Abstract
The human sodium channel family includes seven neuronal channels that are essential for the initiation and propagation of action potentials in the CNS and PNS. In view of their critical role in neuronal firing and their strong sequence conservation during evolution, it is not surprising that mutations in the sodium channel genes are responsible for a growing spectrum of channelopathies. Nearly 700 mutations of the SCN1A gene have been identified in patients with Dravet's syndrome (severe myoclonic epilepsy of infancy), making this the most commonly mutated gene in human epilepsy. A small number of mutations have been found in SCN2A, SCN3A and SCN9A, and studies in the mouse suggest that SCN8A may also contribute to seizure disorders. Interactions between genetic variants of SCN2A and KCNQ2 in the mouse and variants of SCN1A and SCN9A in patients provide models of potential genetic modifier effects in the more common human polygenic epilepsies. New methods for generating induced pluripotent stem cells and neurons from patients will facilitate functional analysis of amino acid substitutions in channel proteins. Whole genome sequencing and exome sequencing in patients with epilepsy will soon make it possible to detect multiple variants and their interactions in the genomes of patients with seizure disorders.
Miriam Meisler is a geneticist at the University of Michigan who uses mouse mutants to characterize gene function and identify candidate genes for human disorders. In 1997 her lab developed the Q54 mouse expressing a mutant Scn2a sodium channel with elevated persistent current. The observation of seizures in the Q54 mouse, together with the linkage of human GEFS+ to chromosome 2q24 containing a sodium channel cluster, led the lab to identify the first mutations of SCN1A in 2000. Janelle O’Brien, a graduate student in Human Genetics, and Lisa Sharkey, a neuroscientist and postdoctoral fellow, are studying mutations of SCN8A in the Meisler lab.
 |
Introduction
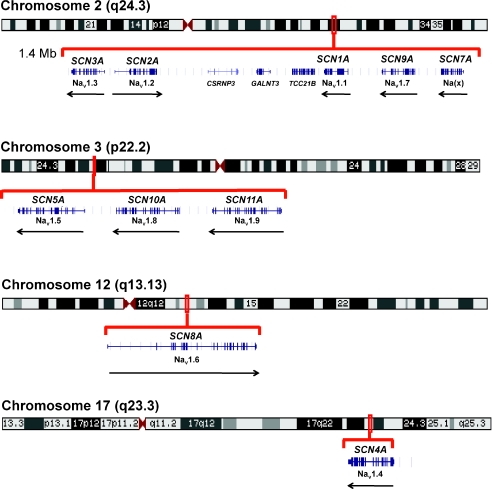
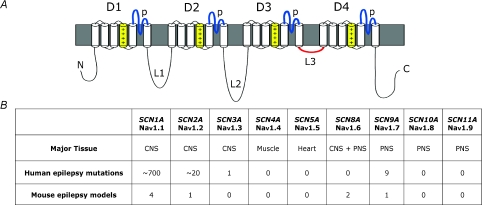
The human genome contains ten paralogous genes encoding the large transmembrane protein that constitutes the pore-forming α-subunit of the voltage-gated sodium channel. Two clusters of tandemly duplcated genes are located on chromosomes 2q24 and 3p22, and two single genes are located on chromosomes 12q13 and 17q23 (Fig. 1). All of these genes except SCN7A encode proteins that exhibit sodium channel activity in exogenous expression systems. During the evolution of this gene family its members have diverged with respect to tissue-specific expression; the major expression site for each gene is shown in Fig. 2. The basic architecture of the four-domain protein with its 24 transmembrane segments (Fig. 2) is conserved in all family members, and the amino acid sequence identity varies from 50% to 85% between channels. In this review we address the roles of these channels and their mouse orthologues in epilepsy.
Figure 1. Genomic arrangement of 10 paralogous human genes encoding α subunits of the voltage-gated sodium channels.
Protein nomenclature is indicated below the gene symbol. Arrows represent the direction of transcription. Data from the UCSC genome browser at http://www.genome.ucsc.edu
Figure 2. Tissue specificity and number of reported epilepsy mutations in the 9 functional voltage-gated sodium channel genes.
A, conserved domains of the sodium channel α-subunit protein. D, homologous domains; p, pore loops; L, inter-domain loops; +, positively charged transmembrane segments contributing to voltage sensing. B, major sites of expression of the sodium channel genes and numbers of mutations identified in patients with epilepsy.
