarge to keep force per area constant14,15. However, where and when forces across vinculin occur on the sub-cellular level is unknown. Indeed, direct evidence that vinculin bears mechanical force is absent. Estimates from traction force microscopy suggest that tension across molecules in FAs is in the pN range14,16, a factor of 10–50 below the resolution of existing methods to measure forces across proteins within cells17. We therefore developed a genetically encoded vinculin tension sensor with single pN sensitivity for use in living cells.
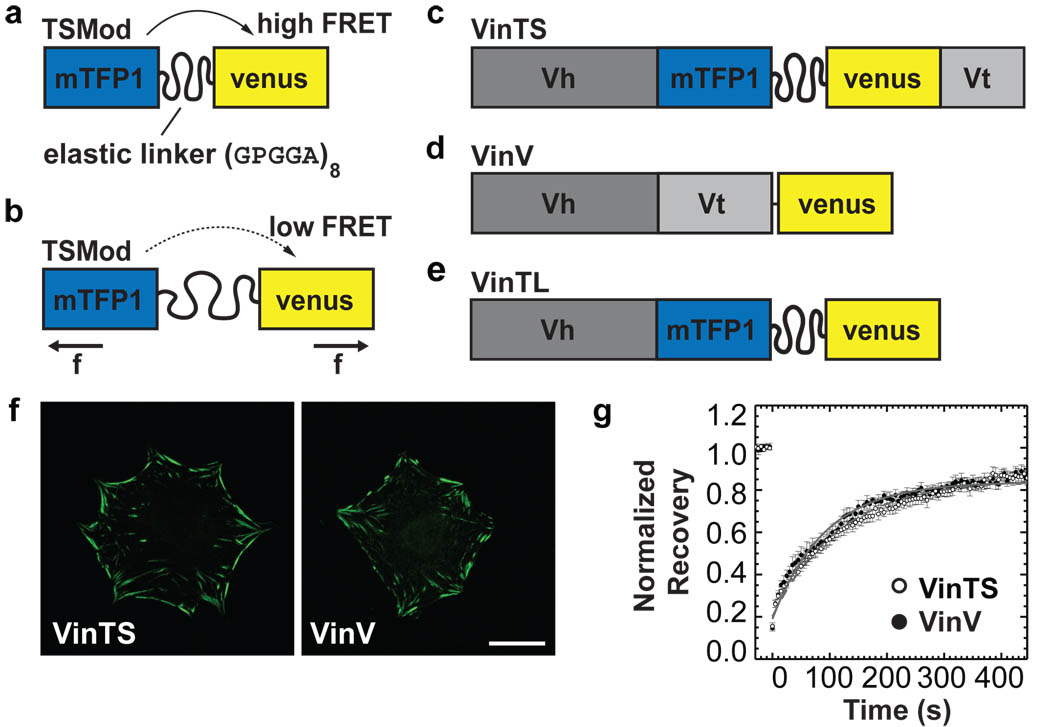
We designed a tension sensor module (TSMod) in which a 40 amino acid (aa)-long elastic domain was inserted between two fluorophores (mTFP1 and venusA206K) that undergo efficient fluorescence-resonance energy transfer (FRET) (Fig.1a)18. The elastic domain was derived from the spider silk protein flagelliform, which is composed of repetitive aa motifs that form entropic nano-springs suitable for measuring pN forces19. Since FRET is highly sensitive to the distance between the fluorophores, it should decrease under tension (Fig.1b). The vinculin tension sensor contains the sensor module between the Vh and Vt domain of vinculin after aa 883 (VinTS, Fig.1c). Controls include a C-terminally tagged vinculin-venus (VinV, Fig.1d) and a tail-less mutant, which cannot bind F-actin or paxillin (VinTL, Fig.1e). Thus, tension cannot be applied to the VinTL construct.
Figure 1. Vinculin tension sensor (VinTS) construct.
