Summary
Apoptosis is a genetically controlled process of cell suicide that plays an important role in animal development and in maintaining homeostasis. The nematode Caenorhabditis elegans has proven to be an excellent model organism for studying the mechanisms controlling apoptosis and the subsequent clearance of apoptotic cells, aided with cell-biological and genetic tools. In particular, the transparent nature of worm bodies and eggshells makes C. elegans particularly amiable for live cell microscopy. Here we describe a few methods for identifying apoptotic cells in living C. elegans embryos and adults and for monitoring their clearance during embryonic development. These methods are based on Differential Interference Contrast microscopy and on fluorescence microscopy using GFP-based reporters.
Keywords: C. elegans, Apoptosis, Programmed cell death, Engulfment, Phagosome maturation, CED-1, PI(3)P, Time-lapse recording, GFP, mRFP, Differential interference contrast microscope
1. Introduction
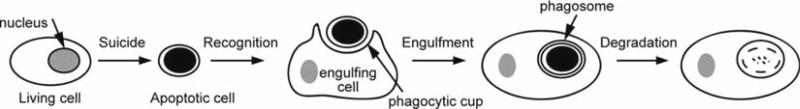
During an animal's development and adult life, a large number of unwanted cells are eliminated by programmed cell death, or apoptosis. Apoptotic cells are rapidly engulfed (via phagocytosis) by phagocytes, or engulfing cells, from the body and are degraded inside a membrane-bound structure referred to as a “phagosome” (Fig. 1) (1). Apoptosis plays important roles in sculpting structures; maintaining homeostasis, and eliminating abnormal, nonfunctional, or harmful cells (2). Efficient removal of dying cells is the necessary last step of apoptosis; in addition, it actively prevents harmful inflammatory and autoimmune responses (1).
Fig. 1.
A diagram describes the fate of an apoptotic cell in metazoans.
1.1. A Review of Published Methods for Detecting Distinct Features of Apoptotic Cells in C. elegans
The nematode C. elegans, a small free-living round worm, has been established as an excellent model organism for studying the mechanisms of apoptosis and the engulfment of apoptotic cells due to its simple anatomy, known cell lineage, well-established genetics, and easily distinguishable apoptotic cell morphology (3, 4). During the development of the C. elegans hermaphrodite, 131 somatic cells and approximately 300–500 germ cells undergo apoptosis (5–7). In the soma, due to the fixed cell lineage, both the identity of the cells that undergo apoptosis and the timing of death are invariable in C. elegans (5, 6). Apoptotic cells are rapidly engulfed and degraded by neighboring cells (5–7). Multiple types of cells can function as engulfing cells, including hypodermal cells, gonadal sheath cells, intestinal cells, and pharyngeal muscle cells (6–8). One particularly useful feature of C. elegans is that animals at all developmental stages are transparent. Apoptotic cells are thus easily recognized within living animals under the Nomarski differential interference contrast (DIC) optics as highly reflective, button-like objects that are referred to as “cell corpses”(Fig. 2a) (5–7). DIC microscopy is thus commonly used to detect cell corpses in C. elegans (4). DIC microscopy, however, is unable to distinguish engulfed cell corpses from unengulfed ones because the plasma membrane of an engulfing cell is typically not visible under DIC microscope.
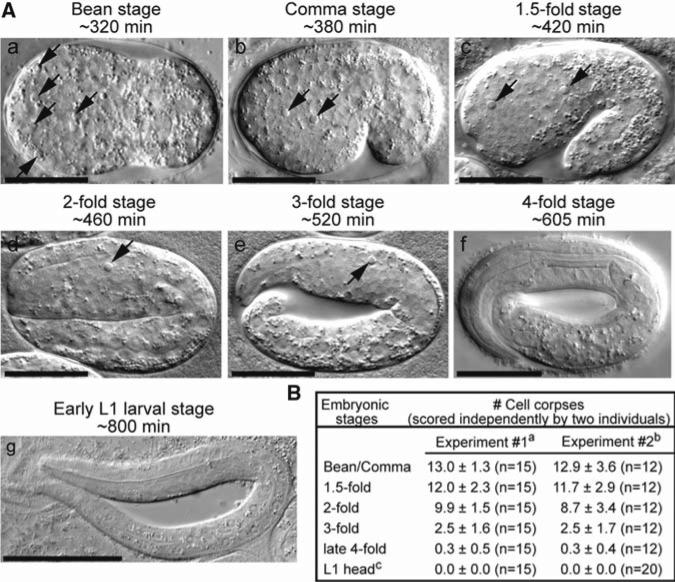
Fig 2.
