Abstract
The biliary bile salts of the medaka, the Japanese rice fish (Oryzias latipes) were isolated and identified. Only bile acids were present, and all were N-acyl amidated with taurine. Three bile acids, constituting 98% of total bile acids, were isolated by chromatography and their structure inferred from their properties compared to those of synthetic standards when analyzed by liquid chromatography-tandem mass spectrometry. The dominant bile acid was the 25R-epimer (82%) of 3α,7α,12α-trihydroxy-5β-cholestan-27-oic acid. The 25S-epimer was also present (11%), as was cholic acid (5%). Complete 1H and 13C NMR signal assignments of the C-25 epimers were made by using a combination of several 1D- and 2D-NMR techniques. The 1H and 13C NMR chemical shifts and spectral patterns of the hydrogen and carbon atoms, being close to the asymmetric centered at C-25, provided confirmatory evidence in that they distinguished the two epimeric diastereomers. The medaka is the first fish species identified as having C27 biliary bile acids as dominant its major bile salts.
Keywords: cholic acid, trihydroxycholestanoic acids, higher bile acids, peroxisomes, Acyl CoA-racemase, fish
Introduction
Bile salts, multifunctional, amphipathic end products of cholesterol metabolism, vary considerably in structure between species (Hagey, 1992; Haslewood, 1967; Hofmann et al., in press; Hoshita, 1985; Une and Hoshita, 1994). Most bile salts may be subdivided into three major classes -- C27 bile alcohols, C27 bile acids, and C24 bile acids -- based on the length of the alkyl side chain and whether the terminal polar group is a primary alcohol or a carboxylic acid (Hofmann et al, in press; Hofmann and Hagey, 2008). Additional structural variation arises from the configuration of the A/B ring juncture (cis or trans), and the number and orientation of hydroxy groups on the steroid nucleus and side chain. After their synthesis, bile alcohols are esterified with sulfate; C27 bile acids are N-acylamidated (conjugated) with taurine; and C24 bile acids, with taurine or glycine. Such conjugation converts bile acids and bile alcohols to bile salts that are soluble at intestinal pH and renders them impermeant to cell membranes, thereby promoting a high, micellar intraluminal concentration which facilitates lipid absorption (Hofmann and Hagey, 2008).
In a recent review compiled by Hofmann et al (in press), available information on biliary bile salt structures from about 700 vertebrate species-fish, reptiles (turtles, crocodilians, squamates), birds, and mammals was tabulated. In early evolving fish and amphibians, C27 bile alcohols dominated; in most later evolving fish the common C24 5β- bile acids, chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid) and cholic acid (CA; 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid) were the major bile acids. C27 bile acids prevailed in reptiles and early evolving birds (emus, kiwis, ostriches, tinamous). C24 bile acids occurred in all vertebrate classes, but were most predominant in mammals, including humans. Bile salt composition showed significant variation between orders and not between the smaller divisions of families, genera, or species. Thus biliary bile salt composition appears to be a biochemical trait that provides clues to evolutionary relationships, complementing anatomical and genetic analyses.
One unexpected finding in the survey of bile salts in fish was that the biliary bile salts of the “Japanese rice fish” (Oryzias latipes), often referred to by its Japanese name “medaka”, differed other fish species in having a high proportion of C27 bile acids.
The Japanese rice fish is a small, Southeast Asian pond fish that has been a popular aquarium fish for several centuries (Parenti, 2008). Its genome has been sequenced (Kasahara et al., 2007), and the fish is widely used as a model organism in many areas of biological research (Matsumoto et al., 2009). We judged it of interest to determine the exact structure of the C27 bile acids occurring in the medaka, not only because it is the only fish species identified to data in whom C27 acids predominate, but also because the bile acid composition of the medaka might be interpreted in light of the genes involved in bile acid evolution. A photograph of this fish species, which in this paper will be referred to as the medaka, is shown in Figure 1.
Fig. 1.
Photograph of the Japanese rice fish, the medaka (Oryzias latipes).
We report here that the major biliary bile acids of the medaka consist of the taurine conjugates of 25R- and 25S-epimers of 3α,7α,12α-trihydroxy-5β-cholestan-27-oic acid. We also show that the 1H and 13C NMR spectra of the two epimers are sufficiently different that the two taurine-conjugated epimers can be distinguished using this analytical technique.
Material and methods
Biological material
Gallbladder bile of the Japanese rice fish (Oryzias latipes subsp.) was collected by excising the gallbladder from about 1000 medaka fish. Bile samples were dispersed in 4 volumes of reagent-grade isopropanol and kept at 4°C until analysis.
Material and reagents
Authentic reference compounds of the taurine conjugates of CA and (25R)- and (25S)-3α,7α,12α-trihydroxy-5β-cholestan-27-oic acid (THCA) were synthesized in our laboratory (Goto et al., 1989b; Une et al., 1984).
RP-HPLC analysis of gallbladder bile of the medaka
The RP-HPLC apparatus used was a Jasco LC-2000 plus HPLC system (two PU-2085 high-pressure pumps, an MX-2080-32 solvent mixing module, and a CO-2060 column heater) equipped with a ChromNAV data-processing system (Tokyo, Japan). A Capcell Pack type AQ C18 column (3.0 mm × 150 mm I.D.; particle size, 3 µm; Shiseido) was employed and kept at 37°C. An Alltech 2000ES evaporative light-scattering detector (ELSD) (Deerfield, IL, USA) was used under the following conditions; the flow rate of purified compressed air used as a nebulizing gas was 2.0 L/min, and the temperature of the heated drift was 82°C. The mobile phase used was 20 mM-ammonium acetate/acetic acid buffer solution (pH 3.8)-methanol mixture (7:3, v/v); the flow rate was kept at 400 µL/min during the analysis.
Isolation of major bile salts from the bile of Japanese rice fish
The isopropanol solution of medaka bile was ultracentrifuged for 10 min (20,000 rpm) and the supernatant liquid was filtered with Mini-Uni Prep membrane filter (pore size, 0.45 µm; Whatman, NJ, USA). The filtrate was passed through a pre-conditioned Sep-Pak tC18 cartridge (5 g; Waters, Milford, MA). After the cartridge was washed successively with water (5 mL) and 5% methanol (5 mL), the bile salt fraction was eluted with methanol (5 mL). The methanol eluate was concentrated under a nitrogen stream at 40°C and the major bile salts were isolated by preparative RP-HPLC.
The preparative RP-HPLC apparatus consisted of a Jasco Gulliver series HPLC system with two PU-980 high-pressure pumps, an HG-980-31 solvent mixing module, and an HG-980-50 degasser. RP-HPLC was carried out by stepwise gradient elution using an Inertsil ODS-3 column (5 µm, 250 mm × 20 mm I.D.; GL Science, Tokyo, Japan) using a gradient of methanol in 5 mM-ammonium acetate as the mobile phase. The methanol composition was gradually increased at a flow rate of 7.2 mL/min using the following HPLC conditions: 5% (0–15 min) → 40% (15.1–30 min) → 50% (30.1–45 min) → 60% (45.1–60 min) → 74~92% (150.1–210 min). The 70, 72, and 74–80% methanol fractions, which contained compounds A, C, and D, respectively, were collected. Each of the fractions was evaporated under reduced pressure and then applied to a pre-conditioned Sep-Pak tC18 cartridge (360 mg; Waters, Milford, MA). After the cartridge was washed with water (7 mL), each of the isolated compounds was eluted with methanol (5 mL), and the eluate was evaporated under a nitrogen stream at 35°C. Three compounds, termed A, C, and D, were isolated and then analyzed using LC/ESI-MS/MS and 1D- and 2D-NMR techniques.
