Abstract
Histone lysine acetylation and methylation are important during gene transcription in a chromatin context1,2. Our knowledge about the types of protein modules that can interact with acetyl-lysine has so far been limited to bromodomains1. Recently, a tandem PHD (plant homeodomain) finger3 (PHD12) of human DPF3b, which functions in association with the BAF chromatin remodelling complex to initiate transcription in the heart and muscle development, was reported to bind histones H3 and H4 in an acetylation sensitive manner4, making it a first alternative to bromodomains for acetyl-lysine binding5. Here, we report the structural mechanism of acetylated histone binding by the double PHD fingers of DPF3b. Our three-dimensional solution structures and biochemical analysis of DPF3b illuminate the molecular basis of the integrated tandem PHD finger, which acts as one functional unit in the sequence-specific recognition of lysine 14-acetylated histone H3 (H3K14ac). Whereas the interaction with H3 is promoted by acetylation at lysine 14, it is inhibited by methylation at lysine 4, and these opposing influences are important during transcriptional activation of DPF3b target genes Pitx2 and Jmjd1c. Binding of this tandem protein module to chromatin can thus be regulated by different histone modifications during the initiation of gene transcription.
We characterized the tandem PHD12 of human DPF3b (Fig. 1a; Supplementary Fig. 1) binding to histone H3 and H4 peptides containing known modifications using nuclear magnetic resonance (NMR) spectroscopy (Fig. 1b; Supplementary Fig. 2), fluorescence polarization (FP) (Supplementary Fig. 3) and isothermal titration calorimetry (ITC) (Fig. 1c) methods. These complementary biophysical measurements yield consistent conclusions on DPF3b’s binding preferences to H3 and H4 that were not resolved previously4. The salient features of these studies are summarized as follows (Supplementary Tables 1 and 2). The DPF3b PHD12 binds to an unmodified H3 peptide (residues 1–18) with a dissociation constant Kd of 2.3 µM; this affinity is increased by 4-fold (Kd of 0.5 µM) when H3 is acetylated at lysine 14 (H3K14ac). The latter is achieved by favorable contributions of binding enthalpy (ΔH = −6.6 kcal/mol) and entropy (TΔS = 2.0 kcal/mol) to the free energy of binding (ΔG = −8.6 kcal/mol) (Fig. 1c; Supplementary Table 2) that represents a 2.2 kcal/mol gain in enthalpy from binding to acetylated H3K14. Strikingly, acetylation or methylation at H3K4 causes a 15 or 20-fold reduction in affinity to the PHD12, respectively, whereas modifications at H3K9 do not affect the binding. Notably, the PHD12 also preferentially recognizes an amino-terminal acetylated H4 peptide (H4-NAc, residues 1–22) with Kd of 7.4 µM, as measured by ITC.
Figure 1. 3D structure of the tandem PHD finger of human DPF3b.
(a) Domain organization of human DPF3 proteins, including 2/3 domain (light brown), nuclear localization signal (NLS, red), nuclear receptor interaction domain (NID, pink), C2H2 Krüppel-like zinc finger (purple), PHD finger 1 (green) and PHD finger 2 (blue). (b) 2D 1H-15N-HSQC NMR spectra showing DPF3b PHD12 binding to histone peptide H3K14ac (residues 1–18) or H4-NAc (residues 1–22), as illustrated by change of protein resonances between the free (black) and peptide bound (red) states. (c) Isothermal titration calorimetry measurements of PHD12 binding to different H3 and H4 peptides. (d) Ribbon diagram depicting the average minimized NMR structure of the PHD12 bound to an H3K14ac peptide (yellow) in front and top views. The zinc atoms are highlighted as red spheres. (e) Space filled PHD12 structure, highlighting the H3K14ac peptide (yellow) bound across the unified structure of PHD1 (green) and PHD2 (blue).
We solved the three-dimensional structure of the DPF3b PHD12 bound to the H3K14ac peptide using NMR methods6. The structure of the complex is well defined (Supplementary Table 3) and shows that each module consists of a two-strand anti-parallel β-sheet followed by a C-terminal α-helix (Fig. 1d; Supplementary Fig. 3c). This typical PHD fold7–13 is stabilized by two zinc atoms coordinated by the Cys4-His-Cys3 motif in a cross-brace topology. The two PHD fingers are pressed against one another in a face-to-back direction with the H3 peptide bound across the unified structure. The acetylated K14 and the amino-terminal residues of H3 interact with PHD1 and PHD2, respectively (Fig. 1e). Binding of lysines 4 and 9 of H3 to acidic residues at the tandem domain interface creates an outward facing loop in the peptide.