SCN1A
Mutations in SCN1A encoding the sodium channel Nav1.1 are the most common genetic cause of inherited and sporadic epilepsy. SCN1A mutations were first identified in families with the mild inherited disorder generalized epilepsy with febrile seizures plus (GEFS+) (Escayg et al. 2000) and shortly thereafter in sporadic patients with the early onset, severe progressive disorder severe myoclonic epilepsy of infancy (SMEI), also known as Dravet's syndrome (Claes et al. 2001). More than 650 heterozygous mutations of SCN1A have been identified in patients with SMEI (∼85% of patients tested), and approximately 20 mutations in patients with GEFS+ (∼10% of patients tested). Half of these mutations result in protein truncation, clearly demonstrating haploinsufficiency of SCN1A (Meisler & Kearney, 2005; Kearney & Meisler, 2009). The remaining mutations are missense mutations which may cause either gain-of-function or loss-of-function, and only a few have been tested functionally. Interestingly, the GEFS+ mutation R1648H exhibits qualitative abnormalities in exogenous test systems but behaves like a null allele in knock-in mice (Martin et al. 2010). Two databases describing published patient mutations are available at http://www.scn1a.info (Lossin, 2009) and http://www.molgen.ua.ac.be/SCN1AMutations/Home (Claes et al. 2009).
The extreme heterogeneity of mutations in SCN1A is striking. Only a few sites of re-mutation have been observed (Kearney et al. 2006a); these include demethylation of certain CpG residues in arginine codons and duplication of short runs of simple sequence. The ‘common disease/common variant’ hypothesis clearly does not apply to SCN1A and epilepsy.
To address the mechanism of hyperexcitability resulting from loss of sodium channel activity in an in vivo system, four mouse models with null or missense mutations of Scn1a have been developed (Table 1). Both null alleles were generated by targeted mutagenesis in embryonic stem (ES) cells (Yu et al. 2006; Ogiwara et al. 2007). Homozygous null mice exhibit spontaneous seizures and die during the third week of postnatal life. Heterozygous null mice also exhibit spontaneous seizures, but approximately 50% survive to adulthood. Recorded sodium currents from −/− and +/− mice revealed substantial reduction of current density in inhibitory bipolar neurons and cerebellar Purkinje cells, but much smaller effects in excitatory neurons (Yu et al. 2006; Kalume et al. 2007). This difference between inhibitory and excitatory neurons was also observed in cortical neurons from mice carrying the GEFS+ mutation R1648H (Martin et al. 2010). The data from these mouse models indicate that Nav1.1 accounts for a larger proportion of total sodium channel activity in inhibitory neurons than in excitatory neurons and as a consequence the net effect of Scn1a mutations is reduced inhibitory signalling. This topic is discussed in detail in the review by Catterall et al. in this issue. Independent evidence for the critical role of inhibitory signals in epileptogenesis is provided by the discovery of heterozygosity for null alleles of the GABAA receptor in epilepsy patients (Kang & Macdonald, 2009).
Table 1.
Mouse models of sodium channel mutations and epilepsy
| Gene | Mutation | Seizure phenotype | Reference |
|---|---|---|---|
| Scn1a | Targeted knock-out | Spontaneous tonic and tonic-clonic | Yu et al. (2006) |
| Scn1a | Targeted knock-out | Spontaneous tonic and tonic-clonic | Ogiwara et al. (2007) |
| Scn1a | R1648H BAC transgene | Reduced threshold to kainic acid | Tang et al. (2009) |
| Scn1a | R1648H knock-in | Spontaneous generalized, febrile, flurothyl-induced | Martin et al. (2010) |
| Scn2a | GAL879-881QQQ transgene (‘Q54’) | Temporal lobe, spontaneous | Kearney et al. (2001, 2006b) |
| Scn3a | Targeted knock-out | Not reported | Nassar et al. (2006) |
| Scn8a | V929F/+, –/+ | Spike wave discharge, resistance to induced seizures | Papale et al. (2009),Martin et al. (2007) Blumenthal et al. (2009) |
| Scn9a | N641Y knock-in | Reduced threshold, increased corneal kindling | Singh et al. (2009) |
Haploinsufficiency of SCN1A
The unique haploinsufficiency of Scn1a compared with the other sodium channels appears to be a direct consequence of the development of severe seizures in heterozygotes. The dependence of inhibitory neurons on the levels of this specific sodium channel could be a consequence of its subcellular localization, for example its function at the axon initial segment. Deletions that disrupt two or more of the genes in the cluster on chromosome 2q24 have been observed in patients with SMEI (Table 2). The phenotypes of patients with multi-gene deletions are not more severe than patients with loss-of-function of SCN1A alone, supporting the view that SCN1A is the only channel that is haploinsufficient. This view is also supported by the normal phenotypes of mice that are heterozygous for null alleles of Scn2a (Planells-Cases et al. 2000), Scn3a (Nassar et al. 2006) and Scn9a (Nassar et al. 2004).