LC/ESI-MS/MS analysis of major components A, C, and D
Negative ion LC/ESI-MS/MS analysis of the isolated compounds was performed using an API 5000 LC-MS/MS system (Applied Biosystems, Inc., CA) equipped with a Nanoscope HPLC system (Shiseido, Tokyo, Japan). Chromatographic separation was carried out with a Capcell Pak C18 type MGII column (5 µm, 100 × 2.0 mm ID) using 10 mM-ammonium acetate (pH 7.0)/ methanol mixture (1:1, v/v) as the mobile phase at a flow rate of 200 µL/min. The mass detector was set to the following conditions: curtain gas (N2) flow, 20 psi; nebulizer gas (N2) flow, 40 psi; turbo gas (zero grade air) flow, 70 psi; turbo ion spray temperature, 600°C; declustering potential, −70 V; collision energy, −90 eV; collision gas (N2) pressure, 8 × 10−3 mbar.
1H and 13C NMR analyses of the major isolated compounds C and D
NMR spectra were recorded at 23°C in CD3OD in a 5 mm tube on a JEOL ECA-600 instrument using 600 MHZ for 1H and 149.4 MHz for 13C. 1H and 13C resonance assignments were made using a combination of two-dimensional (2D) homonuclear (1H-1H) and heteronuclear (1H-13C) shift-correlated techniques, which include 1H-1H correlation spectroscopy (COSY), long-range COSY, 1H-1H nuclear Overhauser effect spectroscopy (NOESY), 1H detected heteronuclear multiple quantum correlation (HMQC), and 1H detected heteronuclear multiple bond correlation (HMBC) experiments. These 2D-NMR spectra were recorded using standard pulse sequences and parameters recommended by the manufacturer. The 13C distortionless enhancement by polarization transfer (DEPT; 135°, 90°, and 45°) spectra were also measured to determine the exact 1H signal multiplicity and to differentiate between CH3, CH2, CH, and C based on their proton environments.
Results
Isolation and identification of major bile acids present in medaka bile
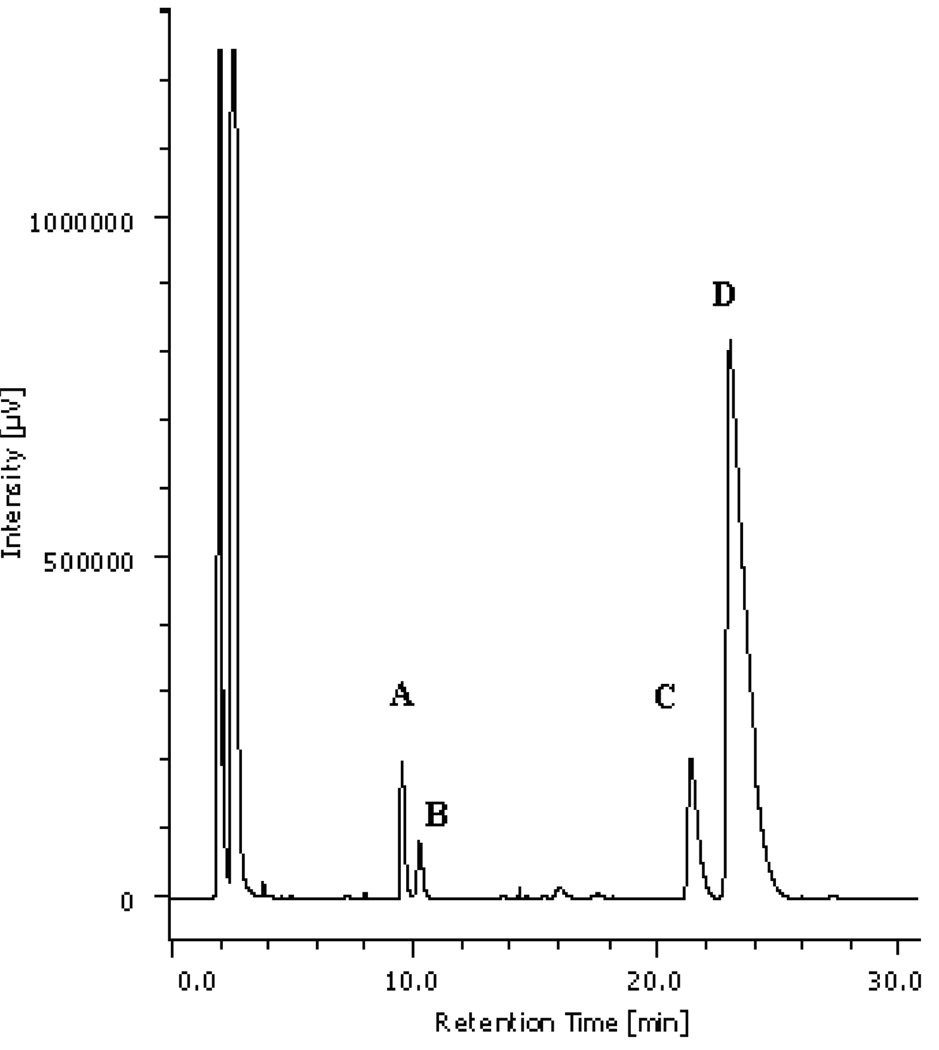
As shown in Figure 2, RP-HPLC analysis of the bile salts present in the gallbladder bile of the medaka showed three major peaks, which were designated as compounds A (constituting 5% of total biliary bile salts, C (constituting 11%), and D (constituting 82%). An additional component B was in much lower proportion (less than 2%) and could not be identified. The three major components A, C, and D, were isolated by preparative RP-HPLC.
Fig. 2.
RP-HPLC (with an ELSD) profile of the bile acids of the medaka.
Peak A, 5% of biliary bile acids, was identified as cholyl taurine; its retention time (RT) was 9.6 min; peak B, 2%, was not identified, RT 10.3 min; Peak C, 11%, was identified as (25S)-3α,7α,12α-trihydroxy-5β-cholestan-27-oyl taurine, RT 21.3 min; and peak D, 82%, was identified as (25R)-3α,7α,12α-trihydroxy-5β-cholestan-27-oyl taurine, RT 23.1 min.
