SUMMARY
In an effort to find new pharmacological modalities to overcome resistance to ATP-site inhibitors of Bcr-Abl, we recently reported the discovery of GNF-2, a selective allosteric Bcr-Abl inhibitor. Here, using solution NMR, X-ray crystallography, mutagenesis and hydrogen exchange mass spectrometry we demonstrate that GNF-2 binds to the myristate binding site of Abl, leading to changes in the structural dynamics of the ATP-binding site. GNF-5, an analog of GNF-2 having improved pharmacokinetic properties, when utilized in combination with the ATP-competitive inhibitors imatinib or nilotinib, suppressed the emergence of resistance mutations in vitro, displayed additive inhibitory activity in biochemical and cellular assays against T315I Bcr-Abl and displayed in vivo efficacy against the recalcitrant T315I Bcr-Abl mutant in a murine bone-marrow transplantation model. These results demonstrate that therapeutically relevant inhibition of Bcr-Abl activity can be achieved using inhibitors that bind to the myristate binding site and that combining allosteric and ATP-competitive inhibitors can overcome resistance to either agent alone.
INTRODUCTION
Chronic myelogenous leukemia (CML) is a hematological malignancy caused by a chromosomal rearrangement that generates a fusion protein, Bcr-Abl, with deregulated tyrosine kinase activity. Although clinical remission is usually achieved in early-stage disease with the ATP-site targeting drug imatinib, advanced-stage patients may relapse due to the emergence of clones expressing inhibitor-resistant forms of Bcr-Abl. Two recently approved drugs nilotinib (AMN107)1 and dasatinib (BMS-354825)2,3 address the majority of the imatinib resistance mutations with the exception of the “gatekeeper” T315I mutation, which is situated in the middle of the ATP-binding cleft4,5,6.
GNF-2 is a highly selective non-ATP competitive inhibitor of oncogenic Bcr-Abl activity (IC50 = 0.14 µM)7. Using nuclear magnetic resonance spectroscopy (NMR) and X-ray crystallography, we identify the myristoyl pocket located near the C-terminus of Abl kinase domain as the precise binding site of GNF-2 to Bcr-Abl. By selecting for