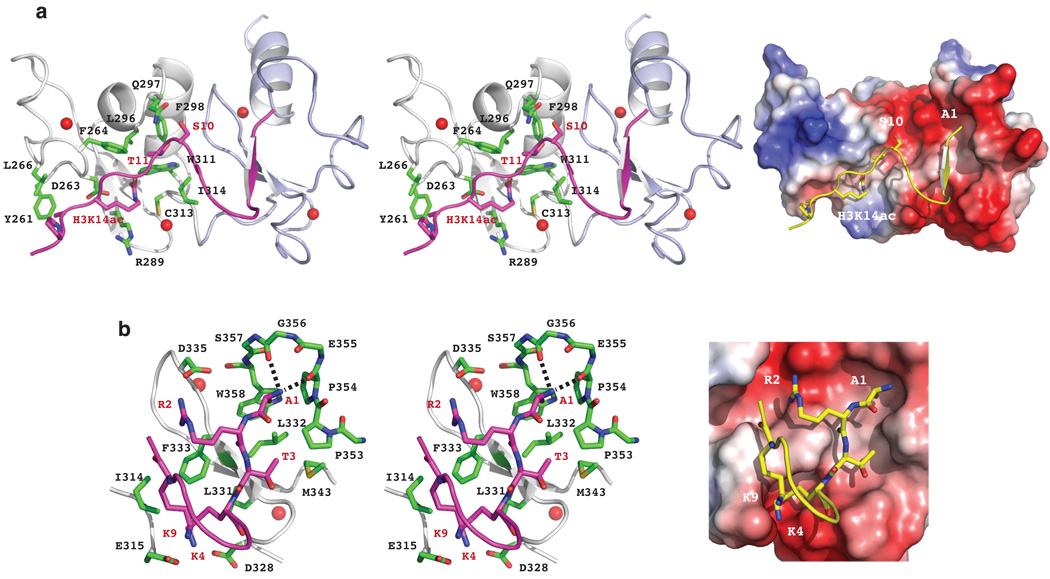
The H3 residues T11-P16 flanking H3K14ac form an extended conformation bound in an elongated groove nearly perpendicular to the β-sheet of PHD1 (Fig. 2a). The acetylated H3K14 intercalates into a hydrophobic pocket bottomed on the β-sheet of PHD1 and lined with Phe264, Leu296, Trp311 and Ile314 (Fig. 2a). The rim of the binding pocket is clustered with Asp263, Arg289, and zinc-coordinating Cys313. The acetyl amide of H3K14ac is positioned within a hydrogen bond distance to the side-chain carbonyl oxygen of Asp263 and the sulfur hydryl of Cys313, whereas the acyl chain of H3K14ac interacts with Arg289 side chain. Further, H3T11’s methyl group binds to Phe264, Leu296 and Ile314 and hydroxyl group forms a hydrogen bond to the side-chain carbonyl oxygen of Gln297 (Fig. 2a). Individual mutation D263A or F264A, which does not cause structural perturbations to the protein, results in a nearly complete loss of acetyl-lysine recognition, but does not affect unmodified H3 binding (Supplementary Figs. 4–8, and Table 2). These DPF3b residues for acetyl-lysine binding are either structurally conserved (Trp311 and Cys313) or functionally distinct (Asp263, Phe264 and Arg289) in the PHD family (Supplementary Fig. 1).
Figure 2. Structural mechanism of acetyl-lysine recognition by human DPF3b PHD12.
(a) DPF3b PHD1 recognition of H3K14ac, as illustrated in the PHD12/H3K14ac complex structure with stereo ribbon view (left) and electrostatic potential surface of the protein (right). (b) DPF3b PHD2 recognition of the N-terminal residues of the H3K14ac peptide, depicted in stereo ribbon structure (left) and protein electrostatic potential surface representation (right). The zinc atoms are highlighted as red spheres.
The amino-terminal NH3+ group of H3A1 is recognized by the PEGSWSC hairpin loop (the residues in bold are conserved in PHD fingers) in a negatively charged cavity above the β-sheet in PHD2 through hydrogen bonding with backbone carbonyl oxygen atoms of Pro354 and Gly356 (Fig. 2b). H3 binding is secured with its amino-terminal R2-T3-K4 that forms an antiparallel β-strand to Leu331-Leu332-Phe333 in PHD2. Side chains of the N-terminal H3 residues establish a network of interactions with PHD2 residues such as those between H3A1 and Trp358, H3R2 and Asp335, as well as H3K4 and Asp328 and Glu315. Notably, Glu315 projects from PHD1 to interact with H3K9. This cluster of inter-molecular interactions forces H3 to adopt a bulge confirmation belted down at K4 and K9 (Fig. 2b). The H3 peptide is further stabilized by H3Q5 interacting with Asp329 and Gln330, and H3S10 that stacks its hydroxyl group between the side-chain amide of Gln297 of PHD1 and the guanidinium group of H3R2.
