Abstract
The third variable domain (V3) of HIV-1 gp120 envelope glycoprotein is critical for HIV-1 entry and represents an attractive target for vaccine design. There are three conserved N-glycans within or around the V3 loop. The N295 and N332 glycans at the base of V3 are usually characterized as high-mannose type in gp120 and the N301 glycan is a complex type. We report in this paper the expression and characterization of glycosylated, full-size V3 domain derived from HIV-1Bal strain as an IgG1-Fc fusion protein, including its binding to two broadly HIV-neutralizing antibodies, 2G12 and 447-52D. It was found that expressing the V3-Fc fusion protein in the HEK293T cells resulted in the production of a glycoform in which all the N-glycans were complex type, in contrast to the glycosylation pattern of V3 in the context of gp120, where the N295 and N332 glycans are high-mannose type. Controlling the glycosylation to restore an epitope of antibody 2G12 was achieved by using an inhibitor of glycan processing enzymes. Mutational studies indicate that the glycan at N301 slightly decreases the binding of V3-Fc to antibody 447-52D, but it can significantly enhance the binding of the V3-Fc to antibody 2G12 when it is changed to a high-mannose type N-glycan. The high-mannose type V3-Fc fusion protein that includes both the 2G12 and 447-52D epitopes represents an interesting immunogen that may be able to raise anti-HIV neutralizing antibodies.
Keywords: HIV-1, V3 domain, glycoforms, IgG-Fc, antibody 2G12, antibody 447-52D
INTRODUCTION
Neutralizing antibodies are an important component of protective immunity against HIV-1 infection (1–3). So far, a few human monoclonal antibodies (MAbs) have been characterized, suggesting the presence of conserved neutralizing epitopes in the HIV-1 envelope glycoproteins. Thus, characterization and reconstitution of the neutralizing epitopes for these broadly neutralizing antibodies constitute an important step for HIV-1 vaccine design. The third variable domain (V3) of HIV-1 gp120 envelope glycoprotein was once considered “the principal neutralization determinant (PND)” for vaccine development (4, 5). While the sequence variation resulted in the production of most anti-V3 antibodies that are strain-specific and neutralize only T-cell line adapted viruses or a very narrow range of primary isolates, there is accumulating evidence showing that broadly reactive anti-V3 antibodies capable of neutralizing HIV-1 primary isolates across clades do exist (2). For example, the anti-V3 monoclonal antibody (mAb) 447-52D is able to neutralize HIV-1 primary isolates of clade A, B, and F, regardless the sequence variability of the V3 loop (6–8). In fact, despite the sequence variations flanking the tip of the loop, V3 domains from different viral strains do share conserved sequence and structural features in order to function as a key determinant in recognizing chemokine coreceptor (CCR5 or CXCR4) during HIV-1 infection (9–12). In addition to those conserved elements, such as a fixed size (30–35 amino acids) of the loop, a conserved disulfide bond at the base, and a β-turn structure at the conserved tip (GPGR or GPGQ) (5, 13), V3 domains from many different strains also carry three conserved N-glycans, within or adjacent to the loop, at the N295, N301, and N332 glycosylation sites (HXB2 numbering, http://hiv-web.lanl.gov). In the context of gp120, the N301 glycosylation site within the V3 loop usually carries a complex type N-glycan, while the N295 and N332 sites at the base normally bear high-mannose type N-glycans (14, 15). The two high-mannose type N-glycans at N295 and N332 sites are particularly interesting, as they were proposed to be an essential component of the epitope of the broadly neutralizing antibody 2G12 (16, 17). The human mAb 2G12 is the only broadly neutralizing antibody so far discovered that recognizes a novel cluster of high-mannose type N-glycans with terminal Manα1,2Man motifs in gp120 (16–19).
As part of our research program on HIV-1 vaccine, we are interested in reconstituting a V3 domain immunogen that integrates both the carbohydrate and peptide epitopes of the broadly neutralizing antibodies 2G12 and 447-52D. V3 domain has been previously expressed in the context of different fusion proteins for biochemical and immunological studies (20–24). But most of the V3 fusion proteins previously described were not glycosylated, as they were usually overproduced in E. coli that lacks protein glycosylation machinery (20–23). A large outdomain (aa 251–481) of a clade C gp120, which consists of V3 and other domains (C3, V4, C4, V5, and C5), was expressed in baculovirus infected insect cells as a glycosylated Fc-fusion protein (24). This study confirms the importance of the N-glycans around the V3 domain for 2G12 recognition, but the nature of the glycans (complex type vs. high-mannose type) attached to V3 and other domains in the recombinant protein is yet to be characterized. We describe in this paper the expression of a glycosylated V3 domain as an IgG1-Fc fusion protein in the human embryonic kidney 293T (HEK293T) cell line. A V3 domain corresponding to the sequence aa291–336 (46 amino acid residues) of HIV-1Bal gp120 was chosen, which was fused to the Cterminus of the human IgG1-Fc domain (232 amino acid residues). The resulting V3-Fc fusion protein contains four glycosylation sites: three (N295, N301, and N332) are in the V3 domain and one is in the Fc domain (Fc-N297) (Figure 1). The advantage of making the IgG1-Fc fusion protein was several fold. In addition to the easiness of purification of the Fc-tagged fusion protein via protein A affinity chromatography, fusing the small V3 polypeptide to the Fc is likely to stabilize the resulting recombinant protein and prevent degradation, as previously demonstrated for the Fc-fused gp41 ectodomain (25). Moreover, it has been suggested that the Fc domain could enhance the immunogenicity of the Fc-fusion protein, by facilitating the uptake of antigens via Fc-mediated interactions with Fcγ receptors on antigen presenting cells (24–26). We report here the expression, characterization, and antibody-binding of different glycoforms of the recombinant V3-Fc fusion protein. Our results suggest that the high-mannose type V3-Fc fusion protein, which constitutes the epitopes of both the neutralizing antibody 2G12 and 447-52D, may be a valuable immunogen for raising broadly neutralizing antibodies against HIV-1.