Bcr-Abl alleles resistant to GNF-2 in vitro, we identified residues both within and outside of the myristate cleft that are required for drug efficacy. Simultaneous binding to Bcr-Abl of a myristoyl mimic and an ATP-competitive inhibitor reduces the appearance of resistance-conferring mutations and results in inhibition of both wild-type and T315I Bcr-Abl kinase activity and cell growth. Hydrogen exchange mass spectrometry demonstrates that binding of GNF-5 to the myristate pocket results in alterations to the conformational dynamics of the ATP-site and provides a possible mechanism for allosteric communication between these sites.
GNF-2 binds to the C-terminal myristate pocket of Abl
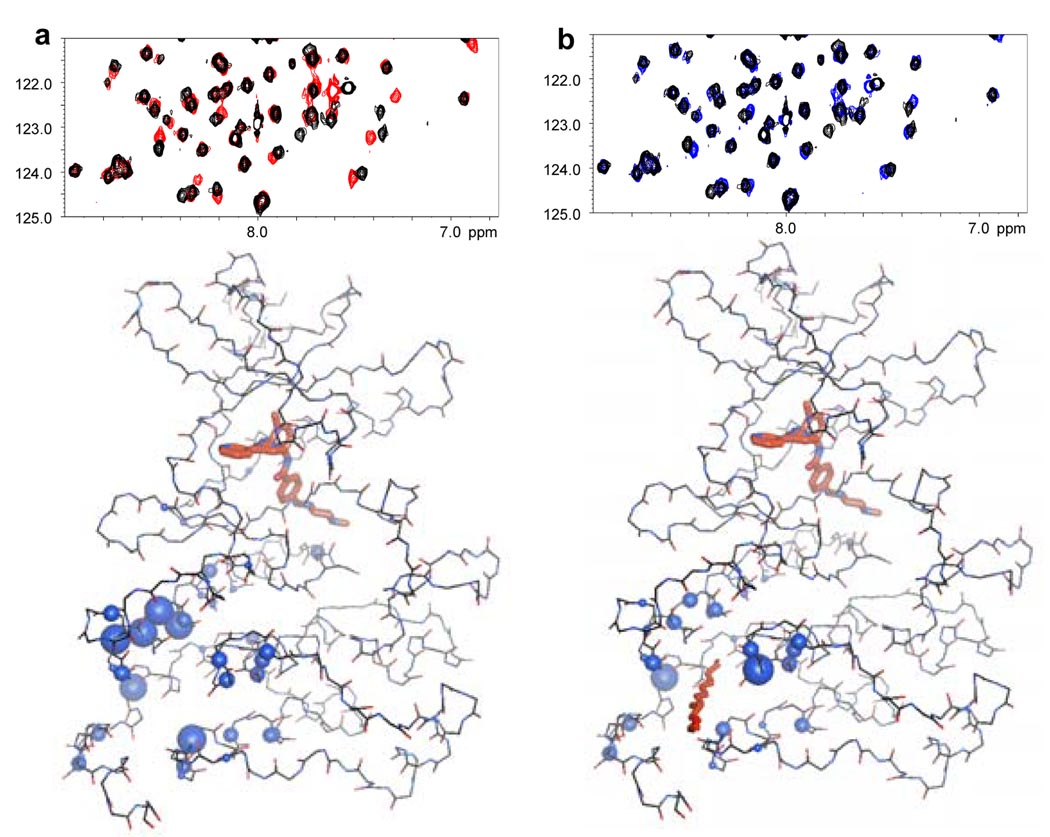
GNF-2 had previously been suggested to bind in the myristate binding pocket of Abl based upon the observation that engineered mutations located at the entrance (A337N) and rear (A344L) of the myristoyl cleft conferred resistance to GNF-2, but not to imatinib7. In order to establish the GNF-2 binding site by an independent biophysical method, we used solution NMR on the Abl/imatinib/GNF-2 complex 10 11, to demonstrate that GNF-2 induces chemical shift changes that cluster around the myristate binding pocket (Fig. 1a). No significant chemical shift perturbations were observed for the ATP pocket, indicating that GNF-2 does not interfere with imatinib for binding at the ATP-site. Myristic acid was found to induce qualitatively the same pattern of chemical shift perturbations (Fig. 1b), providing additional evidence that GNF-2 and myristate share the same binding site.
Figure 1. NMR spectroscopy provides evidence for GNF-2 binding to the c-terminal myristate pocket of Abl.
a, An HSQC spectrum of the Abl/imatinib complex with (red) and without (black) GNF-2 (top) showing chemical shift changes induced by ligand binding. Mapping of the chemical shift changes to the structure of the Abl/imatinib complex (PDB entry 1OPK8 bottom) identifies the myristate pocket as the GNF-2 binding site. The size of the spheres is proportional to the magnitude of the chemical shift changes. b, Same as a except myristic acid used instead of GNF-2.