Ala mutation of Glu315 or Asp335 causes an 80–250-fold reduction in affinity to the H3K14ac peptide (Supplementary Fig. 4c,d, and Table 2), confirming their importance for H3 binding at K4 and K9, and R2, respectively. Consistent with the structure, acetylation or methylation of H3K4 results in a major drop in affinity to the PHD12 much more so than H3K9 (Supplementary Tables 1 and 2). The structure of the PHD12 bound to a H3 peptide (residues 1–20) reveals that the mode of DPF3b recognition of the H3 N-terminal residues remains the same when H3 is not acetylated at K14, albeit with a 4-fold reduction in affinity (Supplementary Fig. 9 vs. Fig. 1d,e). Taken together, these results show how sequence-specific histone H3 binding by DPF3b is modulated positively by acetylation at H3K14 and negatively by methylation at H3K4.
The structure of PHD12 bound to an H4-NAc peptide (residues 1–20) shows that the key acetyl-lysine binding residues Asp263, Phe264 and Arg289 are also engaged in recognition of the N-terminal acetyl group of H4S1 (Supplementary Fig. 10, and Table 3). A network of electrostatic interactions formed between H4 residues R17-H18-R19-K20 and DPF3b residues of Trp311, Ile314, Asp328, Asp329, Phe333, Asp335, Asp338, and Glu355 located at the tandem PHD interface secure the H4 recognition. This mode of H4 recognition by DPF3b – both for the acetyl group and the positive motif – is further confirmed by the 3D structure of DPF3b PHD12 bound to an H4K16ac peptide (residues 9–23) (Supplementary Fig. 11, and Table 3).
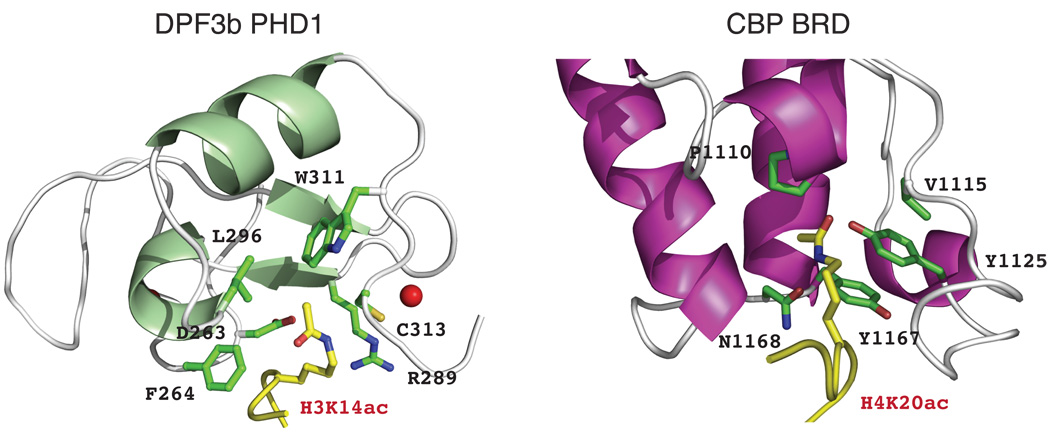
Our structural analyses argue that the DPF3b PHD12 uses a conserved mechanism as an alternative to bromodomains for acetyl group recognition1. The PHD12 employs an Asp263-Phe264 motif for recognizing the N-acetyl group of H3K14ac or H4-Nac whereas bromodomains use the conserved signature motif Tyr-Asn, such as Tyr1167-Asn1168 in the CBP bromodomain to interact with that of H4K20ac5,14 (Fig. 3). In contrast to PHD12, the acetyl group-binding pocket in bromodomains is constructed with different structural elements, i.e. formed by residues from the inter-helical ZA and BC loops of the 4-helix bundle. However, judged from the extent of histone interactions with these modules, PHD12 has more restricted binding preferences for acetylated histone sequences than that of the bromodomains.
Figure 3. Distinct modes of acetyl-lysine and H3 recognition by DPF3b PHD12 and other protein modules.