Apoptotic cells display the same distinct morphology detectable by DIC microscopy in different embryonic development stages. (A) DIC images of a wild-type embryo at different developmental stages. Time (labeled in min) represents the time point the embryo enters the corresponding stage after the first cleavage. (e) and (f) were captured at mid-3-fold (~550 min) and late 4-fold (~770 min) stages, respectively. Dorsal is up and anterior is to the left. Scale bars: 20 μm in (a–f) and 50 μm in (g). Arrows indicate cell corpses. (B) The number of cell corpses in wild-type embryos and newly hatched L1 larvae scored under the DIC microscope by two different individuals in independent experiments. Data are presented as mean ± SD. n, number of animals scored. aData published in (15). bData published in (16). cScored in the head of L1 larvae hatched within 1 h.
Besides DIC microscopy, a number of methods have been used to recognize apoptotic cells at all developmental stages in C. elegans based on their distinct cellular features. These include the transmission electron microscopy (TEM) for detecting cell corpses in larvae and adults, the TUNEL (terminal transferase dUTP nick end labeling) assay that detects DNA ends generated during apoptosis in embryos, and the staining of larvae and adults with SYTO dyes. For an excellent review of these methods, please see ref. 9. Recently, several methods have been developed to detect the exposure of phosphatidylserine (PS), a membrane phospholipids kept in the inner leaflet of the plasma membrane of living cells, on the outer surface of C. elegans apoptotic cells using PS-binding proteins, such as MFG-E8 and annexin V, as reporters (10–12). In addition, apoptotic cells undergo chromatin condensation (13). A chromatin-associated histone H3 reporter (HIS-72::GFP) (14), which allows us to detect the distinct condensed chromatin morphology in apoptotic cells in C. elegans embryos, is another cell corpse-specific marker (15).
Recently, we developed CED-1::GFP and 2xFYVE::mRFP, two fluorescent markers that label the surface of phagocytic cups and that of maturing phagosomes (Fig. 1). These two markers not only offer new methods for distinguishing cell corpses but also enable us to determine whether a cell corpse is engulfed (8, 15–17). Using these markers, we further established time-lapse recording methods to monitor the processes of engulfment as well as degradation of individual apoptotic cells in developing embryos, a procedure that enables us to dissect the steps of apoptotic cell clearance on a subcellular level (15–17).
1.2. The Basis for Using CED-1 and 2xFYVE to Identify Apoptotic Cells
CED-1 is a single-pass transmembrane protein expressed in engulfing cells and acts on cell surfaces as a phagocytic receptor for neighboring apoptotic cells (8). CED-1 recognizes the cell-surface features of cell corpses, clusters on the phagocytic cup and then transiently on nascent phagosomes (Figs. 3B and 4B) (8, 17). This feature enables a CED-1::GFP reporter (Fig. 3A) to specifically label cell corpses that are in the process of being engulfed (Fig. 4B). In addition, CED-1::GFP is particularly useful for detecting unengulfed or partially engulfed cell corpses in engulfment-defective mutants (except the ced-7 mutants), because the blockage or delay of pseudopod extension around cell corpses do not affect the ability of CED-1 to recognize cell corpses and cluster on phagocytic cups (8, 10). As a consequence, in these mutants, CED-1::GFP is observed as bright, distinct partial circles around cell corpses, which represent not-enclosed phagocytic cups (8, 10).