Crystal structure of GNF-2/Abl/imatinib complex
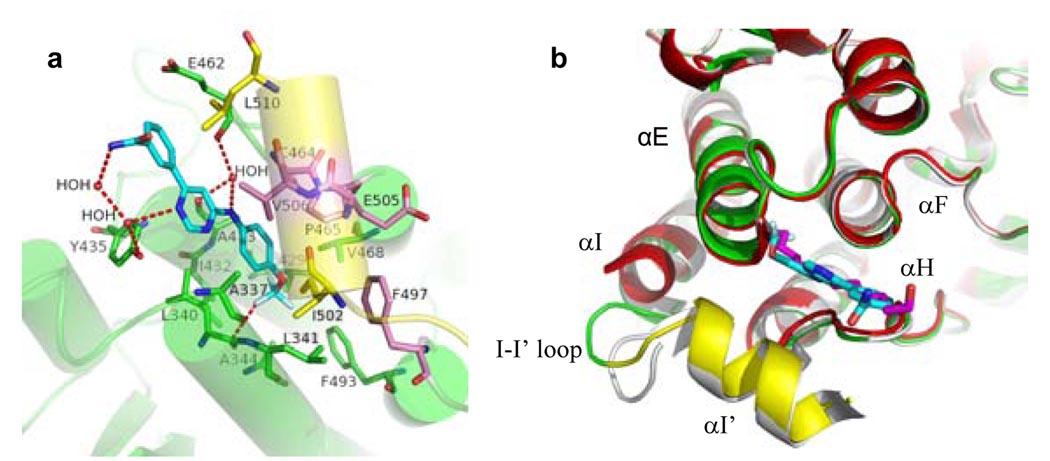
The binding of GNF-2 to the myristoyl pocket of Abl was further confirmed by X-ray crystallography. The structure of the Abl/imatinib/GNF-2 complex was obtained by soaking crystals of Abl/imatinib/myristate, obtained as described by Nagar et al8, in an excess of GNF-2. As judged by the shape of the electron density (Fig. S1), GNF-2 replaces the myristoylated peptide in the crystals. There are two molecules in the asymmetric unit and the myristate site is fully occupied by GNF-2 in one and partially in the other. GNF-2 binds in an extended conformation in the myristate pocket with the CF3-group buried at the same depth as the final two carbons of the myristate ligand (Fig. 2). There is a favorable, but probably weak, polar interaction between one fluorine atom and the main chain of L340 (similar to that observed between nilotinib and D381 of Abl)12, and there are water-mediated hydrogen bonds, but no direct hydrogen bonds with the protein. As expected, the majority of the interactions between GNF-2 and the protein are hydrophobic. As discussed below, mutation of three residues near the mouth of the myristate site (C464Y, P465S, and V506L) is found to cause resistance to the binding of GNF-2, presumably for steric reasons. The overall structure of the Abl kinase domain complexed to GNF-2 is similar to that of the myristate complex (Fig. 2b) with some small differences that likely result from crystal contacts, but no changes in the ATP-site.
Figure 2. Crystal structure of GNF-2 bound to the Abl myristoyl pocket.
a, Abl kinase is indicated in green (helices indicated with transparent cylinders) with the bent part of the I-helix in yellow, GNF-2 resistance mutations in pink, and GNF-2 carbons in cyan. H-bonding and other polar interactions are indicated by dotted red lines. b, Superposition of the Abl–imatinib–myr (white), Abl-imatinib-GNF-2 (green and yellow), Abl-imatinib (red) structures. GNF-2 is colored in cyan and myristic acid in magenta.
GNF-2 analog structure-activity relationships
Following the identification of GNF-2 as a lead compound, a systematic evaluation of the structural features necessary to impart function as a cellular Bcr-Abl inhibitor were investigated through the synthesis of over two hundred analogs and rationalized in the context of binding to the myristate binding site (to be described elsewhere and Fig. S2a). The structure-activity relations are fully consistent with the conformation of GNF-2 observed in the crystal structure: the trifluoromethoxy group of GNF-2 can only be accommodated at the para-position, the aniline NH is required due to formation of a water mediated H-bond to the backbone carbonyls of A433 and E462, water-mediated hydrogen bonds between the carboxamide of GNF-2 and Abl confer enhanced inhibitory activity, and the extended compound conformation is required to fit the cylindrically shaped binding cavity.
