Figure 7.
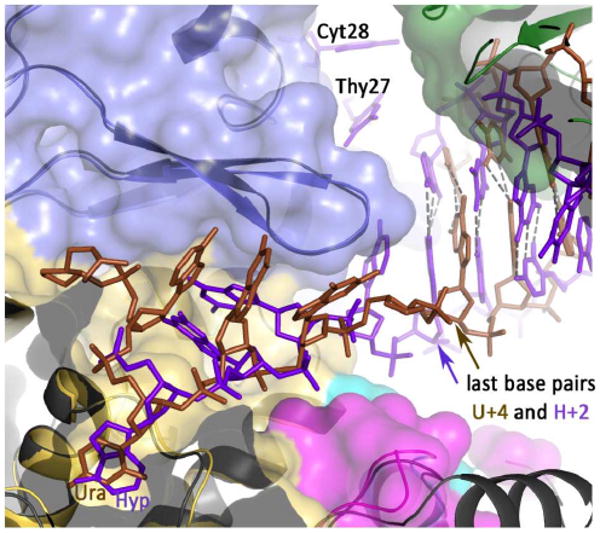
Relative spatial positions of uracil/hypoxanthine, the β-hairpin and the nearest double-stranded base for the U+4 and H+2 structures. The two structures have been superimposed and the space fill for H+2 is shown (thumb domain, green; exonuclease domain, blue; amino-terminal domain, yellow). The uracil-containing DNA is brown and the hypoxanthine-containing DNA blue, with both the deaminated bases labeled. The β-hairpin (β-strand/loop/β-strand) is illustrated for both structures with that of uracil in slightly darker blue. The positions of the base pairs (last base pairs) nearest the deaminated bases and the β-hairpin are arrowed, showing the relative disposition of these three elements are unchanged between he two structures. However, with U+4 the last base-pair contains the 3′ base of the primer. This is not the case with H+2, as the two bases at the 3′ end of the primer (T27 and C28) become single stranded and positioned in the editing channel of the polymerase.