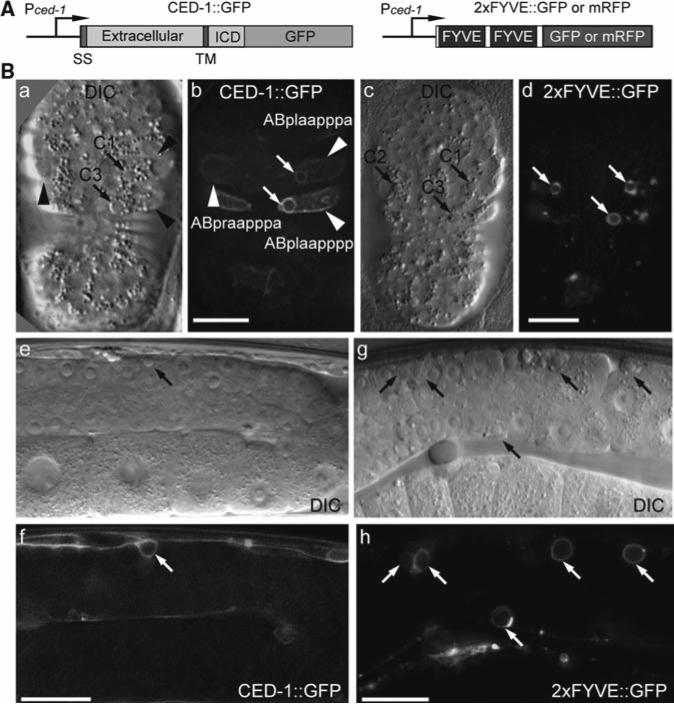
Fig. 3.
CED-1 and 2xFYVE as markers for apoptotic cells. (A) Diagrams of the reporter constructs. Pced-1ced-1 promoter, ICD Intracellular domain, SS Signal sequence, TM transmembrane domain. (B) DIC and fluorescence images illustrating that CED-1:GFP and 2×FYVE:GFP are enriched on the engulfing cell membrane surrounding cell corpses. (a–d) ~330 min-stage wild-type embryos. Anterior is to the top. Ventral faces readers. Scale bars: 10 μm. Arrows indicate phagosomes containing cell corpses. Arrowheads label the three ventral hypodermal cells as engulfing cells for C1, C2, and C3. (e–h) Part of the gonad in wild-type adult hermaphrodites. Mid-body is to the left. Scale bars: 20 μm. Arrows indicate phagosome scontaining germ cell corpses.
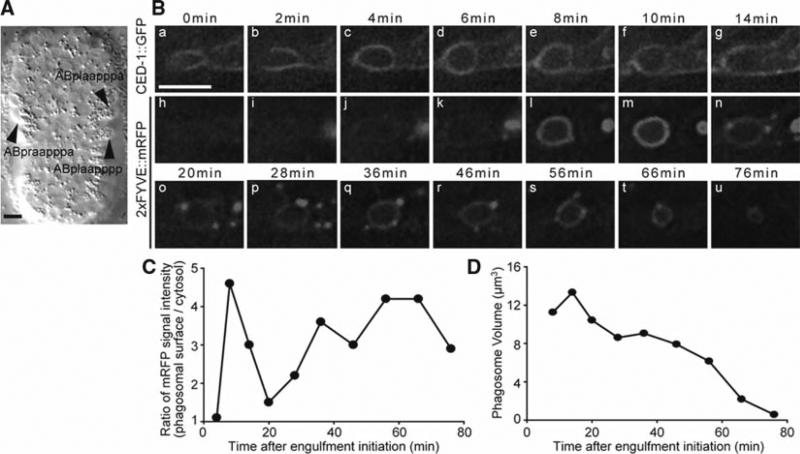
Fig. 4.
Time-lapse recording of the engulfment and degradation of apoptotic cells in C. elegans embryos. (A) DIC image of a ~310 min-stage embryo at which stage the time-lapse recording should begin. Anterior is to the top. Ventral faces readers. Arrowheads indicate the three ventral hypodermal cells that will engulf C1, C2, and C3. C1, C2, and C3 have not displayed the distinct DIC morphology of cell corpses at this time point. (B) Time lapse images of the co-expressed CED-1:GFP (a–g) and 2xFYVE:mRFP (h–u) around cell corpse C3 in a wild-type embryo. 0 min: the time point when the engulfing cell extends pseudopods halfway around C3. The scale bar: 5 μm. (C) The relative PI3P signal intensity on the surface of the phagosome containing C3 measured from images in (B) (h–u) plotted over time. (D) The volume of the phagosome containing C3 measured from images in (B) (h–u) plotted over time.
In wild-type C. elegans embryos, the clustering of CED-1::GFP around a cell corpse is detectable throughout the entire engulfment process (~5 min) and the first 9 min of phagosome maturation, which lasts 50–70 min in total (17). As a result, at any given time point, only a small portion of cell corpses is labeled by CED-1::GFP in animals that display normal engulfment activity. Recently, we established 2xFYVE::GFP as a marker for cell corpses that remains on the phagosomal surface until the complete degradation of the cell corpse inside and therefore labels almost all cell corpses.