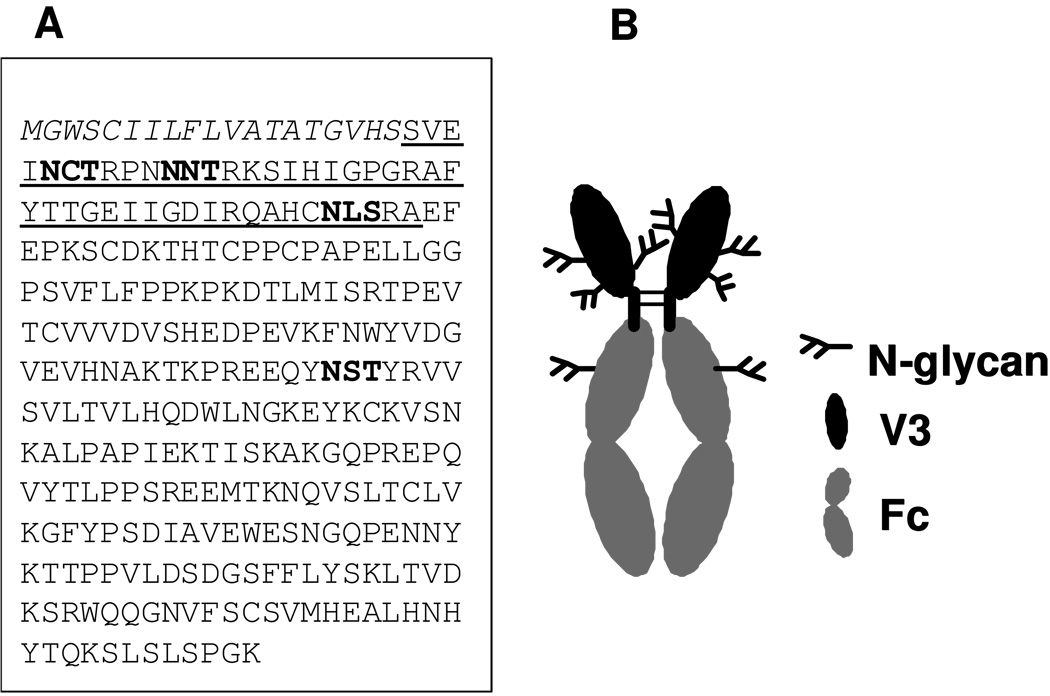
Fig. 1. Schematic presentation of the glycosylated V3-Fc fusion protein.
(A) the amino acid sequence of the V3-Fc fusion protein. The signal peptide sequence was in italic, the V3 domain was underlined, and the glycosylation sites were in bold. (B) a homodimer of V3-Fc, in which the N-glycans at the N295 and N332 glycosylation sites in V3 domain are expected to be high-mannose type (HM), the glycan at N301 site in V3 domain is expected to be complex type (CT), and the N-glycan at the Fc region is expected to be complex type (CT).
EXPERIMENTAL PROCEDURES
Strains, cell line, media, and enzymes
Escherichia coli DH5α was used for all molecular cloning steps. It was grown in LB broth (1% Casein Peptone, 0.5% yeast extract, and 1% NaCl). Human HEK293T cell line was used for the expression of V3-Fc fusion protein. It was cultured in DMEM medium supplemented with 10% low IgG fetal bovine serum (HyClone, Logan, UT) and 12.5 mM HEPES buffer.
Construction of pcDNA-LV3Fc expression vector and site-directed mutagenesis
A 46 amino acid V3 domain was chosen from the sequence (aa 291–336, HXB2 numbering) of the gp120 derived from the HIV-1 BAL strain (R5 tropic). This domain includes the two glycosylation sites at the base of the V3 loop. The fusion protein was arranged by adding the Fc domain of human IgG1 to the C-terminus of the V3 domain. The human CD5 antigen leader sequence (27) was added to the N-terminus of fusion protein. The whole fusion construct was assembled in pGEM-T vector (Promega, Madison, WI). First, the coding fragments for human IgG1-Fc, V3, and CD5 leader were amplified by PCR with respective primer pairs and cloned into pGEM-T vector. The Fc coding fragment was amplified by a primer pair, (forward) CGA ATT CGA GCC CAA ATC TTG TGA C, (reverse) GTA GCG GCC GCT TAT TTA CCC GGG GAC AGG GAG. V3 fragment was amplified by the following primers, (forward) TCT CGA GCG TGG AGA TCA ACT GCA C and (reverse) TGA ATT CGG CGC GGC TCA GGT TGC. CD5 leader was amplified with (forward) CGG TAC CAT GGG ATG GTC ATG TAT CAT C and (reverse) GCT CGA GTG TAC ACC GGT TGC AGT. Each fragment was inserted into the pGEM-T vector, resulting in three plasmids: pT-Fc, pT-V3, and pT-CD5. To assemble the final expression construct, the Fc fragment was released from pT-Fc with EcoR I and Not I and cloned into corresponding sites of pT-V3, resulting in pT-V3Fc. Subsequently, V3Fc fragment was released with Xho I and Not I from the pT-V3Fc and cloned into corresponding site of pT-L, resulting in pT-LV3Fc. Finally, the assembled LV3Fc fragment was released from pT-LV3Fc and cloned into Kpn I and Not I site of mammalian expression vector pcDNA3.1+ (Invitrogen, Carlsbad, CA), resulting in the expression vector pcDNA-LV3Fc. To generate the single mutants of N-glycosylation sites in fusion protein, site-directed mutagenesis was carried out by using a commercial site-directed mutagenesis kit (QuickChange, Stratagene). All mutants were confirmed by DNA sequencing.
Transfection and expression of V3-Fc in HEK293T cells
The phenol-free pcDNALV3Fc plasmid for transfection was prepared with HiSpeed Plasmid Midi Kit or QIAfilter Maxi Kit (Qiagen). Transient transfection was performed using Fugene6 transfection reagent (Roche) following the manufacture’s protocol. After transfection, the expression of fusion protein was periodically monitored using an anti-human IgG1 Fc ELISA assay kit (Human IgG ELISA Quantitation Kit, BETHYL Laboratories Inc, Montgomery TX). Complex type V3-Fc was expressed without the addition of mannosidase inhibitor. For generation of HM type V3-Fc, kifunensine (Caltech) -the mannosidase inhibitor- was added into the culture system immediately after the transfection.
Purification of the fusion glycoprotein
The culture supernatant was collected 4 days after transfection. The fusion protein was purified by affinity chromatography using a protein Aagarose resin (Pierce). The cell-free culture supernatant was loaded on the protein A column pre-equilibrated with the Binding Buffer (pH8.0, Thermo Scientific, Rockford, IL) After extensive wash with the same buffer, V3-Fc was eluted with IgG Elution Buffer (pH3.0, Thermo Scientific, Rockford, IL). Each elution fraction was immediately neutralized with 1M Tris-HCl, pH 8.8. Subsequently, all fractions were combined and concentrated with centrifugal filtration (Amicon Ultra centrifugal filter, Millipore, Billerica, MA). The integrity and purity of protein was checked by SDS-PAGE. The protein A column was regenerated with the elution buffer.
Size exclusion chromatography (SEC)
The SEC chromatography was performed on an AKTA FPLC system (GE Healthcare) at 4 °C. The purified V3-Fc was loaded on a Superdex 200 10/300 GL size exclusion column (GE Healthcare). The column was eluted with a buffer containing 150 mM NaCl and 20 mM Tris-HCl (pH 8.0) at a flow rate of 0.5 mL/min. The sizes of proteins were estimated by using thyroglobulin (670 kDa) γ-globulin (158 kDa), ovalbumin (44 kDa), and myoglobin (17 kDa) as standards.
Enzymatic de-glycosylation
Purified V3-Fc proteins in various glycoforms were deglycosylated by different glycosidase treatment. PNGase F and Endo H (New England Biolabs) treatments were performed according to the manufacture’s instruction, respectively. Deglycosyaltion of high mannose type V3-Fc was carried out by incubation of the V3-Fc fusion protein with endo-β-N-acetylglucosaminidase from Arthrobacter (Endo-A) (1/100, by weight) in a 25 mM Tris-Cl buffer (pH 8.0) at 37 °C for 2 h. Endo-A was expressed according to the reported method (28).