Comparison of the structural features of acetyl-lysine binding pockets between DPF3b PHD12/H3K14ac (left) and CBP bromodomain/H4K20ac (right) complexes. The protein and peptide residues are color-coded in the same scheme as described in Fig. 2.
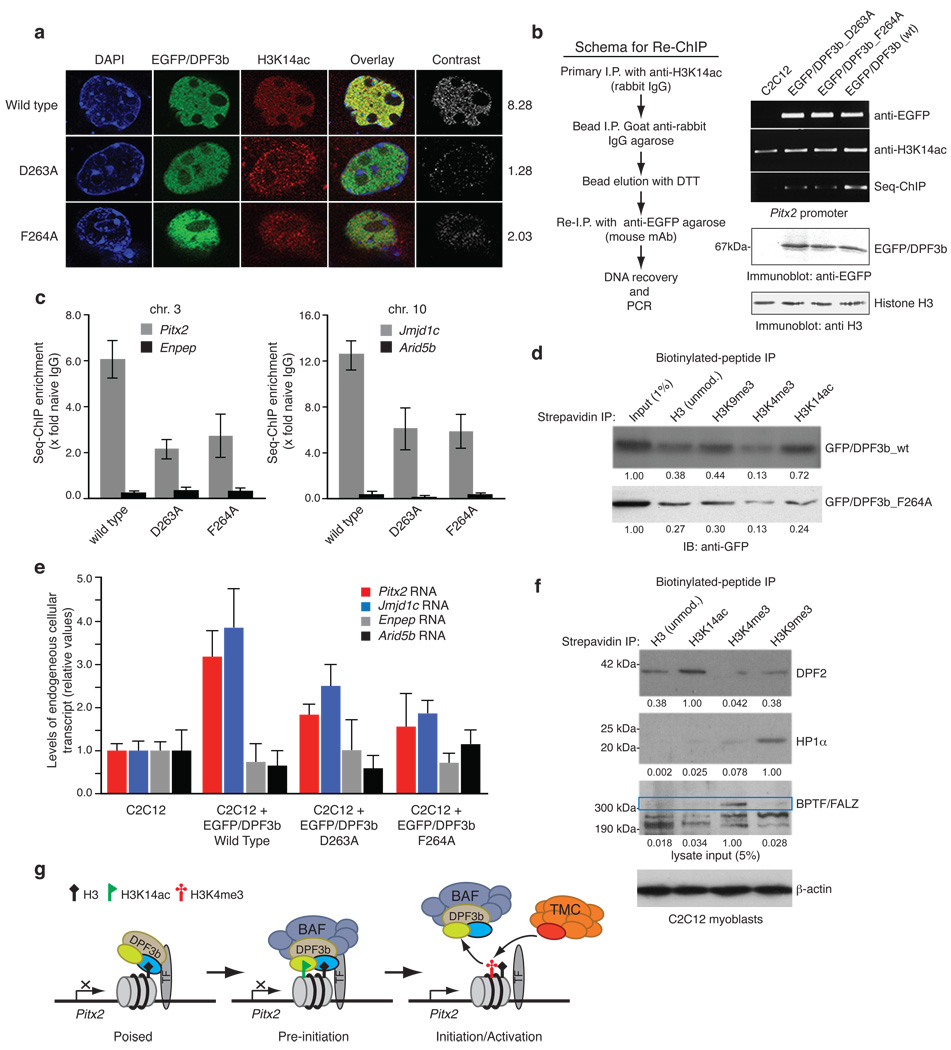
We next assessed the functional importance of DPF3b and H3 interactions in C2C12 cells wherein DPF3b is preferentially expressed and functions with the BAF remodeling complex in control of gene transcription in the development of heart and muscle4. We observed that the ectopically expressed EGFP-fusion DPF3b is co-localized with H3K14ac sites in the C2C12 cell nucleus as visualized by confocal fluorescence imaging, and this co-localization is dramatically reduced in the cells transfected with the acetyl-lysine binding deficient mutant D263A or F264A (Fig. 4a; Supplementary Fig. 12a and Table 4). We then examined PHD12 binding to H3K14ac for transcription of its target gene Pitx2 located in chromosome 3 using a re-chromatin immunoprecipitation (re-ChIP) assay, in which the cell nuclear extracts were immunoprecipiated sequentially with antibodies directed first against H3K14ac and then against GFP to recover EGFP-DPF3b followed by PCR at the Pitx2 promoter (Fig. 4b, left). The re-ChIP results reveal that DPF3b interacts with H3K14ac at the Pitx2 promoter site, and this DPF3b/H3K14ac co-occupancy at the Pitx2 site was reduced by 2–3-fold in the cells transfected with the D263A or F264A mutant (Fig. 4b,c). This is consistent with F264A’s inability to bind acetylated-K14 of H3 as shown in a peptide pull-down assay (Fig. 4d). Further, the endogenous cellular transcript of Pitx2 was enhanced by more than 3-fold in the C2C12 cells transfected with wild type DPF3b, and the enhancement was reduced to less than 2-fold with the D263A or F264A mutant (Fig. 4e). Similar results were seen with another DPF3b target gene Jmjd1c in chromosome 10 in the C2C12 cells (Fig. 4c,e). Finally, we observed that endogenous DPF2 in the C2C12 cells favors binding to H3 when acetylated at K14 over unmodified H3, and does not bind to H3 when K4 is trimethylated. As positive controls, we confirmed preferential association of endogenous BPTF15 and HP1α16 with H3K4me3 and H3K9me3, respectively. Together, these results demonstrate that DPF3b/H3K14ac binding plays an important role in gene transcriptional activation.