Phosphatidylinositol-3-phosphate (PI3P) is a phosphoinositide species that is specifically enriched on the surface of endosomes and phagosomes and that acts as a signaling molecule for vesicle trafficking events (18). The FYVE domain of C. elegans EEA-1, in a tandem repeat, specifically associates with PI3P (19). The 2xFYVE::GFP and 2xFYVE::mRFP fusion proteins, which are expressed in engulfing cells under the control of the ced-1 promoter Pced-1 (Fig. 3A) (8), are localized to the cytoplasmic puncta. Immediately after the closure of a phagocytic cup, these markers are enriched on the surface of nascent phagosomes and persist on phagosomal surfaces until the complete degradation of the cell corpse (17). In embryos co-expressing CED-1::GFP and 2xFYVE::mRFP, the entire cell-corpse removal process can be monitored in real time (Fig. 4B).
In addition, the 2xFYVE::GFP or 2xFYVE::mRFP reporters are excellent tools for scoring the number of cell corpses at all stages of C. elegans development. We found that in wild-type embryos, the disc-like DIC morphology of a cell corpse appears when engulfment starts, and disappears ~30 min after the initiation of cell-corpse degradation (N. L. and Z. Z., unpublished observation). Compared to the DIC morphology, the 2xFYVE::GFP signal persists on the surface of a phagosome until its complete disappearance, it is thus able to detect cell corpses that partially or totally lose their distinct DIC morphology.
2. Materials
The materials and methods described here are specific for the detection of apoptotic cells in C. elegans. For general materials and methods for raising and handling C. elegans, please see ref. 20. For general introduction of using DIC microscopy in C. elegans, please see ref. 21.
2.1. General Materials
4% agarose solution, prepared by heating 2 g agarose in 50-mL autoclaved deionized water until agarose is completely melted. After usage, the solidified solution can be stored at room temperature and melted in a microwave oven again.
M9 Buffer (1 L): 3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 mL of 1 M MgSO4 dissolved in 850 mL H2O, add H2O to 1 L, autoclave.
30 mM sodium azide (NaN3) in M9 buffer.
Microscope slides, cover slips (22 × 22 mm), Pasteur pipette and bulb, high vacuum grease (Dow Corning), DeltaVision immersion oil N = 1.514 (Applied Precision), a platinum wire mounted on a Pasteur pipette functioning as a worm pick.
2.2. Equipment and Software
Nikon SMZ645 Stereomicroscope or any stereomicroscope from other manufactures for handling of C. elegans.
An Olympus IX70-DeltaVision microscope (Applied Precision) equipped with 20×, 63×, and 100× Uplan Apo objectives, DIC microscopy accessories, motorized stage, a Coolsnap digital camera (Photometrics), and the SoftWoRx software (for the deconvolution and processing of images) (Applied Precision).
A temperature control chamber mounted over the DeltaVision microscope that maintains the temperature of the stage at 20°C. Alternatively, the DeltaVision microscope can be kept in a room where the temperature is maintained at 20°C.
A PC computer for image processing and analysis.
The ImageJ software (downloaded from (http://rsb.info.nih.gov/ij/index.html) for quantitative image analyses.
3. Methods
3.1. Using DIC Microscopy to Score the Number of Cell Corpses
3.1.1. Determining Which Developmental Stages to Score
Cell corpses can be recognized as reflective, disc-like objects in living animals using DIC microscopy (Fig. 2a). As the execution of cell death and the clearance of apoptotic cells are dynamic processes, it is critical to score the number of cell corpses at defined developmental stages and within the defined regions of an animal for meaningful comparison of results obtained from different genetic backgrounds. To assay for the pattern of the apoptosis events of somatic cells during embryonic development, we score the number of cell corpses in the entire embryo at the following stages: bean, comma, 1.5-fold, 2-fold, 3-fold, and late 4-fold stages. Embryos at these stages, which correspond to ~320, ~380, ~420, ~460, ~520 – ~605, and ~700 – ~790 min after the first cleavage (the first cytokinesis), respectively (6, 16), are easily recognized using DIC microscopy by their distinct body morphology (Fig. 2A). Two independent sets of data, obtained from the same wild-type strain (N2) (20) by two different individuals in the laboratory, are similar to each other (Fig. 2B) (15, 16). These results are consistent with the invariable embryonic cell lineage described in (6).