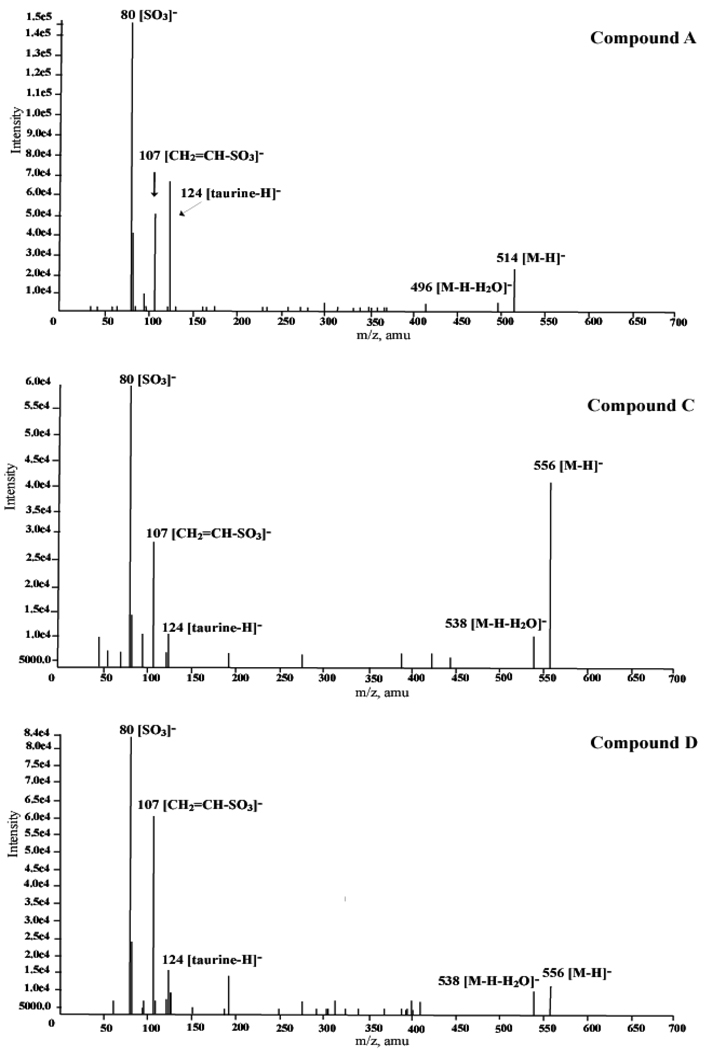
The individually isolated components A, C, and D were then examined by LC/ESI-MS/MS (Figure 3). In the first ESI-MS spectra, the deprotonated molecules [M-H]− and [M-H-H2O]− were as follows: peak A, m/z 514 and 496, indicating a taurine-conjugated C24 trihydroxy bile acid; and peaks C and D, m/z 556 and 538, indicating taurine-conjugated C27 trihydroxy bile acids. In the collision induced dissociation (LC/ESI-MS/MS) spectra obtained by selecting the deprotonated ions as a precursor ion, all three components afforded the characteristic fragment ions at m/z 124 [taurine-H]−, 107 [CH2=CH-SO3]−, and 80 [SO3]−, indicating the presence of an N-acylamide linkage with taurine on the side chain.
Fig. 3.
LC/ESI-MS/MS spectra of the isolated compounds A, C, and D.
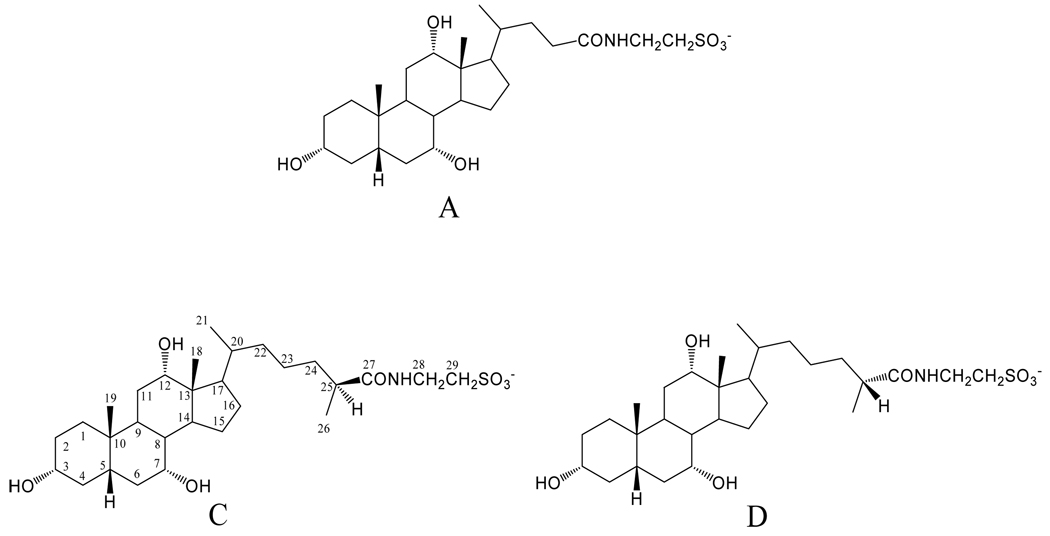
Based on their RP-HPLC retention times and the LC-MS fragmentation patterns compared to those of the synthetic reference standards (Goto et al., 1989; Une et al., 1984), the three components A, C, and D were identified as the taurine-conjugate of cholic acid, (peak A), the taurine conjugate of (25S)-THCA (peak C) and the taurine conjugate of (25R)-THCA (peak D). The assigned structures were confirmed by NMR (see below). Figure 4 shows the steric structures of the three isolated compounds (A, C, and D). Carbon atoms have been numbered.
Fig. 4.
Structures of the C24 and C27 trihydroxy bile acids isolated from the medaka: cholyl taurine (compound A); (25S)-3α,7α,12α- trihydroxy-5β-cholestan-27-oyl taurine (compound C); and (25R)- 3α,7α,12α-trihydroxy-5β-cholestan-27-oyl taurine (compound D).
Characterization of the epimeric (25R)-/(25S)-THCAs by NMR
To establish the location and orientation of functional groups present in the isolated compounds, we examined their 1D- and 2D-NMR spectra. We wished to determine whether NMR could be used to distinguish the two epimers of THCA, both of which are key intermediates in C24 bile acid biosynthesis. Table 1 shows the complete 1H (600 MHz) and 13C (149.4 MHz) signal assignments for the isolated compounds C and D. Carbon signals were grouped according to their multiplicity using DEPT experiments. Subsequently, individual 1H and 13C signal assignments were made from a combination of several 2D-NMR techniques, including COSY, long-range COSY, NOESY, HMBC, and HMQC. Both C-25 epimers showed essentially identical 1H and 13C chemical shifts and signal multiplicities for protons and carbons C-1 to C-19 in the 5β-steroid nucleus (Ijare et al., 2005; Nagane Gowda et al., 2006), because these carbon atoms are far from the chiral C-25.
Table 1.