(a) The tension sensor module (TSMod) consists of two fluorophores separated by a flagelliform linker sequence (GPGGA)8. (b) When force across TSMod extends the elastic linker, FRET efficiency decreases (f: force). (c) The vinculin tension sensor (VinTS) consists of TSMod inserted after aa 883 of vinculin. (d) Vinculin-venus control (VinV). (e) Vinculin tail-less control (VinTL). (f) Localization of VinTS and VinV in vinculin−/− cells. Scale bar: 20µm. (g) Normalized average fluorescence recovery rates of VinTS (open circles, n=10) and VinV (closed circles, n=8). Error bars represent standard error of the mean (s.e.m.). (Recovery half-time VinTS: 87.6s +/− 6.6s, VinV: 68.3s +/− 13.1s, mean ± s.e.m., p=0.205).
In transiently transfected vinculin−/− cells, VinTS was properly recruited to FAs. FA shape and size, and F-actin organization were indistinguishable from cells expressing VinV (Fig.1f and Supplementary Fig.1a, b, e). Cells expressing VinTL displayed significantly enlarged FAs, consistent with previous studies20 (Supplementary Fig.1c, e). TSMod localized to the cytoplasm and the nucleus (Supplementary Fig.1d). All constructs produced stable proteins with the expected molecular size (Supplementary Fig. 2a). Expression of VinTS in vinculin−/− cells was comparable to the level of endogenous vinculin in murine embryonic fibroblasts (MEFs) or bovine aortic endothelial cells (BAECs) (Supplementary Fig.2b).
Next, we probed the activation state of VinTS. In solution, binding of Vh to Vt induces a closed, auto-inhibited conformation that does not bind F-actin. However, F-actin and the bacterial protein IpaA bind cooperatively to vinculin to induce its activation21. High speed centrifugation of hypotonic cell lysates from cells expressing VinTS or VinV to sediment F-actin, co-sedimented only a small amount of either vinculin construct even when actin was supplemented. Addition of IpaA alone modestly increased sedimentation, presumably due to vinculin binding to endogenous actin in cell lysates. IpaA and actin together induced nearly complete sedimentation of both vinculin constructs (Supplementary Fig.2c). Furthermore, fluorescence recovery after photobleaching (FRAP) in live cells showed that recovery rates in FAs were similar between VinTS and VinV, indicating normal vinculin dynamics22 (Fig.1g). Thus, insertion of TSMod does not significantly affect vinculin’s localization to FAs, its activation state, actin binding, or intracellular dynamics.
We next tested for two potential confounding factors, inter-molecular FRET and effects of vinculin conformation on FRET (FRET metrics are explained in Supplementary Note II). When cells were transfected with both vinculin-mTFP1 and vinculin-venus (fluorophore insertion after aa883), the FRET index in FAs was very low compared to cells expressing VinTS, indicating that inter-molecular FRET is negligible (Supplementary Fig.3a–c). To examine effects of conformational changes during vinculin activation, we used IpaA and actin to activate vinculin. As a positive control, we generated a mTFP1-venusA206K version of a previously described vinculin conformation FRET probe21 (VinCS, Supplementary Fig.3d). Spectrofluorimetric analysis of hypotonic cell lysates containing these constructs showed that, as expected, adding neither actin nor IpaA alone affected FRET efficiency of VinCS, whereas IpaA plus actin decreased FRET efficiency. In contrast, VinTS showed no change in FRET efficiency under identical conditions (Supplementary Fig.3e). Thus, vinculin’s conformational changes do not affect FRET measurements of VinTS.