To study the effect of mutants defective in cell-corpse removal, the number of persistent cell corpses is often scored in the head (the area between the anterior end of the worm and the anterior boundary of the intestine) of L1 Larvae. In the head of newly hatched wild-type L1 larvae (hatched within 1 h), no cell corpses are observed (Fig. 2B); on the other hand, mutations that induce extra cell deaths or block cell-corpse removal result in the persistent presence of cell corpses in the head (22, 23).
Germ cells that undergo apoptosis during germline development or are induced to die by DNA damaging agents can be scored in the adult hermaphrodite gonad using DIC microscopy (7, 24). Again, to obtain reproducible results, it is critical to score in animals of defined age. The most commonly used samples are adult hermaphrodites that are aged 48 h post the mid-L4 larval stage.
3.1.2. Mounting Animals on an Agar Pad
Melt the 4% agarose solution by heating it in a microwave oven.
Dispense a drop of agarose solution on a glass microscope slide and flatten the drop immediately with another glass slide. Wait until agarose solidifies, then gently separate the two slides by sliding one against the other. An agarose pad provides support to the cover slip so that the living specimens are not squashed.
Cut the round agarose pad into an approximately 12 × 12 mm2 with the edge of a glass slide. Place 3 μL of 30 mM NaN3 in M9 buffer at the center of the pad (see Note 1).
Under the Nikon SMZ 645 Stereomicroscope, transfer animals at the stage of choice with a worm pick from a plate to the drop of 30 mM NaN3 in M9 buffer, gently disperse eggs with a worm pick.
Gently place a cover slip over the drop of liquid. Remove any solution outside the cover slip with tissue paper.
3.1.3. Observation Under the DIC Microscope
Align the DIC light path carefully for optimal DIC effect according to the manufacturer's instruction (http://www.appliedprecision.com).
Under the 63× or 100× objective, identify cell corpses and score the number. As C. elegans is transparent under the light microscopy, by focusing from the bottom to the top of the animal, cell corpses in the z-axis of the entire desired region can be scored.
Alternatively, instead of scoring directly from the eyepiece, serial z-section DIC images could be captured (see below for z-sectioning) and the number of cell corpses could be scored later by replaying the serial images on the computer. This method allows a longer period of time for scoring and avoids the long-term effect of NaN3 in altering the DIC appearance of cell corpses (see Note 1).
3.2. Using CED-1::GFP and 2xFYVE::mRFP1 as Reporters to Monitor the Clearance of Apoptotic Cells in Real Time in Embryos
The DeltaVision Deconvolution Microscope is a white-light microscope that relies on specially designed computer deconvolution algorithm to achieve high resolution (25). Comparing with conventional confocal microscope, the DeltaVision results in less photobleaching of images and less photodamage to living specimens and offers comparable, under some conditions even superior, resolution and sensitivity. Here we described a specific protocol for image capture and time-lapse recording that we developed using the DeltaVision. For step-by-step operation of the DeltaVision microscope and the SoftWoRx software, see the manufacturer's instruction (http://www.appliedprecision.com).
3.2.1. C. elegans Strain
ZH814 is an unc-76(e911) mutant strain carrying reporter constructs Pced-1ced-1::gfp and Pced-12xfyve::mrfp as well as pUNC-76(+), a plasmid containing the wild-type unc-76 gene, in the same transgenic array. Transgenic animals are normal for locomotion, whereas nontransgenic animals are Unc (Uncoordinated). To cross the transgenic array to the strains of your interest, follow standard genetic operation (20).
3.2.2. Mounting Embryos on a Microscope Slide
Follow the description of Subheading 3.1.1 to prepare an agarose pad on a microscope slide. Spot 3 μL M9 buffer in the center of the pad, transfer eggs to the pad, disperse eggs in M9 buffer (see Note 2).
Gently squeeze a thin line of high vacuum grease around agarose pad and cover the pad gently with a cover slip. Avoid air bubbles. Vacuum grease prevents the drying of the agarose pad and allows air exchange. No more than 50 eggs should be loaded onto one slide, and eggs should be sufficiently dispersed in M9 solution (see Note 3).