Table 2.
Multi-gene deletions of the sodium channel cluster on chromosome 2q24 identified in patients with SMEI
| Size of deletion | Genes deleted | No. of patients |
|---|---|---|
| 3 to 9 Mb | SCN1A, SCN7A, SCN9A, SCN2A, SCN3A | 4 |
| 0.6 to 6 Mb | SCN1A, SCN7A, SCN9A | 9 |
| 0.2 to 0.6 Mb | SCN1A, SCN9A | 2 |
For details see Pereira et al. (2004), Madia et al. (2006), Suls et al. (2006) and Marini et al. (2010).
SCN2A
Approximately 20 missense mutations of SCN2A have been detected in patients with mild seizure disorders including benign familial neonatal-infantile seizures (BFNIS) and GEFS+. More recently, three missense mutations of SCN2A were identified in patients with SMEI (Ogiwara et al. 2009; Shi et al. 2009). The SMEI mutations E211K and I1473M result in hyperpolarizing shifts in voltage dependence of activation, consistent with premature channel opening and hyperactivity (Ogiwara et al. 2009). In contrast, the BFNIS mutation R1319Q results in a depolarizing shift in voltage dependence of activity, consistent with reduced activity under physiological conditions (Misra et al. 2008). The three BFNIS mutations all exhibited reduced current density, consistent with reduced channel activity (Misra et al. 2008). One nonsense mutation in SCN2A was identified in a patient with intractable epilepsy and mental decline (Kamiya et al. 2004). This mutation, R102X, truncates the channel protein within the cytoplasmic N-terminal domain. The authors reported a dominant negative effect of the N-terminal fragment on activity of the wild-type channel. Taken together, the functional tests of SCN2A mutations suggest that the severity of the biophysical effect is correlated with clinical severity. In the mouse, heterozygotes for the Scn2a knockout allele were not reported to exhibit seizures (Planells-Cases et al. 2000).
A model of temporal lobe epilepsy was generated in the ‘Q54’ mouse strain that carries a transgene with the gain-of-function missense mutation GAL879QQQ. This three amino acid substitution is located in an intracellular linker in domain 2 and results in elevated persistent current in vivo (Kearney et al. 2001). Mice carrying the Q54 transgene on a (B6XSJL)F1 strain background exhibit severe spontaneous seizures with onset at 1 month and lethality by 6 months. However, on a pure C57BL/6J background, onset is delayed and the phenotype is mild. Two modifier loci responsible for the difference in severity between strains C57BL/6J and SJL have been mapped, and the evidence points to the voltage-gated potassium channel gene Kcnv2 as one modifier (Bergren et al. 2005; Bergren et al. 2009). The Q54 mutation also interacts with another potassium channel, Kcnq2. Double heterozygous mice inheriting mild alleles of Scn2a and Kcnq2 exhibit severe myoclonic seizures (Kearney et al. 2006b). The ‘modifier’ effect of these potassium channel mutations may be a model for gene interactions underlying human polygenic epilepsies.
SCN3A
A single patient with partial epilepsy and a mutation in SCN3A has been described (Holland et al. 2008). The proband was heterozygous for the missense mutation K354Q located in the pore loop of domain 1. Functional studies of this lysine to glutamine substitution demonstrated slowing of fast inactivation and increased persistent current. Although SCN3A expression appears to be widespread in human adult brain (Whitaker et al. 2000), in rodents the expression of Scn3a is highest in young animals and is low in adults (Felts et al. 1997). In a mouse with targeted inactivation of Scn3a there were no reported spontaneous seizures (Nassar et al. 2006), but the species difference in expression may limit the relevance of the mouse mutant.
SCN8A
A single human family with a null muation of SCN8A has been described (Trudeau et al. 2006). The four heterozygous carriers of the null allele exhibited cognitive deficits but no history of seizures. EEG studies of the SCN8A heterozygotes were not available. However, three mouse studies implicate Scn8a in seizures. First, a missense mutation of Scn8a was found to ameliorate the seizure phenotype of Scn1a null heterozygotes (Martin et al. 2007). The proposed mechanism was that reduction of Nav1.6 in excitatory neurons reduces firing capacity and compensates for the loss of Nav1.1 in inhibitory neurons. In addition, Scn8a null heterozygotes were found to be resistant to kainate- or fluorethyl-induced seizures (Martin et al. 2007) and were also resistant to kindling by electrical stimulation (Blumenfeld et al. 2009). Finally, Scn8a heterozygous null mice exhibit altered spike wave discharges similar to the interictal patterns seen in seizure-prone mutants (Papale et al. 2009). The discharges were responsive to administration of the drug ethosuximide, leading to the suggestion that SCN8A might play a role in human absence epilepsy.