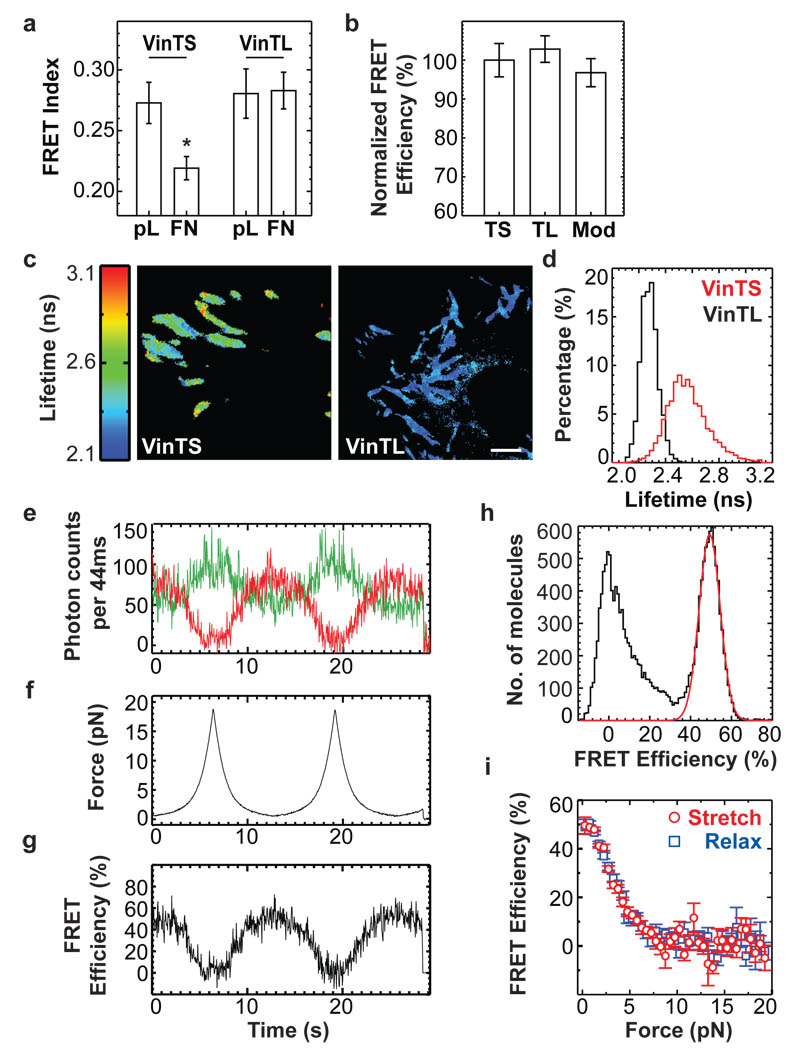
To evaluate responses to cellular forces, vinculin−/− cells expressing VinTS or VinTL were seeded on fibronectin (FN)-or poly-L-Lysine (pL)-coated coverslips. Cells on FN rapidly spread and formed FAs, whereas cells on pL remained round, with the constructs diffusely distributed in the cytoplasm. FRET was high for both constructs on pL (Fig.2a). Spectrofluorimetry of hypotonic cell lysates, another zero force state, also showed no difference between VinTS, VinTL and TSMod in solution (Fig.2b). In contrast, VinTS but not VinTL displayed reduced FRET index in FAs on FN, indicating increased mechanical tension (Fig.2a). These results were confirmed by fluorescence lifetime microscopy (FLIM)23. In adherent cells, VinTS displayed significantly longer lifetimes (corresponding to lower FRET efficiency, see Supplementary Note II). Lifetimes were also distributed over a much wider range than VinTL, indicating that individual molecules are subject to a range of forces (Fig.2c, d).
Figure 2. Responses to mechanical force.
(a) FRET index in vinculin−/− cells expressing VinTS or VinTL seeded on poly-L-Lysine (pL) or fibronectin (FN) (*: p<0.05, Tukey-b test, n=11–18) (b) FRET measured by spectrofluorimetry of lysates containing VinTS, VinTL or TSMod (n=4, p>0.5, Tukey-b test) (c) Fluorescence lifetime images of vinculin−/− cells expressing VinTS or VinTL. Scale bar: 2µm. (d) Fluorescence lifetime histograms from FAs of VinTS (n=11) or VinTL (n=8) expressing vinculin−/− cells. (e–g) Multiple stretch/relax cycles of a single TSModCy using fluorescence-force spectroscopy. (e) Fluorescence intensity time traces for donor (green) and acceptor (red). (f) Applied force vs. time. (g) FRET efficiency vs. time. (h) Single molecule FRET histogram of TSModCy at zero force. The peak marked by a red Gaussian fit represents the TSModCy labelled with both donor and acceptor. (i) Averaged FRET-force curves from n=7 molecules reveal reversible stretching and relaxing of TSModCy between 0.25 and 19 pN. All error bars represent s.e.m.