MALDI-TOF mass spectrometry
For the MALDI-TOF MS analysis of N-glycans, the total N-glycans of V3-Fc were released from 60 µg glycoprotein by 10 U of PNGase F with incubation at 37 °C for 48 h. The reaction mixture was then treated with 10 mU neuraminidase (from Clostridium perfringens, Sigma) and incubation at 37 °C for 2 h for de-sialylation. The released N-glycans were separated with centrifuge filtration (Microcon centrigufal filter, 10 KDa cut-off, Millipore), desalted by treatment with anion-exchange resin (DOWEX 1X2-100, Sigma-Aldrich) and cation-exchange resin (DOWEX 50WX8-100, Sigma-Aldrich), and finally lyophilized. The N-glycans were analyzed using a Bruker Autoflex II linear MALDI-TOF mass spectrometer (Bruker Daltonics) with 2,5-dihydroxybenzoic acid (DHB) as the matrix. The MALDI-TOF analysis of V3-Fc fusion protein was performed with the same machine, with sinapinic acid as matrix.
Surface plasmon resonance (SPR) measurement
The interactions between the glycoforms of V3-Fc fusion protein and antibodies 2G12 and 447-52D were evaluated using a BIAcore T100 system (GE Healthcare, USA). Monoclonal antibodies 2G12 and 447-52D (NIH AIDS Research& Reference reagent program) were immobilized respectively on CM5 sensor chips in an acetate buffer (10 mM, pH 5.0) by use of the amine coupling kit provided by the manufacturer, until the targeted response unit (RU) was reached. The reaction was quenched by the addition of ethanolamine HCl (1 M, pH 8.5). A reference cell without the antibody was prepared by similar procedures. Analyses were performed at 25 °C by injecting a solution of the respective V3-Fc glycoforms in a HBS-P buffer (10 mM HEPES, 150 mM NaCl, and 0.05% surfactant P20, pH 7.4) at a flow rate of 30 µL/min for 2min, followed by wash for 5 min with HBS-P buffer for dissociation. The sensor surface was regenerated through a two-step wash (1st wash, 3.5M MgCl2 for 0.5 min; 2nd wash, HBS-P for 1 min). Data processing was carried out using the BIAcore T100 software.
RESULTS & DISCUSSION
Expression, purification and characterization of the V3-Fc fusion protein
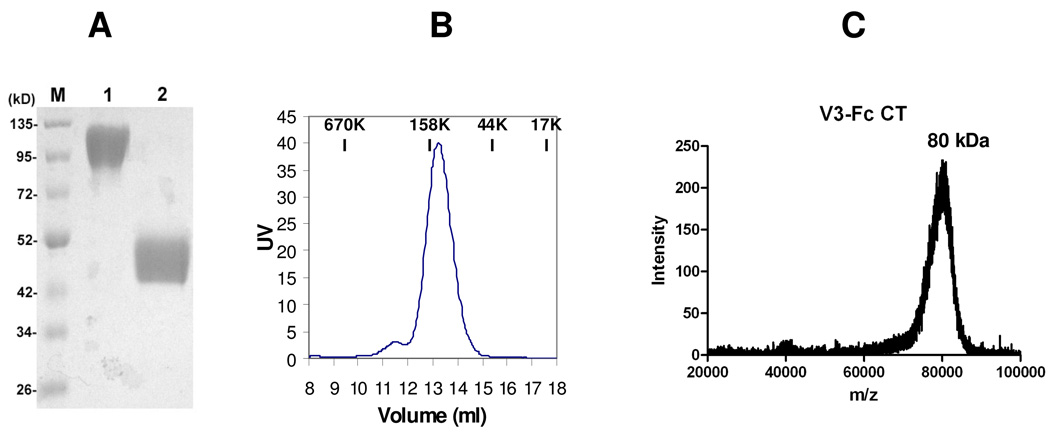
The expression vector encoding the fusion protein was introduced into HEK293T cells by transient transfection. The culture supernatant was collected 4 days post transfection and the V3-Fc fusion protein was purified with protein A affinity chromatography. The fusion protein appeared as a broad band at ca. 49 kDa under reduced conditions (Figure 2A, lane 2) on SDS-PAGE. The broad band implicates a significant heterogeneity in the N-glycans attached. Under non-reducing (native) conditions, the fusion protein migrated on SDS-PAGE as a broad band at 110 kDa (Figure 2A, lane 1). These results suggest that the fusion protein is present as a homodimer in aqueous solution. This is expected, as the hinge-containing IgG-Fc turns to form a covalently linked homodimer through the formation of two pairs of disulfide bonds (29). The dimer format of the fusion protein under native conditions was further confirmed by size-exclusion chromatography. The V3-Fc fusion protein was eluted as a peak at ca. 110 kDa, suggesting that it exists as a homodimer (Figure 2B). However, it was observed that the apparent molecular weight of the fusion protein was significantly bigger than the calculated molecular weight. Based on the assumption that the V3-Fc fusion protein (31 kDa for the protein backbone) carries 4 N-glycans and each N-glycan has an average molecular weight of 2 kDa, the estimated molecular weight should be around 39 kDa. The apparent molecular size (ca. 49 kDa) of the monomeric fusion-protein on SDS-PAGE was clearly larger than the calculated data. The homodimer also showed a much larger size on SDS-PADE (under native condition) as well as in size-exclusion chromatography. One possible explanation is that the heavily glycosylated protein might have an extensive hydration to give a large hydration volume. To verify the actual molecular mass, the native V3-Fc fusion protein was subject to MALDI-TOF MS analysis (Figure 2C). While the fusion protein appeared as a broad signal, indicating the heterogeneity of the attached N-glycans, the average molecular mass is ca. 80 kDa, which is consistent with the expected molecular weight of the heterogeneous dimeric V3-Fc glycoprotein (78–82 kDa).
Fig. 2. Analysis of the expressed V3-Fc fusion protein.
(A) SDS-PAGE of the V3-Fc fusion protein under nonreducing (lane 1) and reducing conditions (lane 2); (B) Gel-filtration chromatography of the V3-Fc under native condition; (C) MALDI-TOF MS analysis of the native V3-Fc.