3.2.3. Identifying Three Particular Cell Corpses C1, C2, C3 and Their Engulfing Cells
Among the 113 cells that undergo apoptosis during embryogenesis (6), we choose to monitor the clearance of three apoptotic cells referred to as C1, C2, and C3 (Fig. 3B). These three cells are located at the ventral surface of an embryo, in approximately the same or adjacent focal planes, and are engulfed at approximately the same time, between 320–330 min post-first cleavage (16). C1, C2, and C3 are each engulfed by a different ventral hypodermal cell, ABplaapppa, ABpraapppa, and ABplaapppp, respectively, while these hypodermal cells extend their cell bodies to the ventral midline (Fig. 3b) (16). These temporal and spatial features make it easy to identify C1, C2, and C3, and their engulfing cells; furthermore, they allow the recording of the clearance of all three cell corpses in the same time-lapse series, using a z-stack containing 8–12 serial z-sections (at 0.5 μm/section) at every time point.
Place the prepared slide on the microscope stage, start microscope operation, prewarm the mercury light source for 10–15 min. During this time period, align the DIC light path for optimal DIC effect. Open the SoftWoRx program.
Using the GFP channel, identify embryos that carry the transgenic array. Under the 100× objective, identify transgenic embryos whose ventral side faces the objective and which are at ~320 min post-first cleavage (Fig. 4A) (see Note 4). Once an appropriate embryo is identified, its exact location on the slide should be recorded using the “point marking” function of the SoftWoRx program.
3.2.4. Time-Lapse Recording
Set up microscope parameters. Use the 100× objective. For capturing DIC images, exposure time is usually set at 0.1 s. For fluorescence imaging, two sets of fluorescence filters, both from Chroma, Inc., are used, including the FITC filter (excitation wavelength 490/20 nm; emission wavelength 528/38 nm) for the GFP signal and the Rhodamine filter (excitation wavelength 555/28 nm; emission wavelength 617/73 nm) for the mRFP signal. The exposure time is 0.1 s for each channel and each z-section (see Note 3). If the signal is weak, 2 × 2 binning is recommended (see Note 3).
Set up the recording program. Serial z-sectioning is performed from the ventral surface of an embryo toward the center. The setting of 8–12 z-sections at 0.5 μm per section is sufficient to include C1, C2, C3 in one z-section series (cell corpses are of 2.5–3 μm in diameter). An image size of 512 × 512 pixels is sufficient for capturing the entire embryo if 2 × 2 binning is performed (see Note 3). For recording the engulfment process, which lasts for ~5 min in a wild-type strain, 30 time points at a 1-min interval is sufficient if recording starts at a time between 310 and 320 min post-first cleavage. For the degradation process, which lasts for ~50–70 min in wild-type embryos but could last much longer in degradation-defective mutants (17), we record for 100–120 min at a 2-min interval. After embryos reach ~460 min post-first cleavage, rapid body movement starts, which interferes with image recording.
Using the “point marking” and “point visiting” functions of the software to record multiple embryos in the same program. Using the parameters described above, at least three embryos can be recorded in the same program in a time interval of 2 min.
Keep observing images from time to time. Adjust the starting focal plane during the interval of recording if any change of focal plane occurs. Abort recording if an embryo slows down or stops its development due to photodamage (see Note 3).
After recording is completed, deconvolve images using SoftWoRx.
Open deconvolved files with softWoRx, save desired images as tiff or jpeg files for quantitative analysis using the ImageJ software and for further processing using Adobe Photoshop.
3.2.5. Quantitative Image Analysis
Measuring Signal Intensity on Phagosomal Surfaces
The dynamic changes of the signal intensity of CED-1 and PI3P indicate the progress of engulfment and phagosome maturation; in addition, alteration of the dynamic pattern of these signals in mutant backgrounds suggests specific defects in phagosome formation and maturation (15, 17). The signal intensity of CED-1 and PI3P on phagosomal surfaces is quantified by measuring the fluorescence intensity of CED-1::GFP and 2xFYVE::mRFP, respectively. The absolute fluorescence signal intensity, however, varies from embryos to embryos due to the different expression levels of the transgene. Thus, we use the relative signal intensity represented by the ratio of the intensity on phagosomal surface to that in an adjacent area inside the cytosol to indicate the enrichment of CED-1 or PI3P on phagosomal surfaces. We use the software ImageJ to quantify fluorescence signal intensity.