To calibrate the tension sensor, we used single molecule fluorescence-force spectroscopy, which combines confocal scanning fluorescence microscopy with optical tweezers2. Because fluorescent proteins’ low photostability precludes single molecule FRET measurements, we generated a version of TSMod using the organic fluorophores Cy3 and Cy5 (TSModCy, Supplementary Fig.4a). The flagelliform linker (F40) was connected to a polymer-coated glass surface via 18 bp double-stranded (ds) DNA and to a microsphere held in tweezers through ~50 kbp dsDNA. DNA tethers presented the fluorophores in close proximity to terminal cysteine residues of F40, allowing estimation of the linker end-to-end distance as a function of force from FRET measurements (Supplementary Fig.4a). FRET efficiency changes over multiple force cycles showed that TSModCy reached conformational equilibrium rapidly and displayed no hysteresis, indicating reversibility (Fig.2e–g). The zero force FRET efficiency of ~50 % determined separately (Fig.2h) matched the FRET value at the lowest force analyzed (0.25 pN, Fig.2i) indicating no adverse effects due to linkers or the optical tweezer. Together, these experiments showed that TSModCy is most sensitive at 1–6 pN (Fig.2i).
These measurements were used to estimate the force sensitivity of TSMod (Supplementary Fig.4b and Supplementary Note III) and to calculate forces across vinculin in living cells using FLIM microscopy data. This analysis showed that the average force in stationary FAs is ~2.5 pN (Supplementary Fig.4c–e). The assumptions underlying this analysis and the inherent limitations are discussed in Supplementary Note III.
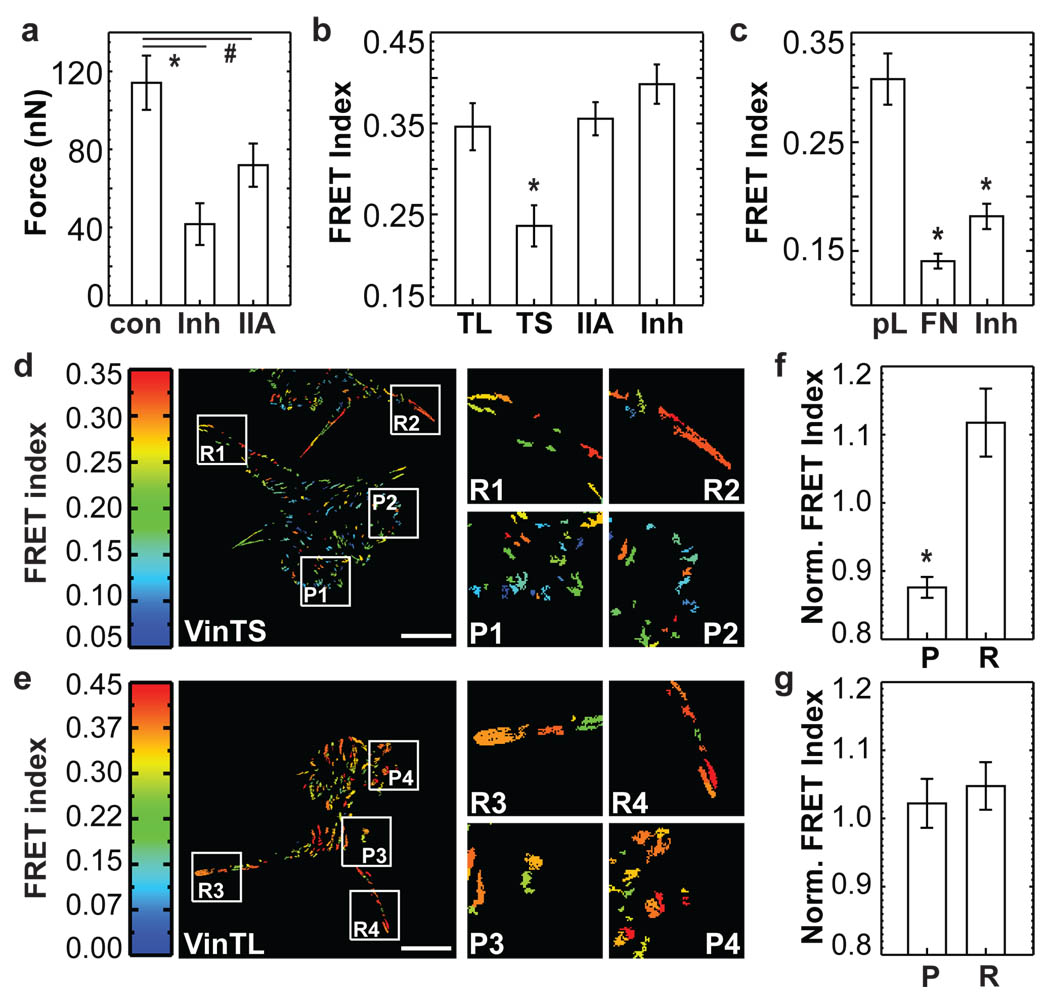
Vinculin recruitment to FAs is force-dependent7,8. To test whether recruitment correlates with force transmission across vinculin, we treated cells with a ROCK inhibitor (Y-27632) to reduce myosin-dependent contractility. Alternatively, we depleted myosin IIa (MIIa) by RNA interference (Supplementary Fig.5). Both treatments reduced FA size and