Complete 1H and 13C chemical shifts of the isolated taurine conjugates of (25R)/ (25S)-THCAs.a
| Compound C (25S) | Compound D (25R) | Difference in the chemical shifts between C and D |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon no. | Type | 13C |
1H |
Type | 13C |
1H |
13C |
1H |
|||
| α | β | α | β | α | β | ||||||
| 1 | CH2 | 36.52 | 1.80 | 0.96 | CH2 | 36.52 | 1.79 | 0.96 | 0.00 | 0.01 | 0.00 |
| 2 | CH2 | 31.21 | 1.42 | 1.58 | CH2 | 31.20 | 1.41 | 1.58 | 0.01 | 0.01 | 0.00 |
| 3 | CH | 72.93 | 3.35 (brm) | CH | 72.92 | 3.35 (brm) | 0.01 | 0.00 | |||
| 4 | CH2 | 40.50 | 2.26 | 1.64 | CH2 | 40.50 | 2.26 | 1.64 | 0.00 | 0.00 | 0.00 |
| 5 | CH | 43.25 | 1.36 | CH | 43.24 | 1.36 | 0.01 | 0.00 | |||
| 6 | CH2 | 35.83b | 1.50 | 1.94 | CH2 | 35.81b | 1.50 | 1.94 | 0.02 | 0.00 | 0.00 |
| 7 | CH | 69.10 | 3.78 (m) | CH | 69.11 | 3.78 (m) | −0.01 | 0.00 | |||
| 8 | CH | 41.07 | 1.54 | CH | 41.08 | 1.54 | −0.01 | 0.00 | |||
| 9 | CH | 27.89 | 2.24 | CH | 27.87 | 2.24 | 0.02 | 0.00 | |||
| 10 | C | 35.92 | C | 35.92 | 0.00 | ||||||
| 11 | CH2 | 29.57 | 1.56 | 1.56 | CH2 | 29.57 | 1.56 | 1.56 | 0.00 | 0.00 | 0.00 |
| 12 | CH | 74.05 | 3.94 (m) | CH | 74.09 | 3.94 (m) | −0.04 | 0.00 | |||
| 13 | C | 47.47 | C | 47.47 | 0.00 | ||||||
| 14 | CH | 42.97 | 1.96 | CH | 42.95 | 1.95 | 0.02 | 0.01 | 0.00 | ||
| 15 | CH2 | 24.24 | 1.72 | 1.09 | CH2 | 24.22 | 1.73 | 1.09 | 0.02 | −0.01 | 0.00 |
| 16 | CH2 | 28.86 | 1.86 | 1.26 | CH2 | 28.82 | 1.86 | 1.26 | 0.04 | 0.00 | 0.00 |
| 17 | CH | 48.34 | 1.84 | CH | 48.30 | 1.82 | 0.04 | 0.02 | |||
| 18 | CH3 | 12.99 | 0.70 (s) | CH3 | 13.00 | 0.70 (s) | −0.01 | 0.00 | |||
| 19 | CH3 | 23.16 | 0.91 (s) | CH3 | 23.16 | 0.91 (s) | 0.00 | 0.00 | |||
| 20 | CH | 37.15 | 1.38 | CH | 37.12 | 1.38 | 0.03 | 0.00 | |||
| 21 | CH3 | 18.04 | 0.98 (d, 6.6) | CH3 | 18.08 | 0.98 (d, 6.6) | −0.04 | 0.00 | |||
| 22 | CH2 | 37.05 | 1.06, 1.38 | CH2 | 37.08 | 1.04, 1.38 | −0.03 | 0.02, 0.00 | |||
| 23 | CH2 | 25.18 | 1.38 | CH2 | 25.07 | 1.20, 1.34 | 0.11 | 0.04 | |||
| 24 | CH2 | 35.80b | 1.26, 1.52 | CH2 | 35.78b | 1.34, 1.52 | 0.02 | −0.08, 0.00 | |||
| 25 | CH | 42.33 | 2.24 | CH | 42.35 | 2.26 | −0.02 | −0.02 | |||
| 26 | CH3 | 18.31 | 1.08 (d, 6.6) | CH3 | 18.08 | 1.08 (d, 6.6) | 0.23 | 0.00 | |||
| 27 | C | 179.40 | C | 179.50 | −0.10 | ||||||
| 28 | CH2 | 36.49 | 3.58 (t, 7.2) | CH2 | 36.48 | 3.58, 3.59 (each t, 7.2) | 0.01 | 0.00, −0.01 | |||
| 29 | CH2 | 51.58 | 2.95, 2.96 (each t, 7.2) | CH2 | 51.59 | 2.95 (t, 6.9), 2.96 (t, 7.2) | −0.01 | 0.00, 0.00 | |||
Measured in CD3OD at 600 MHz in 1H-NMR and at 149.4 MHz in 13C-NMR; chemical shifts were expressed as δ ppm relative to Me4Si; abbreviations used: s, singlet; d, doublet; m, multiplet; brm, broad multiplet; t, triplet; values in parentheses refer to signal multiplicity and coupling constant (J in Hz).
Assignment down a vertical column may be interchanged.
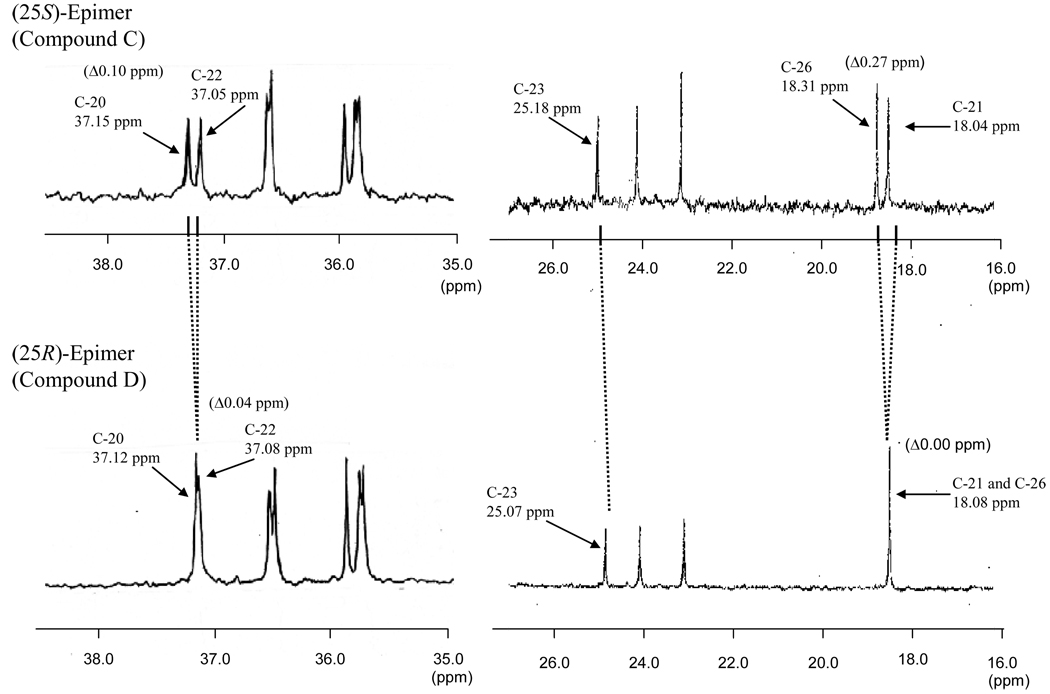
The assignments of the individual 1H signals to the side chain carbons (C-20~C-29) were based on our previous 13C signal assignments for the taurine conjugate of (25R)-THCA (Hagey et al., 2009). The resonance position of these 1H and 13C signals generally gave chemical shifts that were similar for the two epimers. However, careful comparison of both the spectra revealed that some of the side chain protons and carbons, being close to the asymmetric center at C-25, had small, but distinct and significant differences in the chemical shifts and/or signal patterns. Thus, the 13C signals due to C-23 and C-26 exhibited downfield shifts of 0.11 and 0.23 ppm, respectively, for the 25R-epimer (compound D) compared to the 25S-epimer (compound C), whereas the oxygenated carbon at C-27 showed an upfield shift of 0.10 ppm. Of further interest were the differences in the spectral pattern of the 13C signals arising from C-21 vs. C-26 and C-20 vs. C-22, as shown in Figure 5. The significant differences in the 13C chemical shifts (defined as Δ-values) between the two epimeric pairs are of particularly useful to characterizing each of the compounds: the Δ-values observed for C-21 vs. C-26 is 0.27 ppm in C and 0.00 ppm in D and those for C-20 vs. C-22 is 0.10 ppm in C and 0.04 ppm in D.
Fig. 5.
Partial 13C NMR spectra of the isolated compounds C and D.