Open an epifluorescence image file (in tiff or jpeg format) in the ImageJ program. Increase the magnification of the image until the boundary of phagosome can be clearly distinguished.
Use the freehand selection tool to define a donut-like and closed area with one continuous line that surrounds the surface of a phagosome.
Select the Measure tool from the Analyze menu to display the mean or median value of the fluorescence signal intensity measured in this area (see Note 5).
Use the freehand selection tool to select an area in engulfing cell cytosol adjacent to the phagosome. Repeat step 3 to obtain the mean or median value.
Calculate the ratio of the values obtained from the phagosomal surface and that obtained from the cytosol.
Plot the ratio over time. An example of the results is shown in Fig. 4C.
Measuring the Volume of a Phagosome over Time
During phagosome maturation, the volume of a phagosome decreases as the content is gradually digested, and is a reliable index that reflects the progression of the degradation of apoptotic cells (15, 17).
Among a z-stack of serial optical sections, identify the middle section of a phagosome in the z-axis, which represents the equator plane. Open this image (in tiff or jpeg format) in ImageJ.
Set up the μm/pixel scale (see Note 6) by selecting Set Scales in the Analyze menu and entering the scale for each pixel. As a reference, images obtained from the DeltaVision using the 100× objective and subject to 2 × 2 binning have a scale of 0.133 μm per pixel.
Increase the magnification of the image until the boundary of a phagosome can be clearly distinguished. Use the freehand selection tool to draw a continuous line along the phagosome surface. Always draw along the path that has the brightest signal.
Select the Measure tool from Analyze menu to display the area (A) of the selected shape (the phagosome) in μm2.
Regarding a phagosome as a sphere, calculate the radius (r) of the phagosome using the formula A = πr2. Calculate the volume of the phagosome (V) using the formula V = (4/3) πr3.
Plot the phagosome volume over time. An example is shown in Fig. 4D.
3.3. Scoring the Number of Cell Corpses Using 2xFYVE::GFP
Using the protocols described above, mount embryos on slides and identify embryos at the stage of your choice. Capture serial z-section images of an entire embryo at 40 × 0.5 μm/s optic interval (see Note 7). Other parameters for image recording are the same as described above except a time course for recording is not necessary. Scoring the number of 2xFYVE::GFP(+) rings using deconvolved serial z-section images. The results represent the number of phagosomes, or engulfed cell corpses. As mentioned before (Subheading 1.2), this method is highly sensitive in identifying apoptotic cells, including those that lose their distinct DIC morphology.
Acknowledgments
Z. Z. was supported by NIH (GM067848), the Cancer Research Institute, the Rita Allen Foundation, and a Basil O’ Connor Starter Scholar award from March of Dimes Foundation. X. H. was supported by NIH (GM068676).
Footnotes
NaN3 anesthetizes and immobilizes animals. Larvae and adults are immobilized within a few minutes after incubation with the 30 mM NaN3 solution. It takes NaN3 a much longer time to penetrate eggshells. For scoring embryos younger than the 2-fold stage, it is not necessary to use NaN3, since vigorous body movement of embryos does not start until that stage. Note that after 1-h incubation in the NaN3 solution, the DIC morphology of larvae and adults starts to become abnormal, whereas that of embryos are not affected.
Anesthetization is not necessary since at the particular embryonic stages for recording, there is minimum embryonic body movement. NaN3 stops embryonic development and should be avoided.
How to ensure that embryos develop normally during time-lapse recording? (a) To ensure normal embryonic development in a chamber with limited oxygen supply, load no more than 50 eggs onto the glass slide, disperse eggs thoroughly in the M9 solution, and carry over as little bacteria as possible. (b) To avoid photodamage of embryonic development and photobleach of fluorescence signals, use a highly sensitive CCD camera so that the light exposure time could be minimized, and restrain or avoid direct observation of fluorescent light under the eyepiece. Instead, “snapshots” with the camera should be used for finding and setting the focal plane to begin the recording. For weak fluorescence signals, use the “2 × 2 binning” function to keep the exposure time minimal. In addition, include only the necessary number of z-sections at each time point. As a rule of thumb, for recording of two channels, the exposure time of each channel should be kept below 0.2 s per z-section. (c) Signs of the photodamage of embryonic development. Data obtained from those embryos whose development is arrested due to photodamage are not useful. We rely on a few embryonic morphology changes to determine whether the development is proceeding in the normal time course. For example, the period from the bean- to the comma-stage, lasts for 60–70 min (Fig. 2A)(a, b). During this period, an embryo rotates 90° (Fig. 2A)(a, b). In addition, a period from the comma stage to the 1.5-fold stage lasts for ~40 min. A significant elongation of any of these time intervals is a sign of developmental arrest.