Figure 4. Acetylation and methylation modulated histone H3 binding by DPF3b PHD12 is important for gene transcriptional activation.
(a) Co-localization of EGFP-DPF3b PHD12, or its acetyl-lysine binding deficient mutants, and H3K14ac in the C2C12 cell nucleus visualized by confocal fluorescence imaging. Calculated mean normalized total signal intensity value for each DPF3b is listed on the right (Supplementary Table 4). (b) Sequential chromatin immunoprecipitation (re-ChIP) assay (scheme, left) used to assess EGFP-DPF3b or its PHD12 mutants in interactions with H3K14ac at the mouse Pitx2 promoter site (right). (c) Quantitation of DNA recovered for Pitx2 and Jmjd1c promoters by qPCR following enrichment by immunoprecipitations with anti-sera against H3K14ac and seq-ChIP of EGFP/DPF3b with anti-EGFP agarose. Two non-DPF3b target genes Enpep in chromosome 3 and Arid5b in chromosome 10 were used in qChIP as negative controls to Pitx2 and Jmjd1c, respectively. (d) Assessing wild type DPF3b and mutant F264A binding to C-terminal biotinylated histone H3 peptides of different modifications in a peptide pull-down assay. Immunoblots depict H3 peptides binding to DPF3b from nuclear extracts of C2C12 cells transfected with the corresponding GFP/DPF3b plasmid. Signals in the immunoblots were quantified using NIH’s ImageJ software, and normalized to the input (1%) as 1.00. (e) Relative transcript levels of endogenous Pitx2 and Jmjd1c in C2C12 cells 2 days following transfections with wild type and mutant DPF3b cDNAs as indicated. (f) Evaluation of endogenous DPF2, HP1α and BPTF/FALZ in nuclear extracts of C2C12 cells binding to histone H3 peptides of different modifications. A single filter was used to determine retention of specific endogenous protein for each of the H3 peptides as shown. Separately, 5% of overall input for the peptide pull-down was monitored with β-actin. Blots were scanned and densitometry measurements were performed as described in d. (g) Schematic diagram illustrating modulation of human DPF3b PHD12 binding to histone H3 by site-specific lysine acetylation and methylation during poised, pre-initiation and initiation/activation stages of gene transcription. PHD1 and PHD2 of DPF3b are color-coded in yellow and blue, respectively. TF stands for a gene specific transcription factor, and TMC (orange) represents the transcriptional machinery complex.
Functional versatility of the PHD fold, held by two zinc atoms, is underscored by the extraordinary ability of some PHD fingers to recognize histone H3 that is reinforced positively or negatively by methylation at H3K4 (ref. 2)2. Structural cooperativity of PHD modules in tandem enriches functional diversity17–21. In this study, we show that the tandem PHD12 of DPF3b work as one functional unit in recognition of H3 that is modulated by two functionally distinct modifications in transcription – promoted by acetylation at H3K14 (by PHD1) and inhibited by methylation at H3K4 (by PHD2). The former marks for recruitment of a chromatin-remodeling complex to target gene locus required for pre-initiation of transcription, whereas the latter leads to DPF3b dissociation, and consequent association of the transcriptional machinery complex that signifies initiation and activation of gene transcription (Fig. 4g). Therefore, the histone H3 interaction modulated by site-specific different modifications is likely crucial for the function of DPF3b to regulate transcription of genes in concert with the BAF remodeling complex during the heart and muscle development. Further study is required to characterize DPF3b’s unique recognition of the commonly acetylated N-terminus of histone H4 (ref. 22)22, which is confirmed in C2C12 cells (Supplementary Fig. 12b). Collectively, our study demonstrates how a tandem protein module functions through lysine acetylation and methylation-modulated histone binding in a cooperative and inter-dependent manner to control the activation of gene transcription.