SCN9A
The major site of expression of SCN9A is in sensory neurons of the PNS, and distinct mutations in this channel are responsible for three inherited pain syndromes (Dib-Hajj et al. 2009). It was therefore surprising when the missense mutation N641Y in SCN9A was identified in a family with febrile seizures linked to the chromosome 2q24 sodium gene cluster (Singh et al. 2009). No mutations in SCN1A were found in this family. Furthermore, a knock-in mouse carrying the N641Y mutation exhibited reduced thresholds to electrically stimulated seizures. Several patients with mutations in both SCN1A and SCN9A were also described, but the relative contributions of the two mutations to the patient phenotype is not yet clear.
Sodium channel β subunit mutations and epilepsy
There are four β subunit genes in the human genome, SCN1B to SCN4B. The β subunits are single transmembrane domain proteins that modulate the subcellular localization and properties of the pore-forming α subunits (Isom et al. 1992, 1995). The extracellular domain of the β subunit also participates in cell–cell interactions (Brackenbury et al. 2008). The recent localization of β subunits at the action initial segment suggests a mechanism for influence on neuronal excitability (Brackenbury et al. 2010). A heterozygous mutation in the extracellular domain of SCN1B encoding the β1 subunit was first identified in a large family with GEFS+ in 1998 (Wallace et al. 1998). Several additional mutations have been described in patients with mild epilepsies (Wallace et al. 2002; Audenaert et al. 2003; Burgess, 2005; Yamakawa, 2005; Scheffer et al. 2007). In 2009, the recessive mutation R125C, which prevents trafficking of the β1 subunit to the cell surface, was identified in a patient with SMEI (Patino et al. 2009). Homozygous Scn1b knockout mice also exhibit spontaneous seizures (Chen et al. 2007), confirming the role of β1 in epilepsy.
Genetic interaction between sodium channel mutations and variants in other channels
It is evident that the firing pattern of a neuron is determined by the net ‘channelome’ expressed in that cell, both qualitative and quantitative. Several examples of interactions between channel variants have recently been described. Kearney et al. (2006b) combined the Q54 mutation in Scn2a (described above) with a subclinical mutation of the potassium channel Kcnq2 and observed dramatic exacerbation of seizures. Heterozygosity for a null mutation of Scn8a can compensate for haploinsufficiency of Scn1a (Martin et al. 2007). Interactions between mutations of the calcium channel gene Cacna1a and the potassium channel gene Kcna1 have also been described (Glasscock et al. 2007). In human epilepsy, a recent report describes modification of the severity of SCN1A-related Dravet's syndrome due to co-segregation of mutations in the closely linked SCN9A gene (Singh et al. 2009). In the future, whole genome sequencing of epilepsy patients will reveal the extent to which gene interaction contributes to common types of human epilepsy.
Low level expression of neuronal channels in non-neuronal tissues
Sensitive methods like RT-PCR and immunostaining have detected a low level expression of sodium channels in unexpected tissues beyond their major sites of expression. For example, expression of brain sodium channels in heart (e.g. Maier et al. 2003) and the heart channel in brain (e.g. Wang et al. 2009). It is not clear whether these low level transcripts have specific functions. Many tissues express a low level of an alternative splice form of SCN8A containing the exon 18N with an in-frame stop codon that is unlikely to encode an active channel protein (Plummer et al. 1997). In situ hybridization detects both the full length- and truncation-encoding transcripts, but the functional implications of the two are quite different. One feasible approach to testing the biological function of low-level transcripts to knock out their expression using CRE recombinase in the mouse, but this has not yet been reported.
Future prospects
In view of their critical role in neuronal excitability, it is not surprising that mutations in the sodium channels genes result in abnormal firing patterns in epilepsy. Although the majority of mutations have been found in the SCN1A gene, the data reviewed here suggest that variants in several other sodium channel genes may have a greater role in human disease than is currently recognized. The application of new methods of whole genome sequencing to patients with seizure disorders is likely to replace the screening of individual candidate genes in the future, increasing the likelihood of detecting multiple variants in individual patients. Genetic variants may also influence individual responses to environmental epileptogenic exposures (Heinzen et al. 2007).