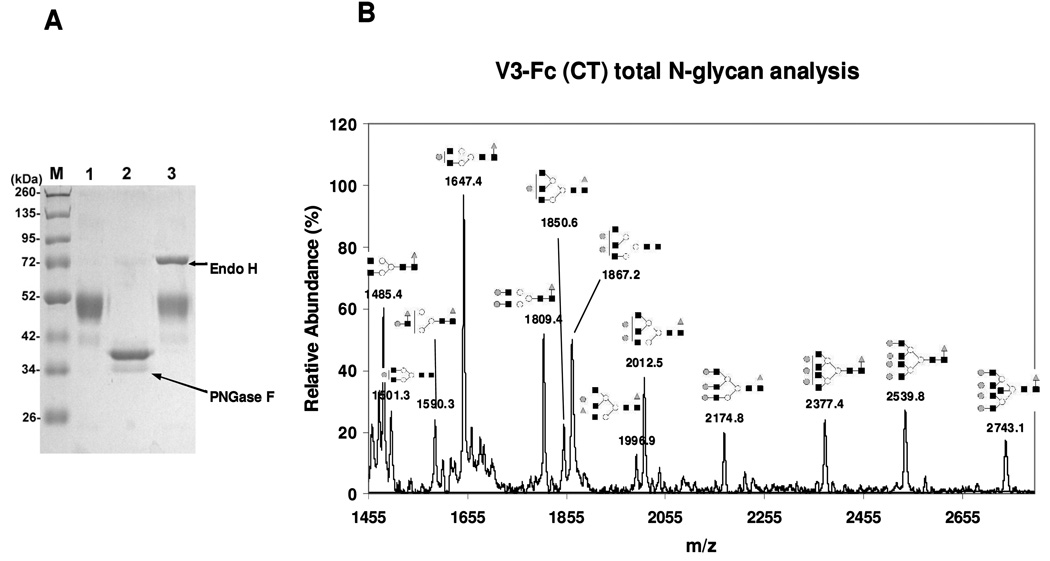
To characterize the N-glycosylation status of the V3-Fc, the fusion protein was treated with PNGase F, which would remove all N-glycans attached. SDS-PAGE showed that the de-glycosylated V3-Fc appeared as a sharp band with a size of ca. 38 kDa (Figure 3A, lane 2). A 11-kDa difference between the glycosylated V3-Fc (ca. 49 kDa) and de-glycosylated V3-Fc (ca. 38 kDa) suggests that the monomeric V3-Fc carries at least four N-glycans. Thus, all the glycosylation sites (3 in the V3 domain and 1 in the Fc domain) in the expressed fusion protein should be glycosylated. Treatment of the fusion protein with Endo-H, an endo-glycosidase that cleaves only high-mannose type or hybrid type N-glycans did not cause any change in the molecular size of the protein (comparing lane 3 and lane 1, Figure 3A). These results suggest that the V3-Fc fusion protein does not contain any high-mannose type or hybrid type N-glycans.
Fig. 3. Characterization of the glycan profile of complex type fusion protein V3-Fc CT.
(A) SDS-PAGE of the fusion proteins before and after enzymatic treatment (Lane 1, untreated V3-Fc; lane 2, treated by PNGase F; lane 3, treated by Endo H); (B) MALDI-TOF MS analysis of total N-glycans released by PNGase F.
To validate the glycan profiles of the expressed fusion protein, the total N-glycans were released by PNGase F treatment. The released N-glycans were subjected to MALDI-TOF MS analysis after desialylation by sialidase, which simplifies the profiles of complex type N-glycans and also enhances the detection sensitivity. It was found that all the N-glycans in the V3-Fc fusion protein were complex type composed of bi-, tri-, and tetra-antennary complex N-glycans, and no high-mannose type or hybrid type sugars were present (Figure 3B). The found molecular mass and proposed structures of the released N-glycans were listed in Table 1. These MS analysis data confirmed the results from the Endo-H treatment. This is an interesting observation. In HIV-1 gp120, the two conserved N-glycans at the base of the V3 loop, N295 and N332, are usually high-mannose type, and the one within the V3 loop (N301) is conserved as a complex type N-glycan (14, 15). We have shown here that when the V3 domain was expressed in the context of Fc-fusion protein, all the N-glycans were processed to form complex type sugars. These results suggest that the two N-glycans at N295 and N332 might be inaccessible for further processing by ER and Golgi α-mannosidases in the context of gp120 but, in the Fc-fusion protein, the N-glycans at the two sites are more exposed, so that further trimming and modifications could be achieved to form complex type sugars. These results also suggest that the microenvironment of the N-glycosylation sites e.g., the steric hindrance around the sites due to the presence of other proximal domains, is critical for determining the nature of the N-glycans attached. This glycoform of the V3-Fc fusion protein, in which all the N-glycans are complex type, is named as V3-Fc-CT.
Table 1.
Molecular mass and proposed structures of the desialylated N-glycans released from the complex type V3-Fc
| Calculated (M+Na+) |
Found (M+Na+) |
Composition | Suggested structures |
|---|---|---|---|
| 1485.5 | 1485.2 | Hex3HexNAc4dHex1 | |
| 1501.5 | 1501.1 | Hex4HexNAc4 | |
| 1590.6 | 1590.3 | Hex4HexNAc3dHex2 | |
| 1647.6 | 1647.2 | Hex4HexNAc4dHex1 | |
| 1809.6 | 1809.0 | Hex5HexNAc4dHex1 | |
| 1850.7 | 1849.7 | Hex4HexNAc5dHex1 | 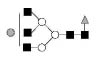 |
| 1866.7 | 1866.3 | Hex5HexNAc5 | 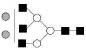 |
| 1996.7 | 1996.9 | Hex4HexNAc5dHex2 | 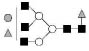 |
| 2012.7 | 2012.1 | Hex5HexNAc5dHex1 | 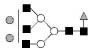 |
| 2174.8 | 2174.0 | Hex6HexNAc5dHex1 | 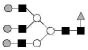 |
| 2377.9 | 2377.2 | Hex6HexNAc6dHex1 | 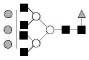 |
| 2539.9 | 2538.9 | Hex7HexNAc6dHex1 | 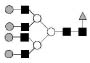 |
| 2743.0 | 2741.9 | Hex7HexNAc7dHex1 | 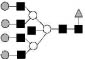 |
Note:  fucose,
fucose,  galactose, ■ N-acetylglucosamine, ○ mannose, Hex, hexose; HexNAc, N-acetylhexosamine; dHex, monodeoxy-hexose
galactose, ■ N-acetylglucosamine, ○ mannose, Hex, hexose; HexNAc, N-acetylhexosamine; dHex, monodeoxy-hexose
Expression and characterization of high-mannose glycoforms of V3-Fc and its selected N-glycosylation mutants
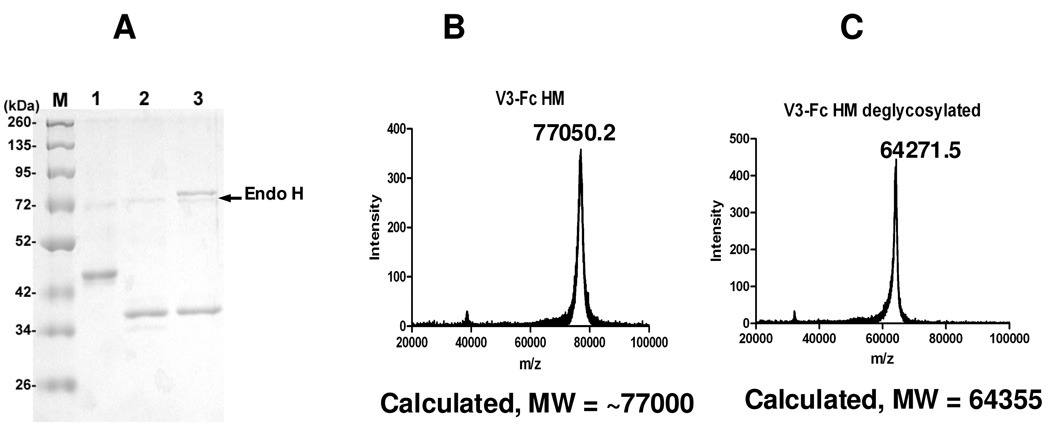
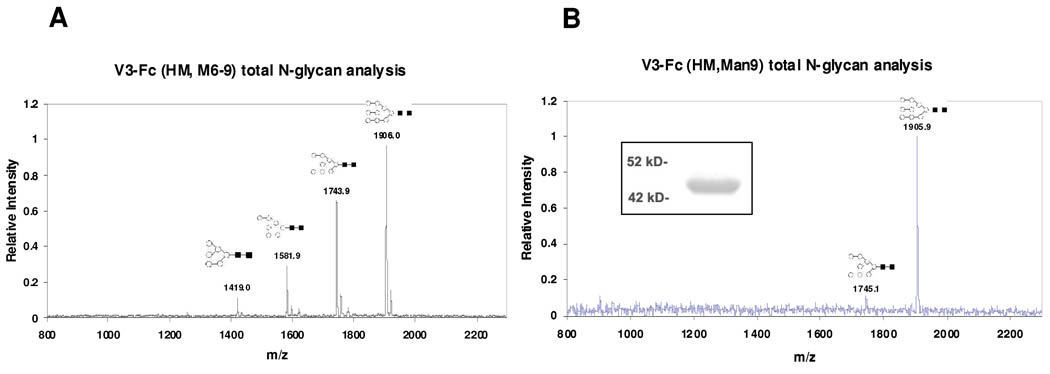