traction forces were moderately reduced, as expected24 (data not shown and Fig.3a), but vinculin was still localized in FAs (Supplementary Fig.5c and data not shown). Interestingly, FRET index of VinTS increased to the level of VinTL, indicating a drastic loss of tension across vinculin (Fig.3b). By contrast, the vinculin conformation sensor (Supplementary Fig.3d) showed only a slight increase in FRET index after treating cells withY-27632, indicating that most of vinculin remained in an open conformation (Fig.3c). These data demonstrate that vinculin activation and recruitment to FAs are separable from transmission of force across vinculin.
Figure 3. Forces across vinculin during cell migration.
(a) Traction forces of VinTS expressing cells (con, n=18), treated with Y-27632 (Inh, n=15) or depleted of myosin IIa (IIa, n=20) (*: p<0.005, #: p<0.05, Dunnet’s test). (b) FRET index of VinTL, VinTS, IIa and Inh (n=30, *: p<0.005, Tukey-b Test). (c) Cells expressing VinCS on pL, FN, or FN treated with Y-27632 (Inh) (n=30, *: p<5×10−6, Tukey-b test). (d) FRET index of VinTS in BAECs. Protruding areas (P), retracting areas (R). (e) FRET index of VinTL in BAECs. (f, g) FRET index of protruding (P) and retracting (R) areas normalized by FRET index of interior FAs. (f) VinTS (n=4, *: p<0.01). (g) VinTL (n=4, p>0.5). Scale bar: 20µm. All error bars represent s.e.m.
We next examined mechanical forces across vinculin during cell migration. To ensure sufficient statistical power for the subcellular analysis, individual FAs were isolated and averaged (Supplementary Fig.6 and Supplementary Note II). In BAECs, vinculin within small FAs near protruding edges and occasionally in the centre of moving cells showed low FRET index indicating high tension, whereas large retracting FAs showed high FRET index indicating lower force per vinculin (Fig.3d, f and Supplementary Mov.1). The VinTL control displayed uniformly high FRET index at all locations (Fig.3e, g and Supplementary Mov.2; data normalization explained in Supplemental Note II).