Another new tool with considerable promise for the study of channelopathies is the generation of neuronal cells from skin fibroblasts taken from patients (Takahashi & Yamanaka, 2006; Vierbuchen et al. 2010). Recent papers describe the generation of neurons from patients with spinal motor atrophy (Ebert et al. 2009), amyotrophic lateral sclerosis (ALS) (Dimos et al. 2008) and Parkinson's disease (Soldner et al. 2009). Applied to epilepsy, this technology will make it possible to assess the effects of channel mutations in the context of all the other variants in the patient's genome, and to record from neuronal cells that will contain tissue-specific accessory proteins and splice factors not available in other test systems. Current limitations include the low yield of some classes of neurons, and the difficulty of assessing circuit-level effects. However, preliminary studies on induced neurons from patients with SCN1A mutations have demonstrated that sodium currents from both excitatory and inhibitory neurons can be recorded (unpublished observations). This approach will make it possible to evaluate the inhibitory-neuron hypothesis for SCN1A mutations in human neurons and to evaluate drugs for correction of the specific channelopathy expressed in each individual patient.
Acknowledgments
We are supported by NIH grants R01 NS034509 and RC1 NS068684 (M.H.M.) and T32 GM 007544 (J.E.O.).
Glossary
Abbreviations
- BFNIS
benign familial neonatal-infantile seizures
- GEFS+
generalized epilepsy with febrile seizures plus
- SMEI
severe myoclonic epilepsy of infancy
Author contributions
All authors contributed to the conception, revision and final approval of this review article.
References
- Audenaert D, De Jonghe P. A deletion in SCN1B is associated with febrile seizures and early-onset absence epilepsy. Neurology. 2003;61:854–856. doi: 10.1212/01.wnl.0000080362.55784.1c. [DOI] [PubMed] [Google Scholar]
- Bergren SK, Chen S, Galecki A, Kearney JA. Genetic modifiers affecting severity of epilepsy caused by mutation of sodium channel Scn2a. Mamm Genome. 2005;16:683–690. doi: 10.1007/s00335-005-0049-4. [DOI] [PubMed] [Google Scholar]
- Bergren SK, Rutter ED, Kearney JA. Fine mapping of an epilepsy modifier gene on mouse Chromosome 19. Mamm Genome. 2009;20:359–366. doi: 10.1007/s00335-009-9193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenfeld H, Lampert A, Klein JP, Mission J, Chen MC, Rivera M, Dib-Hajj S, Brennan AR, Hains BC, Waxman SG. Role of hippocampal sodium channel Nav1.6 in kindling epileptogenesis. Epilepsia. 2009;50:44–55. doi: 10.1111/j.1528-1167.2008.01710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackenbury WJ, Djamgoz MB, Isom LL. An emerging role for voltage-gated Na+ channels in cellular migration: regulation of central nervous system development and potentiation of invasive cancers. Neuroscientist. 2008;14:571–583. doi: 10.1177/1073858408320293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackenbury WJ, Calhoun JD, Chen C, Miyazaki H, Nukina N, Oyama F, Ranscht B, Isom LL. Functional reciprocity between Na+ channel Nav1.6 and β1 subunits in the coordinated regulation of excitability and neurite outgrowth. Proc Natl Acad Sci U S A. 2010;107:2283–2288. doi: 10.1073/pnas.0909434107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DL. Neonatal epilepsy syndromes and GEFS+: mechanistic considerations. Epilepsia. 2005;46(Suppl 10):51–58. doi: 10.1111/j.1528-1167.2005.00359.x. [DOI] [PubMed] [Google Scholar]
- Chen C, Dickendesher TL, Oyama F, Miyazaki H, Nukina N, Isom LL. Floxed allele for conditional inactivation of the voltage-gated sodium channel beta1 subunit Scn1b. Genesis. 2007;45:547–553. doi: 10.1002/dvg.20324. [DOI] [PubMed] [Google Scholar]
- Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes LR, Deprez L, Suls A, Baets J, Smets K, Van Dyck T, Deconinck T, Jordanova A, De Jonghe P. The SCN1A variant database: a novel research and diagnostic tool. Hum Mutat. 2009;30:E904–920. doi: 10.1002/humu.21083. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Dib-Hajj SD, Binshtok AM, Cummins TR, Jarvis MF, Samad T, Zimmermann K. Voltage-gated sodium channels in pain states: role in pathophysiology and targets for treatment. Brain Res Rev. 2009;60:65–83. doi: 10.1016/j.brainresrev.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C, Malafosse A. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- Felts PA, Yokoyama S, Dib-Hajj S, Black JA, Waxman SG. Sodium channel α-subunit mRNAs I, II, III, NaG, Na6 and hNE (PN1): different expression patterns in developing rat nervous system. Brain Res. 1997;45:71–82. doi: 10.1016/s0169-328x(96)00241-0. [DOI] [PubMed] [Google Scholar]
- Glasscock E, Qian J, Yoo JW, Noebels JL. Masking epilepsy by combining two epilepsy genes. Nat Neurosci. 2007;10:1554–1558. doi: 10.1038/nn1999. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Yoon W, Tate SK, Sen A, Wood NW, Sisodiya SM, Goldstein DB. Nova2 interacts with a cis-acting polymorphism to influence the proportions of drug-responsive splice variants of SCN1A. Am J Hum Genet. 2007;80:876–883. doi: 10.1086/516650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland KD, Kearney JA, Glauser TA, Buck G, Keddache M, Blankston JR, Glaaser IW, Kass RS, Meisler MH. Mutation of sodium channel SCN3A in a patient with cryptogenic pediatric partial epilepsy. Neurosci Lett. 2008;433:65–70. doi: 10.1016/j.neulet.2007.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom LL, De Jongh KS, Patton DE, Reber BF, Offord J, Charbonneau H, Walsh K, Goldin AL, Catterall WA. Primary structure and functional expression of the β1 subunit of the rat brain sodium channel. Science. 1992;256:839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- Isom LL, Scheuer T, Brownstein AB, Ragsdale DS, Murphy BJ, Catterall WA. Functional co-expression of the β1 and type IIA α subunits of sodium channels in a mammalian cell line. J Biol Chem. 1995;270:3306–3312. doi: 10.1074/jbc.270.7.3306. [DOI] [PubMed] [Google Scholar]
- Kalume F, Yu FH, Westenbroek RE, Scheuer T, Catterall WA. Reduced sodium current in Purkinje neurons from Nav1.1 mutant mice: implications for ataxia in severe myoclonic epilepsy in infancy. J Neurosci. 2007;27:11065–11074. doi: 10.1523/JNEUROSCI.2162-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya K, Kaneda M, Sugawara T, Mazaki E, Okamura N, Montal M, Makita N, Tanaka M, Fukushima K, Fujiwara T, Inoue Y, Yamakawa K. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. J Neurosci. 2004;24:2690–2698. doi: 10.1523/JNEUROSCI.3089-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JQ, Macdonald RL. Making sense of nonsense GABAA receptor mutations associated with genetic epilepsies. Trends Mol Med. 2009;15:430–438. doi: 10.1016/j.molmed.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JA, Plummer NW, Smith MR, Kapur J, Cummins TR, Waxman SG, Goldin AL, Meisler MH. A gain-of-function mutation in the sodium channel gene Scn2a results in seizures and behavioural abnormalities. Neuroscience. 2001;102:307–317. doi: 10.1016/s0306-4522(00)00479-6. [DOI] [PubMed] [Google Scholar]
- Kearney JA, Wiste AK, Stephani U, Trudeau MM, Siegel A, RamachandranNair R, Elterman RD, Muhle H, Reinsdorf J, Shields WD, Meisler MH, Escayg A. Recurrent de novo mutations of SCN1A in severe myoclonic epilepsy of infancy. Pediatr Neurol. 2006a;34:116–120. doi: 10.1016/j.pediatrneurol.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Kearney JA, Meisler MH. Encyclopedia of Basic Epilepsy Research. London: Elsevier/Academic Press; 2009. Single gene mutations in inherited and sporadic epilepsy. [Google Scholar]
- Kearney JA, Yang Y, Beyer B, Bergren SK, Claes L, Dejonghe P, Frankel WN. Severe epilepsy resulting from genetic interaction between Scn2a and Kcnq2. Hum Mol Genet. 2006b;15:1043–1048. doi: 10.1093/hmg/ddl019. [DOI] [PubMed] [Google Scholar]