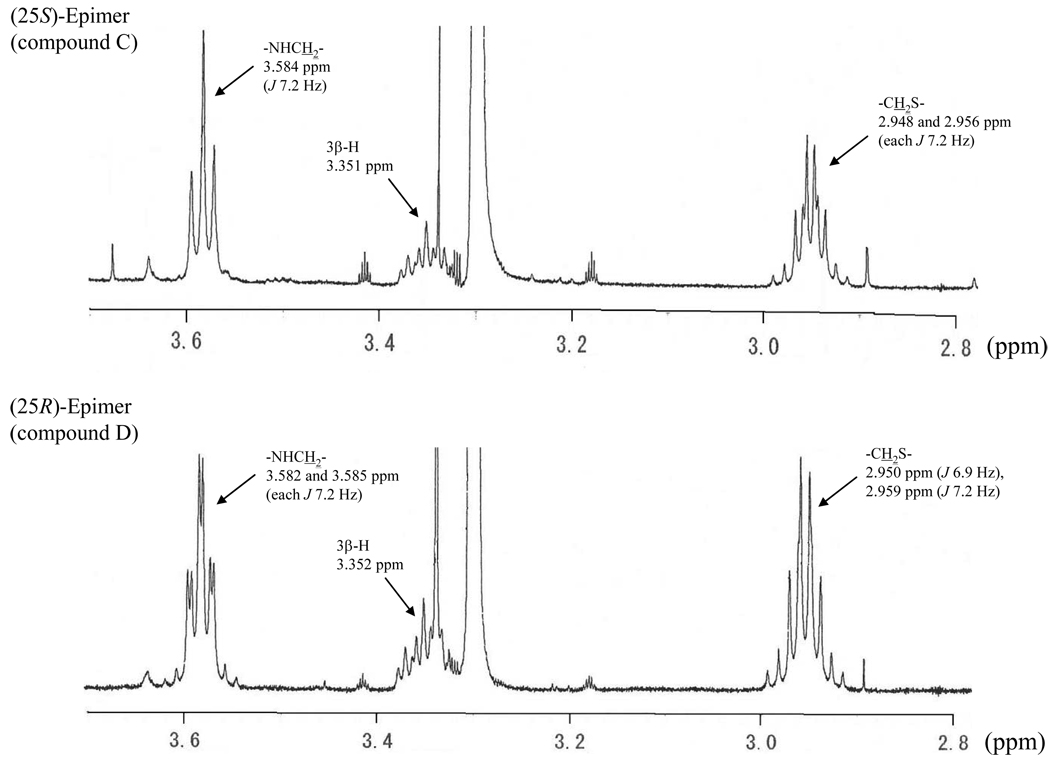
The 1H signals arising from the hydrogens on the taurine moiety (28-H2 and 29-H2) also provided distinct differences in the two 25-epimers. As shown in Figure 6, the 1H signal of 28-H2 in the 25S-epimer (C) occurred at 3.584 ppm as a triplet (J, 7.2 Hz), whereas the corresponding 1H signals in the 25R-isomer (D) appeared at 3.582 and 3.585 ppm as a double triplet (each J, 7.2 Hz). In addition, the 1H signals of 29-H2 in C appearing at 2.948 and 2.956 ppm exhibited a double triplet with the J value of 7.2 Hz, whereas those for D resonated at 2.950 and 2.959 ppm with the J values of 6.9 and 7.2 Hz, respectively. These 1H and 13C NMR characteristics can, therefore, be used to distinguish the two C-25 epimeric THCAs (in the form of their taurine conjugates).
Fig. 6.
Partial 1H NMR spectra of the isolated compounds C and D.
Discussion
Chemistry
The analyses reported here indicate that the major bile salts of the medaka are the 25R- and 25S-epimers of THCA in a ratio of 7 to 1.
In 1939, Kurauti and Kazuno isolated a THCA from the bile of the bullfrog (1939). Two years later, Mabuti (1941) isolated a C27 trihydroxy bile acid from bullfrog bile that was similar, but not identical to that isolated by Kurauti and Kazuno (1939). The bile acid isolated by Mabuti was eventually shown to be the S-isomer of THCA by Bridgwater, working in the laboratory of Haslewood, who synthesized both epimers (Bridgwater, 1956). Others have improved the synthesis (Briggs, 1970; Kurosawa et al., 1995; Starchenkov et al., 2000) or separation (Batta et al., 1991; Goto et al., 1989a, b; Une et al., 1983) of the two epimers. The exact stereochemical configuration of the S-epimer of THCA was established using X-ray crystallography by Batta et al (1979a). The concurrent presence of the two epimers in bullfrog and alligator bile has been reported (Une and Hoshita, 1994), and they are also present in the bile of some children with peroxisomal disease (Une et al., 1987). Nonetheless, the view at present is that the C27 bile acids occurring in crocodiles, turtles, and squamates, and early evolving birds consist mostly of the R-epimer (Une and Hoshita, 1994), although there is little experimental verification of this opinion. We have recently reported that the major C27 bile acids present in the bile of the Red-winged tinamou (Rhynchotus rufescens), an early evolving bird, consist entirely of the R-epimer (Hagey et al., 2009).
A great variety of bile acids with complex side chains occur in amphibians, usually as taurine-conjugates (Une and Hoshita, 1994). Although the amide bond of taurine conjugated THCA can be cleaved enzymatically (Batta et al., 1979b), no such hydrolase is available at present. Concentrated alkali can be used for deconjugation, but may induce artifacts (Une and Hoshita, 1994), although a microwave process has been claimed to effect deconjugation without racemization (Dayal and Ertel, 1998). Our finding that NMR can be used to distinguish the R- and S-epimers of THCA suggests that 1H and 13C NMR will prove to be of value in elucidating the exact chemical structure of bile salts with complex side chains. The observed NMR differences between the R- and S-isomers are probably due to the epimeric diastereomers at C-25.
Bile acid biosynthesis in peroxisomes
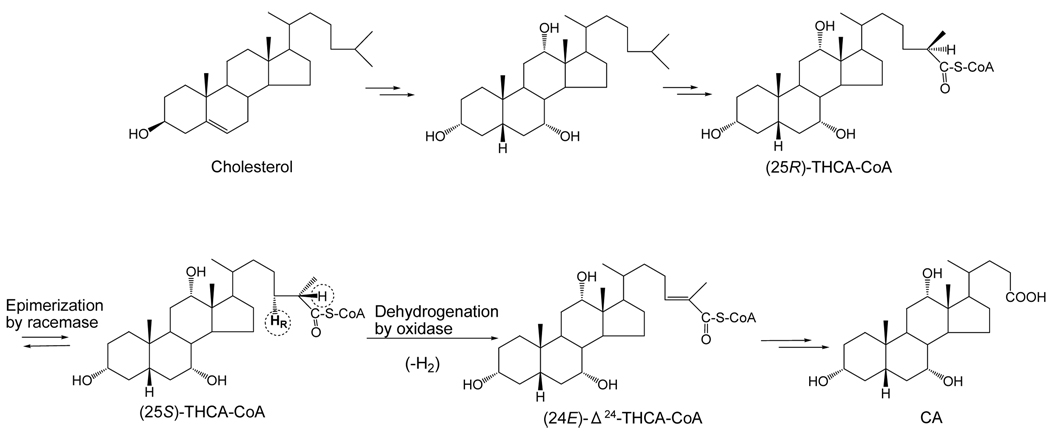
It is well known that in mammals, the oxidative cleavage of the iso-octane side chain of cholesterol proceeds by a pathway that is essentially identical to β-oxidation of fatty acids and occurs in the peroxisomes (Ferdinandusse et al., 2009). In the neutral biosynthetic pathway of C24 bile acids, 5β-cholestane diol or triol undergoes hydroxylation at C-27 (and then oxidation to a carboxyl group) in the mitochondria by sterol 27-hydroxylase (CYP27A). Carboxylation is stereospecific and results in formation of the 25R-epimer (Shefer et al., 1978). The CoA ester is then formed. The next step in the β-oxidation pathway is peroxisomal dehydrogenation (oxidation) of the 5β-cholestanoic acid to form (24E)-5β-cholest-24-enoic acid mediated by acyl-CoA oxidase. In the desaturation step, there is elimination of the HR (pro-R) at C-24 and H at C-25, and this reaction can only occur with the S-epimer of THCA (Ikegawa et al., 1995b, 1998; Van Veldhoven et al., 1996). Therefore, a racemase is required to change the 25R-epimer in part to the 25S-epimer, and this enzyme has been characterized (Cuebas et al., 2002; Ikegawa et al., 1995a, 1997; Lloyd et al., 2008). These early steps in side chain cleavage of cholesterol to form C24 bile acids are illustrated in Figure 7.