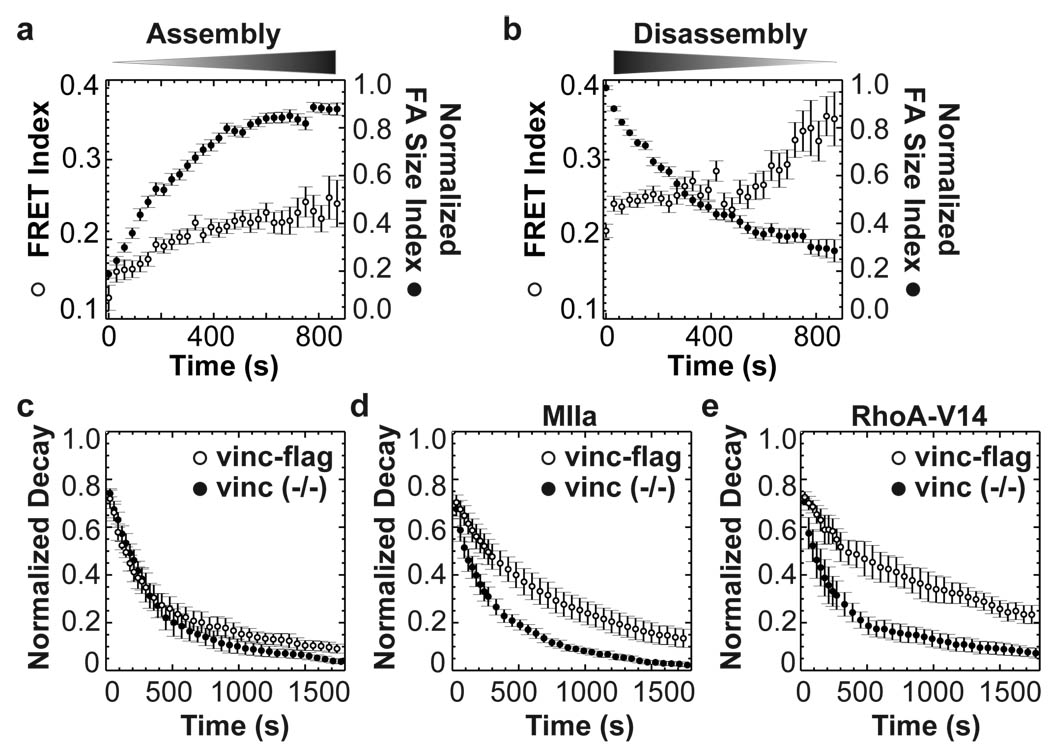
To examine this in more detail, we generated custom software to isolate and track FAs and correlate their dynamics with FRET (Supplementary Note II). Analysis of spreading or migrating vinculin−/− cells expressing VinTS revealed that force on vinculin was very high in small FAs that first appeared at cell edges, but decreased toward average levels as they enlarged (Fig.4a and Supplementary Fig.7). Conversely, as FAs disassembled, forces across vinculin remained low and even slightly decreased (Fig. 4b and Supplementary Fig.7). These effects were not observed with VinTL (Supplementary Fig.8). Previous studies showed that FA sliding or disassembly at retracting edges is associated with high local Rho-25 and myosin-activation and is myosin-dependent26, suggesting that the adhesions in retracting regions are under force. The low force on vinculin is therefore surprising but consistent with predictions based on intracellular force reconstructions from actin speckle analyses13. These data confirm that vinculin recruitment and force bearing are separable, and they suggest that failure of vinculin to bear force is linked to disassembly.
Figure 4. Tension on vinculin in dynamic FAs.
FAs were isolated, tracked and classified as assembling or disassembling. (a) In assembling FAs (n=78), FRET index (open circles) increases with normalized FA size index (defined in Supplemental Note II, closed circles). (b) In disassembling FAs (n=92), FRET index is high and further increases at late stages. (c) Lifetime of FAs visualized with EGFP-paxillin in vinculin−/− cells (n=4 cells, 214 FAs) or cells re-expressing vinculin-flag (n=4 cells, 310 FAs). Difference at 870s was not significant (p=0.24). (d) FA lifetime in vinculin−/− cells (n=7 cells, 408 FAs) and vinculin-flag cells (n=7 cells, 715 FAs) expressing MIIa (difference at 870s: p<0.005). (e) Vinculin−/− cells (n=3 cells, 250 FAs) or vinculin-flag cells (n=3 cells, 192 FAs) expressing RhoA-V14 (difference at 870s: p< 0.05). All error bars represent s.e.m.
We therefore tested whether vinculin is required for FA stabilization under force. Under normal culture conditions, vinculin−/− cells or cells expressing vinculin-flag displayed similar FA dynamics, visualized by EGFP-paxillin (Fig.4c). However, co-expressing either myosin IIa or active RhoA stabilized FAs in cells that contained vinculin but not in vinculin-deficient cells (Fig.4d, e and Supplementary Fig.9). In vinculin−/− cells, increased contractility often led to entire sides of the cell retracting, followed by formation of new protrusions. Indeed, FA behaviour at the periphery of vinculin−/− cells strikingly resembled the trailing edge of migrating cells where force across vinculin was low (Supplementary Fig.9 and Supplementary Mov.3–8). Thus, vinculin is required for FA stabilization under tension.