It is known that antibody 2G12 recognizes a cluster of high-mannose type N-glycans around the V3 domain, and the N-glycans at the N295 and N332 sites of HIV-1 gp120 are the essential components for the epitope (16, 17, 30, 31). The unexpected results that all the N-glycans attached to the V3-Fc fusion protein are complex type sugars when expressed in the HEK293 cell line prompted us to restore high-mannose type N-glycans in the V3-Fc fusion protein by controlling the glycan processing with a specific glycosidase inhibitor. It was previously reported that expression of glycoproteins in HEK293 cell lines in the presence of kifunensine, a potent α-mannosidase inhibitor, could efficiently control the glycosylation at the high-mannose type stage, thus leading to the formation of high-mannose glycoforms of recombinant glycoproteins (32–34). In particular, Scanlan and co-workers have successfully expressed HIV-1IIIB gp120 in CHO cell line in the presence of kifunensine, which produced a novel glycoform of HIV-1IIIB gp120 that carries high-mannose type sugars (33). Thus, we expressed the V3-Fc fusion protein in the HEK293T culture system in the presence of kifunensine (2µg/ml). The expressed fusion protein, named as V3-Fc-HM, appeared at ca. 95 kDa in SDS-PAGE under non-reducing condition (Data not shown) and at ca. 45 kDa under reduced condition as a clear band (Figure 4A, lane 1). This result indicates that the fusion protein expressed is a homodimer. The dimeric form of the fusion protein was also confirmed by size-exclusion chromatography, which appeared as a single peak larger than 90 kDa (Data not shown). In contrast to the complex type fusion protein V3-Fc-CT that appeared as a broad band in SDS-PAGE, the high-mannose type fusion protein V3-Fc HM appeared as a much sharper band, suggesting that the glycans attached are more uniformed. Removal of the V3-Fc-HM by PNGase F gave the de-glycosylated fusion protein that appeared as a single band at 38 kDa (Figure 4A, lane 2), which is at the same size of the de-glycosylated fusion protein from V3-Fc-CT. In addition, treatment of V3-Fc-HM with Endo-H resulted in a single band in SDS-PAGE (Figure 4A, lane 3) that had a similar size of the PNGase-F treated protein. This result indicates that all the glycans attached in V3-Fc-HM are Endo-H-sensitive, high-mannose type N-glycans. The MALDI-TOF MS of the V3-Fc-HM (Figure 4B) showed an average molecular mass of 77050 Da, which is consistent with a V3-Fc homodimer carrying 4 high-mannose N-glycans (calculated, M = 77000 Da, for the Man8GlcNAc2 glycofrom). The observed m/z data from the MALDI-TOF MS of the Endo-H deglycosylated fusion protein (Figure 4C) is also in agreement with the calculated molecular weight of the deglycosylated protein (observed, 64271 Da, calculated, M = 64355 Da). Finally the N-glycans released from the V3-Fc-HM by PNGase F was analyzed by MS. It was found that the majority of glycoforms was Man9GlcNAc2 and Man8GlcNAc2, together with minor fractions of Man7GlcNAc2 and Man6GlcNAc2 glycoforms (Figure 5A and Table 2). Thus, the use of 2 µg/ml of kifunensine in the expression system could efficiently control the glycosylation processing at the high-mannose stage. To produce a homogeneous high-mannose type glycoform of V3-Fc, we increased the concentration of kifunensine in the HEK293 expression system to 10 µg/ml. Interestingly, we found that the fusion protein obtained under this condition was almost the Man9GlcNAc2 glycoform, with only a minor amount of the Man8GlcNAc2 (Figure 5B). No glycans smaller than Man8GlcNAc2 were formed under this expression condition. Thus, a new glycoform, V3-Fc-Man9 was obtained which appeared as a homogeneous glycoprotein as indicated by SDS-PADE (Figure 5B, inlet).
Fig. 4. Characterization of the fusion protein V3-Fc expressed in the presence of α-mannosidase inhibitor kifunensine (2 µg/ml).
(A) SDS-PAGE of the fusion proteins before and after de-glycosylation (Lane 1, untreated V3-Fc; lane 2, treated by PNGase F; lane 3, treated by Endo H); (B) MALDI-TOF MS analysis of intact fusion protein as a homodimer; (C) MALDI-TOF MS analysis of intact fusion protein after de-glycosylation with Endo-H.
Fig. 5. The MALDI-TOF MS profiles of the N-glycans released from the HM type V3-Fc by PNGase F treatment.
(A) N-glycans from the V3-Fc fusion protein expressed in the presence of kifunensine at 2µg/ml; (B) N-glycans from the V3-Fc fusion protein expressed in the presence of kifunensine at 10 µg/ml. Inlet: SDS-PAGE of V3-Fc expressed in the presence of kifunensine at 10 µg/ml.
Table 2.