Fig. 7.
Early steps in the “neutral” biosynthetic pathway of CA from cholesterol in vertebrates. The two epimers of the CoA thioester of THCA are shown.
It is possible that activity of the racemase is rate-limiting for side chain cleavage in the medaka and other vertebrates whose bile acids contain C27 acids. A low racemase activity could explain the predominance of the R-epimer of THCA in the medaka, as also occurs in patients with peroxisomal methylacyl-CoA racemase deficiency (Une et al., 1987). The acyl CoA racemase has been sequenced in the medaka and four other fish species (fugu, freshwater puffer fish, rainbow smelt, and zebrafish), and homology compared to the human racemase. In these 5 fish species, the medaka racemase had a lower homology with human racemase (52%) than any of the four other fish species (range 69–71%) (Hagey et al., in press).
Occurrence of C27 bile acids in fish
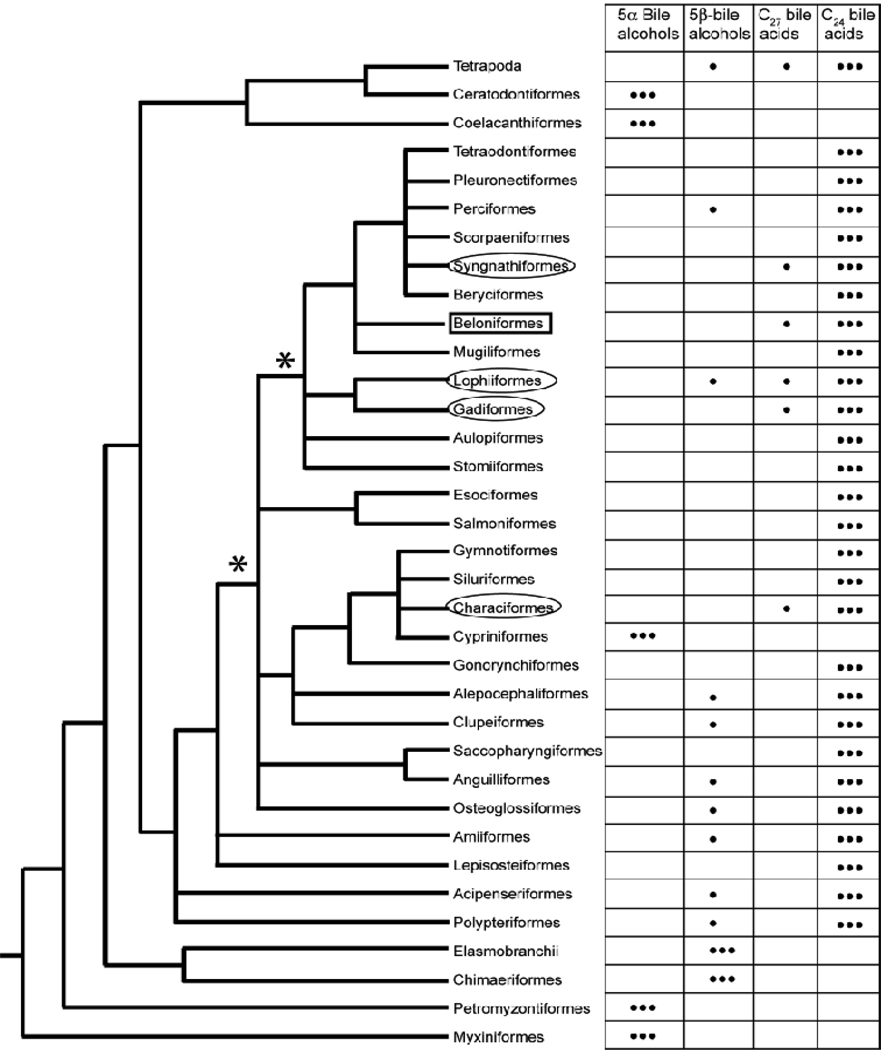
In a recent study, Hagey et al. (2010) determined the biliary bile salt composition of 226 fish species from 38 different orders that included jawless fish (Agnatha), cartilaginous fish, lobe-finned fish, and bony fish. In this survey of fish bile salts, C27 bile acids were uncommon and found in species from only five orders: Beloniformes (which includes the medaka), Syngnathiformes, Lophiformes, Gadiformes, and Characiformes. The medaka, however, was the only species of the 226 surveyed for which C27 bile acids comprised more than 50% of total biliary bile salts.
Current phylogenetic relationships for fish, based on molecular and morphology data (Nelson, 2006; Kawahara et al., 2008; Lavoué et al., 2008), are shown in Fig 8. The occurrence of C27 bile acids in fish from four widely distributed orders in Teleostei suggests the possibility that a common ancestor to these fish used C27 bile acids (see nodes in the phylogenetic tree marked with * in Fig. 8). The phenotype of having C27 bile acids as a significant fraction of biliary bile acids would represent an ‘ancestral’ trait that is now present in only a small number of fish species. If this hypothesis is correct, then most fish now have the ‘derived’ phenotype of having mainly C24 bile acids (the most common bile salt profile in bony fish surveyed so far), with only a small minority of fish species such as the medaka retaining the ancestral phenotype.
Fig. 8.
Bile salt species variation determined by Hagey et al. (2010) overlaid on the fish tree proposed by Nelson (2006) excluding fish orders whose bile salt profiles are unknown. Alepocephaliformes is used instead of Argentiformes following recent results of Lavoué et al. (2008), and Syngnathiformes is used instead of Gasterosteiformes following the results of Kawahara et al. (2008). The major bile salts for each species are indicated by ●●●; minor bile salts are indicated by ●. Medaka is classified in Beloniformes (box). Other than Beloniformes, only four other fish orders (circled) have species which have C27 bile acids comprising greater than 10% of total biliary bile salts. C27 bile acids are not found as major components of biliary bile salts in jawless or cartilaginous fish, or in basal teleost fish (e.g., Polypteriformes, bichirs; Acipenseriformes, including paddlefishes and sturgeons). Ancestral fish that may have used C27 bile acids are indicated by the * at two nodes in the phylogenetic tree.
Alternatively, a less parsimonious but certainly possible explanation would be that the occurrence of C27 bile acids in fish evolved independently in different fish orders. More extensive surveys of biliary bile salt composition in fish species may help in providing evidence for this proposed evolutionary pathway for C27 bile acids in fish. The bile salt composition of the medaka differs markedly from that of zebra fish bile, a fish species also used widely for biological research, whose bile salts are composed entirely of 5α-cyprinol (sulfate), a C27 bile alcohol (Reschly et al, 2008; Hofmann et al., in press).