Taken together, these data reveal an unexpected regulatory mechanism in which the ability of vinculin to bear force determines whether adhesions assemble or disassemble under tension. Although creating new biosensors is always challenging (Supplementary Note III), this genetically encoded tension sensor with pN sensitivity should be applicable to other molecules involved in force transmission and mechanotransduction.
Methods Summary
Generation and characterization of expression constructs
For detailed information on the generation of cDNA constructs see Supplementary Note I. Immunostaining and Western blotting was performed using standard protocols. Expression and purification of GST-IpaA, and the vinculin-sedimentation assay were described elsewhere21.
FRET analysis
Calculations were performed with custom-written programs in IDL (ITT Visual Information Systems). The pFRET algorithm was implemented for non-linear bleed-through corrections27. FRET index is the corrected FRET divided by acceptor intensity. FAs were identified in the venusA206K channel using the water algorithm28, and utilized as masks for local averaging. For more detailed information see Supplementary Note II.
FRAP and FA dynamics analysis
For FRAP analysis, bleached and control FAs were identified by the water algorithm and polygons were automatically moved for slowly moving FAs. Recovery curves were analyzed as described22. FA dynamics were analyzed in vinculin−/− cells expressing EGFP-paxillin. FA decay curves were generated as described29 except that individual FA curves were generated, normalized, and averaged. Particle tracking was performed as previously described30 with particle centres determined by the water algorithm. Trajectory accuracy and selection of dynamic FAs were determined manually (Supplementary Note II).
FLIM analysis
Time-domain FLIM experiments and FLIM data analysis were performed as described previously23 using TRI2 software (developed by Paul Barber, Gray Cancer Institute, UK). Fluorescence lifetime was determined by fitting a single-exponential decay model, as mTFP1 modification was uniform within a pixel. With 890 nm excitation, acquisition times up to 150 seconds achieved sufficient photon statistics.
In vitro force calibration
The 40 aa flagelliform peptide (GPGGA)8 was crosslinked to maleimide-functionalized DNA. The peptide/DNA conjugate was labelled with organic fluorophores for fluorescence-force spectroscopy to determine the force per end-to-end distance of the peptide. For more detailed information see Online Methods and Supplementary Note III.
Supplementary Material
Acknowledgements
We thank Rick Horwitz, Miguel Vicente-Manzanares, Dorothy Schafer, Lukas K. Tamm (University of Virginia, USA) and Kris A. DeMali (University of Iowa, USA) for reagents and Margaret Gardel (University of Chicago, USA) for critical reading of the manuscript. This work was supported by USPHS grant U54 GM64346 to M.A.S. C.G. was supported by a Research Fellowship from the Deutsche Forschungsgemeinschaft (DFG, GR3399/1-1). B.D.H. was supported by USPHS training grant 5T32-HL007284 and an AHA Postdoctoral Fellowship. M.D.B., R.Z. and T.H. were supported by the US National Science Foundation Physics Frontier Center grant 0822613 and by USPHS grant R21 RR025341. T.H. is an investigator with the Howard Hughes Medical Institute. M.P. was supported by a Royal Society University Research Fellowship (UK). M.T.Y. was supported by an IGERT fellowship from the National Science Foundation (DGE-0221664).
Footnotes
Supplementary Information is linked to the online version of this paper at www.nature.com/nature.
Author Contributions. M.A.S. conceived the general idea. C.G. conceived the use of the flagelliform sequence, designed and generated vinculin expression constructs, performed spectrofluorimetry and all cell and imaging experiments. B.D.H. generated analysis tools and analyzed the in vivo data. T.H., M.M. and S.G.S. conceived the sensor calibration scheme. M.D.B. designed and generated the tension sensor construct for the calibration. M.D.B. and R.Z. performed and analyzed the single molecule experiments. T.H., R. Z. M.D.B and B.D.H. developed an algorithm to map in vitro and in vivo tension sensor data. M.P. performed and analyzed FLIM experiments. M.T.Y and C.S.C. generated micropost arrays and analyzed traction force data. C.G., B.D.H. and M.A.S. designed the in vivo experiments, discussed the results and wrote the paper with input from all authors.