Molecular mass and proposed structures of the high-mannose type N-glycans released from the HM-V3-Fc
| Calculated (M+Na+) |
Found (M+Na+) |
Composition | Suggested structures |
|---|---|---|---|
| 1419.5 | 1418.9 | Hex6HexNAc2 | 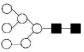 |
| 1581.5 | 1581.8 | Hex7HexNAc2 | |
| 1743.6 | 1743.9 | Hex8HexNAc2 | |
| 1905.6 | 1905.9 | Hex9HexNAc2 |
Note: ■ N-acetylglucosamine, ○ mannose; Hex, hexose; HexNAc, N-acetylhexosamine
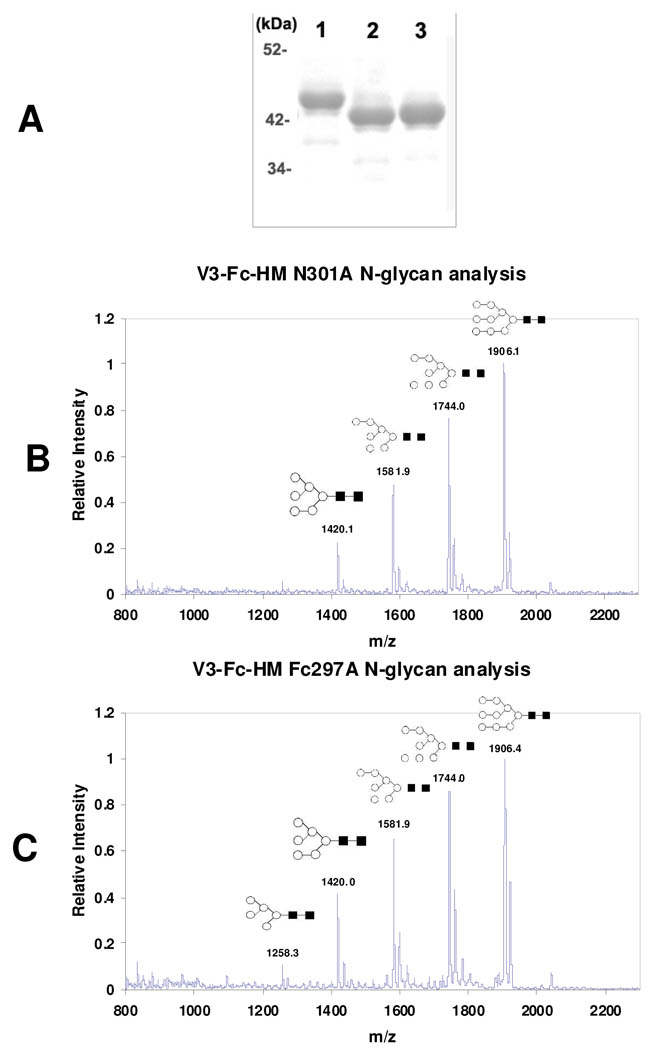
We also produced two mutants, N301A and Fc-N297A, in which the N301 glycosylation site in the V3 and the N297 site in the Fc domain were deleted, respectively. Both mutants were expressed in HEK293T cells in the presence of 2 µg/ml of kifunensine to provide the high-mannose type glycoforms. SDS-PAGE analysis indicated that the two mutants were about 2 kDa smaller in size than that of the high-mannose glycoform of the wild type fusion protein (V3-Fc-HM) (Figure 6A, lane 1, V3-Fc-HM; lane 2, N301A; lane 3, Fc-N297A). These data suggest the deletion of one N-glycan in the two mutants. The N-glycan profiles of the two mutants were shown in Figure 6B and Figure 6C, respectively. Both have very similar profiles as that of the V3-Fc-HM, with Man9GlcNAc2 and Man8GlcNAc2 as the major glycoforms.
Fig. 6. Characterization of V3-Fc mutants and their glycoprofiles.
(A) SDS-PAGE analysis of V3-Fc mutants. Lane 1: Wild type V3-Fc HM); Lane 2: V3-Fc N301A; Lane 3: V3-Fc Fc-N297A. (B) and (C): MALDI-TOF MS analysis of PNGase F released N-glycan from N301A and Fc-N297A mutants, respectively.
Binding of V3-Fc glycoforms with broadly neutralizing antibody 2G12
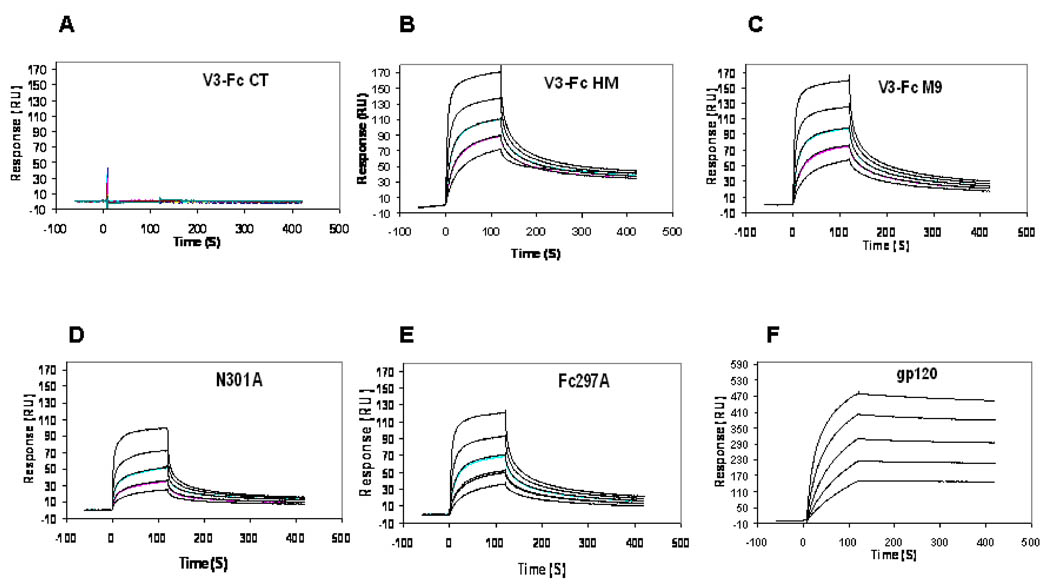
Monoclonal antibody 2G12 is a broadly HIV-neutralizing antibody that recognizes a cluster of high-mannose type N-glycans in HIV-1 gp120 (16–18, 30). We measured the interactions between 2G12 and the various glycoforms of V3-Fc with Biacore surface plasmon resonance (SPR) technology. 2G12 was immobilized on the SPR chip and its real-time association and dissociation with V3-Fc at various concentrations was examined. It was observed that the complex type glycoform V3-Fc-CT did not show measurable interaction with 2G12 (Figure 7A). This result was expected, as 2G12 was previously shown to recognize only those N-glycans that bear terminal Manα1,2Man motifs (16–19). In contrast, the high-mannose type glycoforms of V3-Fc demonstrated significant affinity to 2G12. The binding profiles of the V3-Fc-HM, V3-Fc-M9, and the two mutants (N301A and Fc-N297A) were shown in Figures 7B–7E, respectively. The binding profile of gp120 derived from the HIV-1Bal strain was shown in Figure 7F. It was found that the binding of antibody 2G12 to the four high-mannose glycoforms of V3-Fc fusion protein (including the two mutants) followed a quick association/dissociation kinetic process. Unfortunately, an attempt to fit the SPR data to possible kinetic equations failed to provide an accurate dissociation constant (Kd), making it difficult to have a quantitative comparison of the binding of 2G12 with different V3-Fc glycoforms. Nevertheless, a comparison of the sensorgrams of the various glycoforms indicated that the V3-Fc-HM and V3-Fc-M9 had a comparable affinity to 2G12 and followed a very similar kinetic profile (Figure 7B vs. Figure 7C). Since both glycoforms carry Man9GlcNAc2 and Man8GlcNAc2 as the major N-glycans, these results are consistent with previously observations that Man9GlcNAc2 and Man8GlcNAc2 interacted with 2G12 equally efficiently through the D1 arm contact (18, 19). Interestingly, removal of the N-glycan at N301 site significantly reduced the affinity to antibody 2G12 (Figure 7D). In native HIV-1 gp120, the glycan at the N301 site of V3 is a conserved complex type sugar that did not positively contribute to the binding of 2G12 (14, 15). Thus, changing this N-glycan to a high-mannose type glycan seemed to create a new epitope (likely a cluster of the three Nglycans at N295, N301, and N332) for 2G12 that have a much higher affinity to 2G12 than the epitope formed only by the two glycans at N295 and N332. As to the Fc glycosylation, we also observed that removal of the N-glycan at the Fc domain resulted in the reduction of the binding of the resulting mutant (Fc-N297A) to 2G12 (Figure 7E), albeit to a less extent than the N301A mutant. This result implicated that the Fc domain N-glycan was also involved in the interaction of the V3-Fc fusion protein with 2G12. In contrast to those recombinant V3-Fc fusion proteins, the dissociation between gp120 and 2G12 was found to be extremely slow (Figure 7F), although its association rate was comparable to those of the V3-Fc fusion proteins (Figure 7F). A typical HIV-1 gp120 carries 24 N-glycans, among which 11 are high-mannose type and 13 are complex type N-glycans (14, 15). The extremely slow dissociation rate for the gp120-2G12 interaction suggests that other key elements, e.g., some protein domains or additional high-mannose type N-glycans in the context of gp120, might be also involved in subsequent interactions to strengthen the binding, after initiate recognition with gp120. The difficulty to fit the binding data to any appropriate kinetic equations also implicates a complex process for the 2G12-glycan interactions. It should be pointed out that although the high-mannose N-glycans at the N295 and N332 sites in gp120 is essential for 2G12 recognition, the precise epitope of antibody 2G12 is still not fully characterized.