METHODS SUMMARY
Biochemical and structural analyses of protein and peptide interactions were conducted by using isothermal titration calorimetry, fluorescence polarization/anisotropy and heteronuclear NMR spectroscopy methods as previously described14. Peptide pull-down assay was carried out with the previously described protocol23. Protein/protein interactions at the specific gene loci in the C2C12 cells were assessed in a re-chromatin immunoprecipitation assay. Immunofluorescence imaging was used to assess nuclear co-localization of protein on chromatin.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Sample preparation
A DNA fragment encoding human DPF3b tandem PHD finger (residues 261–372) was amplified from a human heart cDNA library (Invitrogen) with the following two primers: 5'-CGCGGATCCTACTGTGACTTCTGC-3' (coding strand) and 5'-CCGCTCGAGCATTGAGGCTTTCTCTTTGAG-3' (non-coding strand)’ and subcloned into the Bam HI-Xho I sites of the plasmid pGEX 4T-1 vector (GE Healthcare). The pGEX 4T-1 plasmid harboring DPF3b PHD12 gene was transformed into Escherichia coli BL21 PRIL codon plus strain (Stratagene). The E. coli cells containing the DPF3b plasmid were grown in LB medium supplemented with 0.1 mM zinc acetate at 37°C and the protein expression was induced by the addition of IPTG (0.3 mM) at 18°C overnight. Uniformly 15N- and 15N/13C-labeled DPF3b PHD12 were prepared by growing the bacteria in M9 minimal medium containing 15NH4Cl with or without 13C6-glucose. GST-DPF3b PHD12 protein was purified by glutathione Sepharose column, followed by GST cleavage using thrombin (Haematologic Technologies Inc.) and size–exclusion chromatography on a Superdex75 column (GE Healthcare).
NMR spectroscopy
NMR samples contained 0.5 mM DPF3b PHD12 in complex with a H3K14ac or H3K14 peptide (residues 1–20), or a H4-Nac (residues 1–22) or H4K16ac peptide (residues 9–23) (1:1.2–2 molar ratio) were prepared in the 100 mM sodium phosphate buffer, pH 6.5, containing 2.0 mM DTT in 90% H2O/10% 2H2O. NMR spectra were acquired at 25°C on a Bruker 900, 800, 600, or 500 MHz spectrometer. 1H, 15N and 13C resonance assignments of the PHD12 were determined with triple resonance NMR spectra6 collected with 15N/13C- or 15N-labelled protein bound to a unlabelled H3 or H4 peptide. 1H resonance assignments of the free H3 or H4 peptide were obtained with 2D ROESY and TOCSY spectra. The NOE-derived distance restraints were obtained from 15N- or 13C-edited 3D NOESY data collected using protein/peptide samples in 100% 2H2O. 3JHN,Hα coupling constants measured from 3D HNHA data were used to determine ϕ-angle restraints. Other dihedral angle restraints were predicted from TALOS24 based on resonance assignments and hydrogen bond restraints were derived from secondary structure elements as determined by TALOS. Slowly exchanging amide protons were identified from a series of 2D 15N-HSQC spectra recorded after H2O/2H2O buffer exchange. The intermolecular NOEs of the DPF3b PHD12 bound to the H3 or H4 peptide were detected in 13C-edited (F1), 13C/15N-filtered (F3) 3D NOESY spectra.
Structure calculations
Structures of DPF3b PHD12/H3 or H4 complexes were calculated with a distance geometry-simulated annealing protocol with X-PLOR25. Initial structure calculations were performed with manually assigned NOE-derived distance restraints. Hydrogen-bond distance restraints, generated from the H/D exchange data, were added at a later stage of structure calculations for residues with characteristic NOE patterns. The converged structures were used for the iterative automated NOE assignment by ARIA/CNS for refinement25,26. Structural statistics were calculated by CNS and PROCHECK-NMR27.