Physical and biological properties of C27 bile acids
C27 bile acids are difficult to isolate in sizable amounts and with high purity from bile, and their synthesis from their C24 homologues is difficult. Therefore there has been no systematic comparison of the physicochemical properties of the C27 bile acids with those of their corresponding C24 homologues. Based on studies of alkyl sulfates or alkyl sulfonates, one would expect the taurine conjugates of C27 homologues to have a slightly lower critical micellization concentration and a slightly higher critical micellization temperature than those of their corresponding C24 homologues (Shinoda et al, 1963). The presence of branching in the terminal portion of the side chain is likely to diminish the effect of three additional carbon atoms on these parameters. The unconjugated C27 bile acids should also have lower aqueous solubilities of the protonated form, and, as a result, they should have higher critical micellization pH values than those of their corresponding C24 homologues. In addition, the calcium salts of the unconjugated or glycine conjugated C27 bile acid should have lower aqueous solubilities than their C24 homologues were they to be present in bile (Hofmann and Mysels, 1992). The toxicity of C27 bile acids to cultured cells was recently examined by Ferdinandusse et al (2009). Compared to their C24 homologues, C27 bile acids were found to be more cytotoxic, and also to impair mitochondrial function, as evidenced by inhibition of ATP formation as well as the generation of reactive oxygen species.
We conclude that the medaka is the first fish species identified to date whose biliary bile salts contain mostly C27 bile acids. Low activity of the acyl CoA racemase may contribute to this unusual biliary bile salt composition.
Acknowledgements
Dr. Genta Kakiyama contributed able technical assistance. This work was supported by a Grant-in-Aid for Scientific Research (C) (to T.I., Grant 19,510,223) from the Ministry of Education, Sciences, Sports, and Culture of Japan, Nihon University Multidisciplinary Research Grant (to T.I.) for 2007-2008, and the Institute of Natural Science, Nihon University, Joint Research Grant (to T.I.) for 2008-2009. MDK is supported by K08-GM074238 from the National Institutes of Health.
References
- Batta AK, Salen G, Blount JF, Shefer S. Configuration at C-25 in 3α,7α,12α-trihydroxy-5β-cholestan-26-oic acid by X-ray crystallography. J Lipid Res. 1979a;20:935–940. [PubMed] [Google Scholar]
- Batta AK, Salen G, Cheng FW, Shefer S. Cleavage of the taurine conjugate of 3α,7α,12α-trihydroxy-5β-cholestan-26-oic acid by rat fecal bacteria. J Biol Chem. 1979b;254:11907–11909. [PubMed] [Google Scholar]
- Batta AK, Salen G, Arora R, Shefer S, Batta M. High-performance liquid chromatographic separation of bile acids and bile alcohols diastereoisomeric at C-25. J Chromatogr. 1991;542:184–188. doi: 10.1016/s0021-9673(01)88758-6. [DOI] [PubMed] [Google Scholar]
- Bridgwater RJ. Partial synthesis of the two 3α,7α,12α-trihydroxycoprostanic acids and of similar bile acids with extended side chains. Biochem J. 1956;64:593–599. doi: 10.1042/bj0640593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs T. Partial synthesis of 25D- and 25L-cholestanoic acids from some common bile acids. J Org Chem. 1970;35:1431–1434. doi: 10.1021/jo00830a037. [DOI] [PubMed] [Google Scholar]
- Cuebas DA, Phillips C, Schmitz W, Conzelmann E, Novikov DK. The role of α-methylacyl-CoA racemase in bile acid synthesis. Biochem J. 2002;363:801–807. doi: 10.1042/0264-6021:3630801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayal B, Ertel NH. Rapid hydrolysis of bile acid conjugates using microwaves: retention of absolute stereochemistry in the hydrolysis of (25R)-3α,7α,12α-trihydroxy-5β-cholestan-26-oyltaurine. Lipids. 1998;33:333–338. doi: 10.1007/s11745-998-0213-y. [DOI] [PubMed] [Google Scholar]
- Ferdinandusse S, Denis S, Faust PL, Wanders RJA. Bile acids: the role of peroxisomes. J Lipid Res. 2009;50:2139–2147. doi: 10.1194/jlr.R900009-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandusse S, Denis S, Dacremont G, Wanders RJ. Toxicity of peroxisomal C27-bile acids intermediates. Mol Genet Metab. 2009;96:121–128. doi: 10.1016/j.ymgme.2008.11.165. [DOI] [PubMed] [Google Scholar]
- Goto J, Gang S, Miura H, Nambara T, Tazawa Y, Tada K. Separation and characterization of C-25 epimers of unconjugated and conjugated trihydroxycholestanoic acids in urine from a patient with Zellweger syndrome by high-performance liquid chromatography. J Liquid Chromatogr. 1989a;12:1075–1084. [Google Scholar]
- Goto J, Shao G, Miura H, Nambara T. Separation of C-25 epimers of 5β-cholestanoic acids by high performance liquid chromatography with precolumn fluorescence labeling. Anal Sci. 1989b;5:19–22. [Google Scholar]
- Hagey LR. Bile acid biodiversity in vertebrates: chemistry and evolutionary implication. PhD Dissertation. San Diego: University of California; 1992. [Google Scholar]
- Hagey LR, Kakiyama G, Muto A, Iida T, Mushiake K, Goto T, Mano N, Goto J, Oliveira CA, Hofmann AF. A new, major C27 biliary bile acid in the Red-winged tinamou (Rhynchotus rufescens): (25R)-1α,3α,7α-trihydroxy-5β-cholestan-27-oic acid. J Lipid Res. 2009a;50:651–657. doi: 10.1194/jlr.M800521-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagey LR, Møller PR, Hofmann AF, Krasowski MD. Diversity of bile salts in fish and amphibians: evolution of a complex biochemical pathway. Physiol Biochem Zool. doi: 10.1086/649966. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslewood GAD. Bile salt evolution. J Lipid Res. 1967;8:535–550. [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res. doi: 10.1194/jlr.R000042. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol Life Sci. 2008;65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Mysels KJ. Bile acid solubility and precipitation in vitro and in vivo: the role of conjugation, pH, and Ca2+ ions. J Lipid Res. 1992;33:617–626. [PubMed] [Google Scholar]
- Hoshita T. Bile alcohols and primitive bile acids (Chapter 10) In: Danielsson H, Sjövall J, editors. Sterols and Bile acids. Amsterdam, New York, and Oxford: Elsevier; 1985. pp. 279–302. [Google Scholar]