Fig. 7. SPR sensorgram of the binding of different glycoforms of V3-Fc and its mutants with immobilized 2G12 antibody.
V3-Fc was applied to analysis with a serial concentration of 2 fold dilution, with the highest concentration set at 2 µM. The interaction between gp120 and immobilized 2G12 was analyzed in a similar manner, except that the highest concentration of gp120 was set at 1 µM.
Binding of V3-Fc glycoforms with broadly neutralizing antibody 44-52D
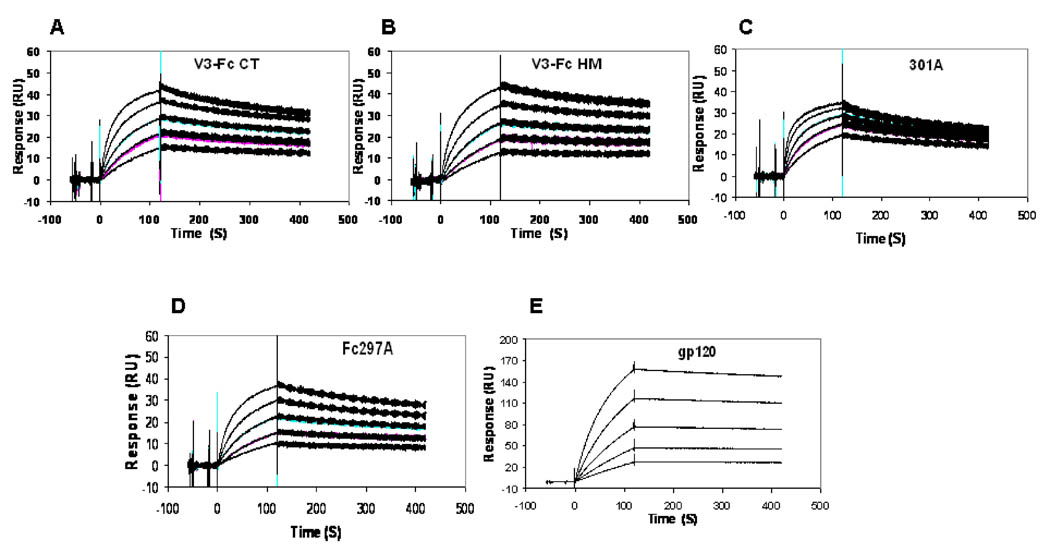
Monoclonal antibody 447-52D is another broadly neutralizing antibody that is specific for the V3 loop. It is a conformation-sensitive antibody. X-ray structural studies indicated that 447-52D Fab recognized the V3 peptide by binding to two domains: the conserved β-turn at the tip (GPGR) of the V3 loop and the peptide amide backbone, rather than the variable side chains of the surrounding sequence (35). Although extensive structural and biochemical studies have been performed for the interactions between 447-52D and various V3 antigens, it is hitherto not clear how the conserved N-glycans within or around the V3 loop affects the antibody-epitope interaction. This is partly attributed to the difficulty to obtain structurally defined, glycosylated full-size V3 domains. To examine the effects of glycosylation on the antibody binding, we tested the interaction of different glycoforms of V3-Fc with antibody 447-52D using SPR technology. Interestingly, both the complex type (V3-Fc-CT) and the high-mannose type glycoforms (V3-Fc-HM) of the fusion protein could bind 447-52D with a high-affinity and with a very similar association/dissociation pattern (Figure 8A and 8B). Fitting of the binding data to a steady-state mode equation gave an estimate Kd of 46 nM and 72 nM for the V3-Fc-CT and V3-Fc-HM, respectively. The data suggest that the nature of the glycosylations at the N295, N301, and N332 sites did not significantly affect the recognition of the antibody to its epitope. On the other hand, the binding of the N301A and F-N297A mutants with 447-52D was assessed by SPR (Figure 8C and Figure 8D) and the dissociation constants (Kd) for N301A and Fc-N297A were estimated to be 21 nM and 67 nM, respectively. Interestingly, the N301A mutant (Kd = 21 nM) showed a 2-fold increase in affinity to 447-52D in comparison with the V3-Fc-CT (Kd = 46 nM), while the Fc-N297A (Kd = 67 nM) had essentially the same affinity as V3-Fc-HM (Kd = 72 nM). These data suggest that an N-glycan at N301 might partially hinder the interaction between antibody 447-52D and the V3 antigen. Nevertheless, All the glycoforms of V3-Fc could still bind antibody 447-52D with very high-affinity at nM concentrations. The affinity of the glycosylated V3-Fc with 447-52D is comparable to that of the non-glycosylated V3 inserted at the surface loop location of E. coli thioredoxin (22). For comparison, we measured the binding of HIV-1Bal gp120 to 447-52D (Figure 8E). The Kd was estimated to be 23 nM. Taken together, these data suggest that the nature and the presence of the three conserved N-glycans at N295, N301, and N332 sites that are near the epitope do not significantly affect the antibody recognition of its epitope, although deletion of the N-glycan at N301 does increase the affinity to some extent. Moreover, the comparable binding affinity among the various Fc- fusion proteins and the HIV-1 gp120 suggest that the fusion protein could present a similar conformational epitope of 447-52D as that existing in native gp120. Thus, the high-mannose type fusion proteins (V3-Fc-HM and V3-Fc-M9), which include both the epitopes of 2G12 and 447-52D, could represent an interesting immunogen that may be used to elicit neutralizing antibodies against HIV-1.
Fig. 8. SPR sensorgram of the binding of different glycoforms of V3-Fc with immobilized 447-52D antibody.