- Lossin C. A catalog of SCN1A variants. Brain Dev. 2009;31:114–130. doi: 10.1016/j.braindev.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Madia F, Striano P, Gennaro E, Malacarne M, Paravidino R, Biancheri R, Budetta M, Cilio MR, Gaggero R, Pierluigi M, Minetti C, Zara F. Cryptic chromosome deletions involving SCN1A in severe myoclonic epilepsy of infancy. Neurology. 2006;67:1230–1235. doi: 10.1212/01.wnl.0000238513.70878.54. [DOI] [PubMed] [Google Scholar]
- Maier SK, Westenbroek RE, Yamanushi TT, Dobrzynski H, Boyett MR, Catterall WA, Scheuer T. An unexpected requirement for brain-type sodium channels for control of heart rate in the mouse sinoatrial node. Proc Natl Acad Sci U S A. 2003;100:3507–3512. doi: 10.1073/pnas.2627986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini C, Scheffer IE, Nabbout R, Mei D, Cox K, Dibbens LM, McMahon JM, Iona X, Carpintero RS, Elia M, Cilio MR, Specchio N, Giordano L, Striano P, Gennaro E, Cross JH, Kivity S, Neufeld MY, Afawi Z, Andermann E, Keene D, Dulac O, Zara F, Berkovic SF, Guerrini R, Mulley JC. SCN1A duplications and deletions detected in Dravet syndrome: Implications for molecular diagnosis. Epilepsia. 2010 doi: 10.1111/j.1528-1167.2009.02013.x. in press. [DOI] [PubMed] [Google Scholar]
- Martin MS, Tang B, Papale LA, Yu FH, Catterall WA, Escayg A. The voltage-gated sodium channel Scn8a is a genetic modifier of severe myoclonic epilepsy of infancy. Hum Mol Genet. 2007;16:2892–2899. doi: 10.1093/hmg/ddm248. [DOI] [PubMed] [Google Scholar]
- Martin MS, Dutt K, Papale LA, Dube CM, Dutton SB, de Haan G, Shankar A, Tufik S, Meisler MH, Baram TZ, Goldin AL, Escayg A. Altered function of the SCN1A voltage-gated sodium channel leads to GABAergic interneuron abnormalities. J Biol Chem. 2010;285:9823–2834. doi: 10.1074/jbc.M109.078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler MH, Kearney JA. Sodium channel mutations in epilepsy and other neurological disorders. J Clin Investig. 2005;115:2010–2017. doi: 10.1172/JCI25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra SN, Kahlig KM, George AL., Jr Impaired NaV1.2 function and reduced cell surface expression in benign familial neonatal-infantile seizures. Epilepsia. 2008;49:1535–1545. doi: 10.1111/j.1528-1167.2008.01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar MA, Baker MD, Levato A, Ingram R, Mallucci G, McMahon SB, Wood JN. Nerve injury induces robust allodynia and ectopic discharges in Nav1.3 null mutant mice. Mol Pain. 2006;2:33. doi: 10.1186/1744-8069-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar MA, Stirling LC, Forlani G, Baker MD, Matthews EA, Dickenson AH, Wood JN. Nociceptor-specific gene deletion reveals a major role for Nav1.7 (PN1) in acute and inflammatory pain. Proc Natl Acad Sci U S A. 2004;101:12706–12711. doi: 10.1073/pnas.0404915101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara I, Ito K, Sawaishi Y, Osaka H, Mazaki E, Inoue I, Montal M, Hashikawa T, Shike T, Fujiwara T, Inoue Y, Kaneda M, Yamakawa K. De novo mutations of voltage-gated sodium channel αII gene SCN2A in intractable epilepsies. Neurology. 2009;73:1046–1053. doi: 10.1212/WNL.0b013e3181b9cebc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara I, Miyamoto H, Morita N, Atapour N, Mazaki E, Inoue I, Takeuchi T, Itohara S, Yanagawa Y, Obata K, Furuichi T, Hensch TK, Yamakawa K. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papale LA, Beyer B, Jones JM, Sharkey LM, Tufik S, Epstein M, Letts VA, Meisler MH, Frankel WN, Escayg A. Heterozygous mutations of the voltage-gated sodium channel SCN8A are associated with spike-wave discharges and absence epilepsy in mice. Hum Mol Genet. 2009;18:1633–1641. doi: 10.1093/hmg/ddp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino GA, Claes LR, Lopez-Santiago LF, Slat EA, Dondeti RS, Chen C, O'Malley HA, Gray CB, Miyazaki H, Nukina N, Oyama F, De Jonghe P, Isom LL. A functional null mutation of SCN1B in a patient with Dravet syndrome. J Neurosci. 2009;29:10764–10778. doi: 10.1523/JNEUROSCI.2475-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S, Vieira JP, Barroca F, Roll P, Carvalhas R, Cau P, Sequeira S, Genton P, Szepetowski P. Severe epilepsy, retardation, and dysmorphic features with a 2q deletion including SCN1A and SCN2A. Neurology. 2004;63:191–192. doi: 10.1212/01.wnl.0000132844.20654.c1. [DOI] [PubMed] [Google Scholar]