- Ijare OB, Somashekar BS, Jadegoud Y, Nagane Gowda GA. 1H and 13C NMR characterization and stereochemical assignment of bile acids in aqueous media. Lipids. 2005;40:1031–1041. doi: 10.1007/s11745-005-1466-1. [DOI] [PubMed] [Google Scholar]
- Ikegawa S, Goto T, Mano N, Goto J. Substrate specificity of THCA-CoA oxidases from rat liver light mitochondrial fractions on dehydrogenation of 3α,7α,12α-trihydroxy-5β-cholestanoic acid CoA thioester. Steroids. 1998;63:603–607. doi: 10.1016/s0039-128x(98)00070-1. [DOI] [PubMed] [Google Scholar]
- Ikegawa S, Goto T, Watanabe H, Goto J. Stereoisomeric bio-inversion of (25R)- and (25S)-3α,7α,12α-trihydroxy-5β-cholestanoic acid CoA thioesters in rat liver peroxisome. Enantiomers. 1997;2:333–342. [Google Scholar]
- Ikegawa S, Goto T, Watanabe H, Goto J. Stereoisomeric inversion of (25R)- and (25S)-3α,7α,12α-trihydroxy-5β-cholestanoic acids in rat liver peroxisome. Biol Pharm Bull. 1995a;18:1027–1029. doi: 10.1248/bpb.18.1027. [DOI] [PubMed] [Google Scholar]
- Ikegawa S, Watanabe H, Goto T, Mano N, Goto J, Nambara T. Stereospecific dehydrogenation of (25R)- and (25S)-3α,7α,12α-trihydroxy-5β-cholestanoic acids by acyl-CoA oxidase in rat liver light mitochondrial fraction. Biol Pharm Bull. 1995b;18:1041–1044. doi: 10.1248/bpb.18.1041. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, Jindo T, Kobayashi D, Shimada A, Toyoda A, Kuroki Y, Fujiyama A, Sasaki T, Shimizu A, Asakawa S, Shimizu N, Hashimoto S, Yang J, Lee Y, Matsushima K, Sugano S, Sakaizumi M, Narita T, Ohishi K, Haga S, Ohta F, Nomoto H, Nogata K, Morishita T, Endo T, Shin-I T, Takeda H, Morishita S, Kohara Y. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Miya M, Mabuchi K, Lavoué S, Inoue JG, Satoh TP, Kawaguchi A, Nishida M. Interrelationships of the 11 gasterosteiform families (sticklebacks, pipefishes, and their relatives): a new perspective based on whole mitogenome sequences from 75 higher teleosts. Mol Phylogenet Evol. 2008;46:224–236. doi: 10.1016/j.ympev.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Kurauti Y, Kazuno T. Tetraoxycholan, Trioxycholen und Trioxy-bis-norsterocholansaure aus der Galle von Rana catesbina Shaw. Z Physiol Chem. 1939;262:53–60. [Google Scholar]
- Kurosawa T, Nakano H, Sato M, Tohma M. Synthesis of 3α,7α,12α-trihydroxy- and 3α,7α-dihydroxy-5β-cholestan-26-oic acids by the use of β-ketosulfoxide. Steroids. 1995;60:439–444. doi: 10.1016/0039-128x(95)00033-m. [DOI] [PubMed] [Google Scholar]
- Lavoué S, Miya M, Poulsen JY, Møller PR, Nishida M. Monophyly, phylogenetic position and inter-familial relationships of the Alepocephaliformes (Teleostei) based on whole mitogenome sequences. Mol Phylogenet Evol. 2008;47:1111–1121. doi: 10.1016/j.ympev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Lloyd MD, Darley DJ, Wierzbicki AS, Thrreadgill MD. α-Methylacyl-CoA racemase – an ‘obscure’ metabolic enzyme takes centre stage. FEBS J. 2008;275:1089–1102. doi: 10.1111/j.1742-4658.2008.06290.x. [DOI] [PubMed] [Google Scholar]
- Mabuti H. Uber Trioxy-bis-nor-sterocholansaure C26H44O5 aus der Galle von Rana catesbina Shaw. J Biochem. 1941;33:117–130. [Google Scholar]
- Matsumoto Y, Oota H, Asaoka Y, Nishina H, Watanabe K, Bujnicki JM, Oda S, Kawamura S, Mitani H. Medaka: a promising model animal for comparative population genomics. BMC Res Notes. 2009;2:88. doi: 10.1186/1756-0500-2-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagane Gowda GA, Ijare OB, Somashekar BS, Sharma A, Kapoor VK, Khetrapal CL. Single-step analysis of individual conjugated bile acids in human bile using 1H NMR spectroscopy. Lipids. 2006;41:591–603. doi: 10.1007/s11745-006-5008-7. [DOI] [PubMed] [Google Scholar]
- Nelson JS. Fishes of the World. Hoboken, NJ: John Wiley and Sons, Inc.; 2006. [Google Scholar]
- Parenti LR. A phylogenetic analysis and taxonomic revision of ricefishes, Oryzias and relatives (Beloniformes, Adrianichthyidae) Zool J Linnean Soc. 2008;154:494–610. [Google Scholar]
- Reschly EJ, Ai N, Ekins S, Welsh WJ, Hagey LR, Hofmann AF, Krasowski MD. Evolution of the Bile Salt Nuclear Receptor FXR in Vertebrates. J Lipid Res. 2008;49:1577–1587. doi: 10.1194/jlr.M800138-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer S, Cheng FW, Batta AK, Dayal B, Tint S, Salen G, Mosbach EH. Stereospecific side chain hydroxylations in the biosynthesis of chenodeoxycholic acid. J Biol Chem. 1978;253:6386–6392. [PubMed] [Google Scholar]
- Shinoda K, Nakagawa T, Tomamushi B, Isemura T. Colloidal Surfactants. New York: Academic Press Inc; 1963. [Google Scholar]
- Starchenkov I, Trapencieris P, Kauss V, Jas G, Kalvinsh I. A convenient synthesis of 5β-cholestan-26-oic and 5β-cholestan-26,27-dioic acids. Steroids. 2000;65:143–147. doi: 10.1016/s0039-128x(99)00099-9. [DOI] [PubMed] [Google Scholar]
- Une M, Hoshita T. Natural occurrence and chemical synthesis of bile alcohols, higher bile acids, and short side chain bile acids. Hiroshima J Med Sci. 1994;43:37–67. [PubMed] [Google Scholar]
- Une M, Morigami I, Kihira K, Yasuhara M, Kuramoto T, Hoshita T. Stereospecific formation of (24E)-3α,7α,12α-trihydroxy-5β-cholest-24-en-26-oic acid and (24R,25S)-3α,7α,12α,24-tetrahydroxy-5β-cholestan-26-oic acid from either (25R)- or (25S)-3α,7α,12α-trihydroxy-5β-cholestan-26 oic acid by rat liver homogenate. J Biochem. 1984;96:1103–1107. doi: 10.1093/oxfordjournals.jbchem.a134927. [DOI] [PubMed] [Google Scholar]
- Une M, Nagai F, Hoshita T. Comparative biochemical studies of bile acids and bile alcohols. High-performance liquid chromatographic separation of higher bile acids. J. Chromatogr. 1983;257:411–415. doi: 10.1016/s0021-9673(01)88201-7. [DOI] [PubMed] [Google Scholar]
- Une M, Tazawa Y, Tada K, Hoshita T. Occurrence of both (25R)-and (25S)-3α,7α,12α-trihydroxy-5β-cholestanoic acids in urine from an infant with Zellweger’s syndrome. J Biochem. 1987;102:1525–1530. doi: 10.1093/oxfordjournals.jbchem.a122200. [DOI] [PubMed] [Google Scholar]
- Van Veldhoven PP, Croes K, Asselberghs S, Herdewijn P, Mannaerts GP. Peroxisomal β-oxidation of 2-methyl branched acyl-CoA esters: stereospecific recognition of the 2S–methyl compounds by trihydroxycoprostanoyl-CoA oxidase and pristanoyl-CoA oxidase. FEBS Lett. 1996;388:80–84. doi: 10.1016/0014-5793(96)00508-x. [DOI] [PubMed] [Google Scholar]