Isothermal titration calorimetry (ITC)
Calorimetric experiments were conducted at 25°C with a MCS-ITC instrument from MicroCal (Northampton, MA). The DPF3b PHD12 was prepared in a 25 mM Tris-HCl buffer of pH 7.5 containing 50 mM NaCl and 2 mM DTT. Histone peptides were dissolved in the same buffer adjusted to pH 7.5. Protein concentrations were determined by UV absorbance, whereas peptide concentrations were calibrated in 1D 1H-NMR with DMSO (0.2 mM) in the sodium phosphate buffer. A control ITC measurement was done by a titration of the H3 peptide into the sample buffer. Dissociation constants and enthalpies and standard deviations were derived after subtracting the raw data from the control measurement.
Fluorescence polarization assay
Fluorescent polarization was performed for DPF3b PHD12 binding to a C-terminal fluorescein isothiocyanate (FITC)-labeled H3K14ac peptide (residues 1–18). Protein and peptide concentration was determined as described above. The fluoresceinated peptide concentration was calculated using absorbance spectroscopy with ε494 of 68,000 M−1cm−1. Fluorescent polarization assay was carried out in polypropylene 96-well plates (Costar) with 10 nM FITC-peptide and varying concentrations of the protein in a 50 mM phosphate-buffer (pH 6.5) containing 100 mM NaCl, 1mM imidazol in 100 µL. Measurements were obtained after 10-minute incubation of the peptide and the protein at 25°C using a fluorescence polarization reader (Tecan). Dissociation constant was determined as a one-site model by fitting the binding data with SigmaPlot to an equation of Fb={(Kd+P+R)-[(Kd+P+R)2-4*P*R]1/2}/(2*P), where Fb is fraction of bound labeled ligand, Kd, dissociation constant, R, total protein concentration and P, total fluorescent peptide concentration. Binding affinity of the other H3 and H4 peptides to the PHD12 was evaluated by their competition against FITC-labeled H3K14ac peptide (10 nM) binding to the protein (5 µM) as a function of concentration of a competing peptide.
Site-directed mutagenesis
Mutants of DPF3b PHD12 were generated using QuikChange site-directed mutagenesis kit (Stratagene). The presence of appropriate mutations was confirmed by DNA sequencing. All the mutants were purified as described for the wild type protein.
Re-Chromatin immunoprecipitation (re-ChIP)
DNA fragment encoding full length human DPF3b was amplied from human heart cDNA library, and cloned into pEGFP-C1 vector. C2C12 cells cultured in DMEM medium supplemented with 10% fetal calf serum were transfected with human EGFP-DPF3b28. Chromatin obtained from approximately 6×107 C2C12 cells was cross-linked with formaldehyde. Re-ChIP was conduced as described previously29. Briefly, immunoprecipitation was conducted first with antisera against H3K14ac (Millipore) and then against EGFP agarose (Santa Cruz Biotechnology) to recover EGFP-DPF3b. The immuno-precipitated DNA with enrichment of H3K14ac and DPF3b was analyzed using the HotStart Polymerase kit (Qiagen). PCR products designed to amplify regions specific to the mouse Pitx2 and Jmjd1c promoters, two target genes of DPF3b, were analyzed on 1.8% agarose/TBE gels with ethidium-bromide stain. PCR of the input DNA prior to immunoprecipitation was used as a control to demonstrate fidelity of PCR. For the re-ChIP, complexes were eluted by incubation for 30 min at 37°C in 50 µL 10 mM DTT. After centrifugation, the supernatant was diluted 12 times with dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl at pH 8.1) and subjected to the ChIP procedure. Quantitative PCR experiments were performed in 25-µL reactions using the 96-well microtiter plate and biological samples were analyzed in triplicate. SYBR GREEN (Qiagen) was used as a marker for DNA amplification. Moreover, two non-DPF3b target genes Enpep and Arid5b, which are located approximately 150 Kb to Pitx2 on the long arm of chromosome 3, and more distal to Jmjd1c on the long arm of chromosome 10 in mouse, respectively, were used as negative controls in the sequential qChIP analysis. Finally, DNA sequences of the primers used in qChIP and RT-qPCR for Pitx2 and Jmjd1c, as well as Enpep and Arid5b are provided in Supplemental Table 5.
Immunofluorescence (IF)
IF was performed using an automated high throughput microscope (Leica TCS SP5DM system, Leica Optics) with a procedure described previously30. Antibodies used for IF were identical to those used for chromatin immunoprecipitation analysis. Secondary antibodies conjugated to Texas Red (Santa Cruz Biotechnology) were used for labeling H3K14ac. Image processing was performed using LAS Image Overlay software (Leica) as described in detail in Supplemental Table 4.