- Planells-Cases R, Caprini M, Zhang J, Rockenstein EM, Rivera RR, Murre C, Masliah E, Montal M. Neuronal death and perinatal lethality in voltage-gated sodium channel αII-deficient mice. Biophys J. 2000;78:2878–2891. doi: 10.1016/S0006-3495(00)76829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer NW, McBurney MW, Meisler MH. Alternative splicing of the sodium channel SCN8A predicts a truncated two-domain protein in fetal brain and non-neuronal cells. J Biol Chem. 1997;272:24008–24015. doi: 10.1074/jbc.272.38.24008. [DOI] [PubMed] [Google Scholar]
- Scheffer IE, Berkovic SF. Temporal lobe epilepsy and GEFS+ phenotypes associated with SCN1B mutations. Brain. 2007;130:100–109. doi: 10.1093/brain/awl272. [DOI] [PubMed] [Google Scholar]
- Shi X, Yasumoto S, Nakagawa E, Fukasawa T, Uchiya S, Hirose S. Missense mutation of the sodium channel gene SCN2A causes Dravet syndrome. Brain Dev. 2009;31:758–762. doi: 10.1016/j.braindev.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Singh NA, Pappas C, Dahle EJ, Claes LR, Pruess TH, De Jonghe P, Thompson J, Dixon M, Gurnett C, Peiffer A, White HS, Filloux F, Leppert MF. A role of SCN9A in human epilepsies, as a cause of febrile seizures and as a potential modifier of Dravet syndrome. PLoS Genet. 2009;5:e1000649. doi: 10.1371/journal.pgen.1000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suls A, Claeys KG, Goossens D, Harding B, Van Luijk R, Scheers S, Deprez L, Audenaert D, Van Dyck T, Beeckmans S, Smouts I, Ceulemans B, Lagae L, Buyse G, Barisic N, Misson JP, Wauters J, Del-Favero J, De Jonghe P, Claes LR. Microdeletions involving the SCN1A gene may be common in SCN1A-mutation-negative SMEI patients. Human Mutat. 2006;27:914–920. doi: 10.1002/humu.20350. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tang B, Escayg A. A BAC transgenic mouse model reveals neuron subtype-specific effects of a Generalized Epilepsy with Febrile Seizures Plus (GEFS+) mutation. Neurobiology of Disease. 2009;35:91–102. doi: 10.1016/j.nbd.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau MM, Dalton JC, Day JW, Ranum LP, Meisler MH. Heterozygosity for a protein truncation mutation of sodium channel SCN8A in a patient with cerebellar atrophy, ataxia, and mental retardation. J Med Genet. 2006;43:527–530. doi: 10.1136/jmg.2005.035667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RH, Wang DW, Singh R, Scheffer IE, George AL, Jr, Phillips HA, Saar K, Reis A, Johnson EW, Mulley JC. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel beta1 subunit gene SCN1B. Nat genet. 1998;19:366–370. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- Wallace RH, Scheffer IE, Parasivam G, Barnett S, Wallace GB, Sutherland GR, Berkovic SF, Mulley JC. Generalized epilepsy with febrile seizures plus: mutation of the sodium channel subunit SCN1B. Neurology. 2002;58:1426–1429. doi: 10.1212/wnl.58.9.1426. [DOI] [PubMed] [Google Scholar]
- Wang J, Ou SW, Wang YJ, Kameyama M, Kameyama A, Zong ZH. Analysis of four novel variants of Nav1.5/SCN5A cloned from the brain. Neurosci Res. 2009;64:339–347. doi: 10.1016/j.neures.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Whitaker WR, Clare JJ, Powell AJ, Chen YH, Faull RL, Emson PC. Distribution of voltage-gated sodium channel α-subunit and β-subunit mRNAs in human hippocampal formation, cortex, and cerebellum. J Comp Neurol. 2000;422:123–139. doi: 10.1002/(sici)1096-9861(20000619)422:1<123::aid-cne8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Yamakawa K. Epilepsy and sodium channel gene mutations: gain or loss of function? Neuroreport. 2005;16:1–3. doi: 10.1097/00001756-200501190-00001. [DOI] [PubMed] [Google Scholar]
- Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, Spain WJ, McKnight GS, Scheuer T, Catterall WA. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006;9:1142–1149. doi: 10.1038/nn1754. [DOI] [PubMed] [Google Scholar]