Two distinct features that can help identify embryos at this stage are (1) the ventral surface slightly invaginates on both sides, and (2) the three soon-to-be engulfing cells are located at the lateral sides, in a edge shape, with the tip of each cell less than halfway extended toward the ventral midline (Fig. 4A).
The Median and mean values are usually very similar. An abnormally bright pixel on phagosome surface, however, is largely ignored in median value, whereas it is counted and significantly increases mean value. Therefore, the median value is more resistant to signal noise and reflects the signal intensity more accurately.
The μm/pixel scale designates the size of each pixel in the image, which can be obtained from the program with which the image is captured.
The average thickness of an embryo is 20 μm (Z. Z., unpublished observation).
References
- 1.Savill J, Fadok V. Corpse clearance defines the meaning of cell death. Nature. 2000;407:784–8. doi: 10.1038/35037722. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–54. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 3.Metzstein MM, Stanfield GM, Horvitz HR. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–6. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Z, Mangahas PM, Yu X. The genetics of hiding the corpse: engulfment and degradation of apoptotic cells in C. elegans and D. melanogaster. Curr Top Dev Biol. 2004;63:91–143. doi: 10.1016/S0070-2153(04)63004-3. [DOI] [PubMed] [Google Scholar]
- 5.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–56. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 6.Sulston JE, Schierenberg E, White JG, Thomson N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 7.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–22. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001b;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz HT. A protocol describing pharynx counts and a review of other assays of apoptotic cell death in the nematode worm Caenorhabditis elegans. Nat Protoc. 2007;2:705–14. doi: 10.1038/nprot.2007.93. [DOI] [PubMed] [Google Scholar]
- 10.Venegas V, Zhou Z. Two alternative mechanisms that regulate the presentation of apoptotic cell engulfment signal in Caenorhabditis elegans. Mol Biol Cell. 2007;18:3180–92. doi: 10.1091/mbc.E07-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Wang J, Gengyo-Ando K, Gu L, Sun CL, et al. C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat Cell Biol. 2007;9:541–9. doi: 10.1038/ncb1574. [DOI] [PubMed] [Google Scholar]
- 12.Zullig S, Neukomm LJ, Jovanovic M, Charette SJ, Lyssenko NN, et al. Aminophospholipid translocase TAT-1 promotes phosphatidylserine exposure during C. elegans apoptosis. Curr Biol. 2007;17:994–9. doi: 10.1016/j.cub.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 13.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 14.Ooi SL, Priess JR, Henikoff S. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2006;2:e97. doi: 10.1371/journal.pgen.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangahas PM, Yu X, Miller KG, Zhou Z. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J Cell Biol. 2008;180:357–73. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X, Odera S, Chuang CH, Lu N, Zhou Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell. 2006;10:743–57. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6(3):e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: aging gracefully. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roggo L, Bernard V, Kovacs AL, Rose AM, Savoy F, et al. Membrane transport in Caenorhabditis elegans: an essential role for VPS34 at the nuclear membrane. EMBO J. 2002;21:1673–83. doi: 10.1093/emboj/21.7.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood WB, Researchers of the C. elegans Community . The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1998. [Google Scholar]
- 21.Shaham S. Methods in cell biology. In: The C. elegans Research Community, editor. WormBook. 2005. [Google Scholar]
- 22.Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, et al. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287:1485–9. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- 23.Mangahas PM, Zhou Z. Clearance of apoptotic cells in Caenorhabditis elegans. Semin Cell Dev Biol. 2005;16:295–306. doi: 10.1016/j.semcdb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–43. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 25.Sibarita JB. Deconvolution microscopy. Adv Biochem Eng Biotechnol. 2005;95:201–43. doi: 10.1007/b102215. [DOI] [PubMed] [Google Scholar]