Both V3-Fc and gp120 were applied to the analysis with a serial concentration of 2 fold dilution. The highest concentration was set at 200 nM.
CONCLUSION
Several glycoforms of the V3-Fc fusion protein were successfully expressed and characterized. Our studies demonstrate that the nature of the V3 glycosylation depends on the protein context where the V3 domain is present. Antibody binding studies revealed that the nature and presence of the glycan at N301 within the V3 loop did not significantly affect the recognition of antibody 447-52D with the V3 domain. On the other hand, controlling the glycosylation at the three sites (N295, N301, and N332) at high-mannose type N-glycans could partially reconstitute the epitope of antibody 2G12. In particular, mutational studies indicate that installing a high-mannose type N-glycan at the N301, which is usually occupied by a complex type N-glycan in gp120, significantly enhances the binding to 2G12. The resulting high-mannose type V3-Fc fusion protein that includes both the 2G12 and 447-52D epitopes thus represents an interesting immunogen that may be able to raise neutralizing antibodies against HIV-1.
ACKNOWLEDGEMENT
We thank Drs. Anthony L. DeVico, George K. Lewis, and Yongjun Guan for helpful discussions during the work. The human antibodies 2G12 and 447-52D were provided by the National Institutes of Health (NIH)’s AIDS Research and Reference Reagent Program. This work was supported by the National Institutes of Health (NIH grant R01 AI067111).
LITERATURE CITED
- 1.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 2.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 2008;6:143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 4.Ghiara JB, Stura EA, Stanfield RL, Profy AT, Wilson IA. Crystal structure of the principal neutralization site of HIV-1. Science. 1994;264:82–85. doi: 10.1126/science.7511253. [DOI] [PubMed] [Google Scholar]
- 5.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 2002;76:9035–9045. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zolla-Pazner S, Zhong P, Revesz K, Volsky B, Williams C, Nyambi P, Gorny MK. The Cross-Clade Neutralizing Activity of a Human Monoclonal Antibody Is Determined by the GPGR V3 Motif of HIV Type 1. AIDS Res. Hum. Retroviruses. 2004;20:1254–1258. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
- 9.Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman TL, Doms RW. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol. Membr. Biol. 1999;16:57–65. doi: 10.1080/096876899294760. [DOI] [PubMed] [Google Scholar]
- 11.Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retroviruses. 2000;16:741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 12.Basmaciogullari S, Babcock GJ, Van Ryk D, Wojtowicz W, Sodroski J. Identification of conserved and variable structures in the human immunodeficiency virus gp120 glycoprotein of importance for CXCR4 binding. J. Virol. 2002;76:10791–10800. doi: 10.1128/JVI.76.21.10791-10800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV's switch-hitter. AIDS Res. Hum. Retroviruses. 2005;21:171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- 14.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 15.Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV- gp120 expressed in CHO cells. Biochemistry. 2000;39:11194–11204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 16.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J. Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 19.Calarese DA, Lee HK, Huang CY, Best MD, Astronomo RD, Stanfield RL, Katinger H, Burton DR, Wong CH, Wilson IA. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13372–13377. doi: 10.1073/pnas.0505763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharon M, Gorlach M, Levy R, Hayek Y, Anglister J. Expression, purification, and isotope labeling of a gp120 V3 peptide and production of a Fab from a HIV-1 neutralizing antibody for NMR studies. Protein Expr. Purif. 2002;24:374–383. doi: 10.1006/prep.2001.1577. [DOI] [PubMed] [Google Scholar]
- 21.Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov C, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 2004;78:2394–2404. doi: 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty K, Durani V, Miranda ER, Citron M, Liang X, Schleif W, Joyce JG, Varadarajan R. Design of immunogens that present the crown of the HIV-1 V3 loop in a conformation competent to generate 447-52D-like antibodies. Biochem. J. 2006;399:483–491. doi: 10.1042/BJ20060588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varona-Santos JT, Vazquez-Padron RI, Moreno-Fierros L. Production of a short recombinant C4V3 HIV-1 immunogen that induces strong anti-HIV responses by systemic and mucosal routes without the need of adjuvants. Viral Immunol. 2006;19:237–249. doi: 10.1089/vim.2006.19.237. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Xu X, Jones IM. Immunogenicity of the outer domain of a HIV-1 clade C gp120. Retrovirology. 2007;4:33. doi: 10.1186/1742-4690-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang MY, Wang Y, Mankowski MK, Ptak RG, Dimitrov DS. Cross-reactive HIV-1-neutralizing activity of serum IgG from a rabbit immunized with gp41 fused to IgG1 Fc: possible role of the prolonged half-life of the immunogen. Vaccine. 2009;27:857–863. doi: 10.1016/j.vaccine.2008.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guyre PM, Graziano RF, Goldstein J, Wallace PK, Morganelli PM, Wardwell K, Howell AL. Increased potency of Fc-receptor-targeted antigens. Cancer Immunol. Immunother. 1997;45:146–148. doi: 10.1007/s002620050418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 28.Fujita K, Takegawa K. Chemoenzymatic synthesis of neoglycoproteins using transglycosylation with endo-beta-N-acetylglucosaminidase A. Biochem. Biophys. Res. Commun. 2001;282:678–682. doi: 10.1006/bbrc.2001.4631. [DOI] [PubMed] [Google Scholar]
- 29.Lund J, Takahashi N, Popplewell A, Goodall M, Pound JD, Tyler R, King DJ, Jefferis R. Expression and characterization of truncated forms of humanized L243 IgG1. Architectural features can influence synthesis of its oligosaccharide chains and affect superoxide production triggered through human Fc-gamma receptor I. Eur. J. Biochem. 2000;267:7246–7257. doi: 10.1046/j.1432-1327.2000.01839.x. [DOI] [PubMed] [Google Scholar]
- 30.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Xu X, Bishop A, Jones IM. Reintroduction of the 2G12 epitope in an HIV-1 clade C gp120. AIDS. 2005;19:833–835. doi: 10.1097/01.aids.0000168980.74713.9e. [DOI] [PubMed] [Google Scholar]
- 32.Chang VT, Crispin M, Aricescu AR, Harvey DJ, Nettleship JE, Fennelly JA, Yu C, Boles KS, Evans EJ, Stuart DI, Dwek RA, Jones EY, Owens RJ, Davis SJ. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15:267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scanlan CN, Ritchie GE, Baruah K, Crispin M, Harvey DJ, Singer BB, Lucka L, Wormald MR, Wentworth P, Jr, Zitzmann N, Rudd PM, Burton DR, Dwek RA. Inhibition of mammalian glycan biosynthesis produces nonself antigens for a broadly neutralising, HIV-1 specific antibody. J. Mol. Biol. 2007;372:16–22. doi: 10.1016/j.jmb.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q, Shankara S, Roy A, Qiu H, Estes S, McVie-Wylie A, Culm-Merdek K, Park A, Pan C, Edmunds T. Development of a simple and rapid method for producing non-fucosylated oligomannose containing antibodies with increased effector function. Biotechnol. Bioeng. 2008;99:652–665. doi: 10.1002/bit.21598. [DOI] [PubMed] [Google Scholar]
- 35.Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure. 2004;12:193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]