Competing financial interest. The authors declare no competing financial interests.
References
- 1.Orr AW, Helmke BP, Blackman BR, Schwartz MA. Mechanisms of mechanotransduction. Dev Cell. 2006;10:11–20. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Hohng S, et al. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 4.Ballestrem C, Hinz B, Imhof BA, Wehrle-Haller B. Marching at the front and dragging behind: differential alphaVbeta3-integrin turnover regulates focal adhesion behavior. J Cell Biol. 2001;155:1319–1332. doi: 10.1083/jcb.200107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakolitsa C, et al. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- 6.Ziegler WH, Gingras AR, Critchley DR, Emsley J. Integrin connections to the cytoskeleton through talin and vinculin. Biochem Soc Trans. 2008;36:235–239. doi: 10.1042/BST0360235. [DOI] [PubMed] [Google Scholar]
- 7.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riveline D, et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- 10.Alenghat FJ, et al. Analysis of cell mechanics in single vinculin-deficient cells using a magnetic tweezer. Biochem Biophys Res Commun. 2000;277:93–99. doi: 10.1006/bbrc.2000.3636. [DOI] [PubMed] [Google Scholar]
- 11.Mierke CT, et al. Mechano-coupling and regulation of contractility by the vinculin tail domain. Biophys J. 2008;94:661–670. doi: 10.1529/biophysj.107.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu K, et al. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 13.Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol. 2008;10:1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balaban NQ, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 15.Shemesh T, Geiger B, Bershadsky AD, Kozlov MM. Focal adhesions as mechanosensors: a physical mechanism. Proc Natl Acad Sci U S A. 2005;102:12383–12388. doi: 10.1073/pnas.0500254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan JL, et al. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng F, Suchyna TM, Sachs F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J. 2008;275:3072–3087. doi: 10.1111/j.1742-4658.2008.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Day RN, Booker CF, Periasamy A. Characterization of an improved donor fluorescent protein for Forster resonance energy transfer microscopy. J Biomed Opt. 2008;13:031203. doi: 10.1117/1.2939094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker N, et al. Molecular nanosprings in spider capture-silk threads. Nat Mater. 2003;2:278–283. doi: 10.1038/nmat858. [DOI] [PubMed] [Google Scholar]
- 20.Humphries JD, et al. Vinculin controls focal adhesion formation by direct interactions with talin and actinl. J Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, et al. Spatial distribution and functional significance of activated vinculin in living cells. J Cell Biol. 2005;169:459–470. doi: 10.1083/jcb.200410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen DM, et al. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J Biol Chem. 2006;281:16006–16015. doi: 10.1074/jbc.M600738200. [DOI] [PubMed] [Google Scholar]
- 23.Parsons M, et al. Quantification of integrin receptor agonism by fluorescence lifetime imaging. J Cell Sci. 2008;121:265–271. doi: 10.1242/jcs.018440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai Y, et al. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys J. 2006;91:3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 26.Kolega J. Asymmetric distribution of myosin IIB in migrating endothelial cells is regulated by a rho-dependent kinase and contributes to tail retraction. Mol Biol Cell. 2003;14:4745–4757. doi: 10.1091/mbc.E03-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Periasamy A, Wallrabe H, Chen Y, Barroso M. Chapter 22: Quantitation of protein-protein interactions: confocal FRET microscopy. Methods Cell Biol. 2008;89:569–598. doi: 10.1016/S0091-679X(08)00622-5. [DOI] [PubMed] [Google Scholar]
- 28.Zamir E, et al. Molecular diversity of cell-matrix adhesions. J Cell Sci. 1999;112:1655–1669. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
- 29.Zaidel-Bar R, Milo R, Kam Z, Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J Cell Sci. 2007;120:137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- 30.Crocker JC, Hoffman BD. Multiple-particle tracking and two-point microrheology in cells. Methods Cell Biol. 2007;83:141–178. doi: 10.1016/S0091-679X(07)83007-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.