Preparation of mouse antisera against NH2-terminal acetylated histone H4 (H4-NAc)
Mice were individually inoculated with 2 mg of an H4-NAc synthetic peptide (Ac-NH2-ARTKQTARKSTGGKAPRKQLA) per mouse and boosted twice. Serum (100 ml) was recovered by submandibular bleeding of mice and serum was titered by ELISA with plated coated with synthetic antigen. Serum was clarified by centrifugation and used for staining.
Biotinylated H3 peptide pull-down
Whole cell extracts from either 75% confluent C2C12 cell cultures, or nuclear extracts from DPF3b plasmid transfected C2C12 cells, were recovered and used in a strepavidin-argarose/biotinylated peptide pull-down assay. Histone H3 peptides used all consist of residues 1–18, and C-terminal biotinylated with no modification, tri-methylation at H3K4 (H3K4me3) or H3K9 (H3K9me3), or acetylation at H3K14 (H3K14ac). Following the peptide pull-down, samples were stringently washed and samples were fractionated by SDS-PAGE and transferred onto filters for Western blot analysis. Immunnoblotting was performed on a single filter first with anti-DFP2 antisera (Aviva Biosystems) and labeled with secondary antisera (AbCam), and then stripped for re-probing for detection of BPTF/FALZ or HP1α with antisera (AbCam) specific for those antigens. Cells transfected with GFP/DPF3b plasmids bearing the wild type or F264A mutation cDNA were prepared as nuclear extracts and used for biotinylated peptide binding study followed by immunoblot analysis with anti-GFP antisera (Santa Cruz Biotech). Filters were exposed to film and signals on the blots were subject to densitometry measurements using NIH’s ImageJ software suite.
Supplementary Material
Acknowledgments
We acknowledge the use of the NMR Facility at the New York Structural Biology Center. We thank S. Hearn of Cold Spring Harbor Lab microscopy facility for help with confocal microscopy study, and A. Buku and G. Gerona-Navarro for technical advice on peptide binding analysis. This work was supported by grants from the National Institutes of Health (M.-M.Z.).
Footnotes
Author Contributions M.J.W. and M.-M.Z. designed the study; Q.Z. generated constructs and protein samples, and contributed to the immunofluorescence study; L.Z. determined the protein structures; A.N.P. contributed to the protein biochemistry study; L.Z. and Q.Z. performed protein and peptide binding study; S.L. and M.J.W. carried out the cell biology experiments; M.-M.Z. supervised the project; and all authors contributed to the manuscript preparation.
Author Information The structural coordinates of human DPF3b PHD12 in complex with the H3K14ac, H3, H4-NAc, or H4K16ac peptide deposited with the Protein Data Bank are under accession codes 2kwj, 2kwk, 2kwn, and 2kwo, respectively. Accessions for the chemical shift assignments of the corresponding NMR structures deposited at the BioMagResBank are 16858, 16859, 16861, and 16865, respectively. Reprints and permissions information is available at www.nature.com/reprints.
Supplementary Information accompanies the paper on www.nature.com/nature.
References
- 1.Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- 2.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Lange M, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22:2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 6.Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- 7.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peña P, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramón-Maiques S, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:8993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1111. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ooi S, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravarty S, Zeng L, Zhou MM. Structure and site-specific recognition of histone H3 by the PHD finger of human autoimmune regulator. Structure. 2009;17:670–679. doi: 10.1016/j.str.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Org T, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 2008;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 16.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 17.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobson R, Ladurner A, King D, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 19.Flanagan J, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, Fang J, Bedford M, Zhang Y, Xu R. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 21.Zeng L, et al. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol. 2008;15:626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phanstiel D, et al. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:4093–4098. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mujtaba S, et al. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 24.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 25.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Rieping W, et al. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 28.Portier GL, Benders AG, Oosterhof A, Veerkamp JH, van Kuppevelt TH. Differentiation markers of mouse C2C12 and rat L6 myogenic cell lines and the effect of the differentiation medium. In Vitro Cell Dev Biol Anim. 1999;35:219–227. doi: 10.1007/s11626-999-0030-8. [DOI] [PubMed] [Google Scholar]
- 29.Furlan-Magaril M, Rincon-Arano H, Recillas-Targa F. Sequential chromatin immunoprecipitation protocol: ChIP-reChIP. Methods Mol Biol. 2009;543:253–266. doi: 10.1007/978-1-60327-015-1_17. [DOI] [PubMed] [Google Scholar]
- 30.Qian C, et al. Structure and chromosomal DNA binding of the SWIRM domain. Nat Struct Mol Biol. 2005;12:1078–1085. doi: 10.1038/nsmb1022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.