GNF-2 and imatinib combinations reduce the emergence of resistant mutants
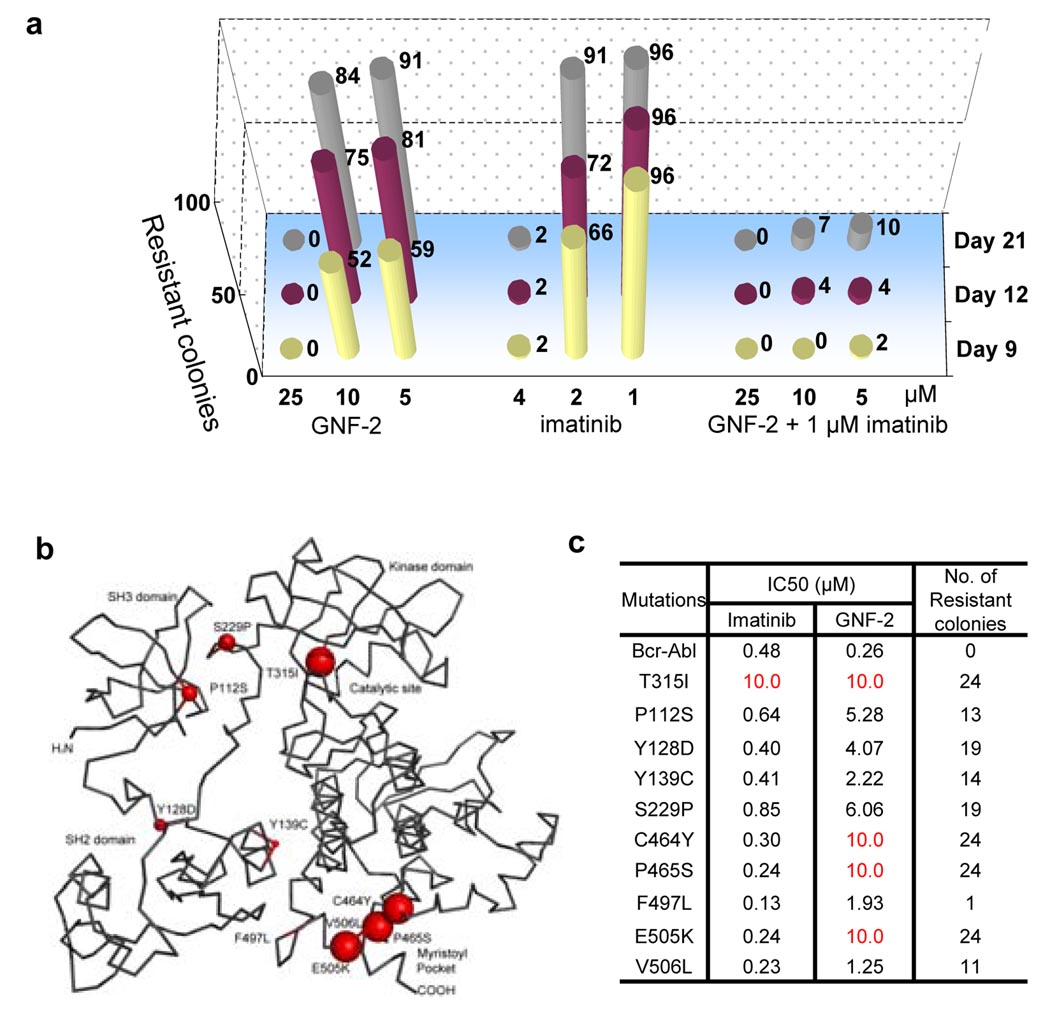
We sought to investigate the frequency with which Bcr-Abl dependent Ba/F3 cells would become resistant to combinations of GNF-2 and imatinib compared to each compound alone. The number of resistant clones that emerged as a result of continuous exposure to 1 µM imatinib was reduced by at least 90% when cells were treated for up to 21 days with 1 µM imatinib combined with 5 or 10 µM of GNF-2 (Fig. 3a). These results demonstrate that combinations of GNF-2 and imatinib can cooperate to suppress the emergence of resistance mutations.
Figure 3. Location and cellular IC50 of Bcr-Abl GNF-2 resistance mutations.
a, Effect of various concentrations of GNF-2, imatinib, or combinations of both on the number of emerging Ba/F3.Bcr-Abl resistant clones (shown on the top of each bar). b, Mutations indicated by red spheres on Abl with size proportional to the degree of resistance (PDB ID 1OPK8, amino acid residues numbering of Abl 1a). c, IC50 for growth inhibition by imatinib or GNF-2 for wild-type and mutant Bcr-Abl transformed Ba/F3 cells. The number of colonies that emerge after 12 days in the presence of 20 µM GNF-2 is indicated.
Identification of the Bcr-Abl mutants resistant to GNF-2
In order to discover the full complement of Bcr-Abl mutants that induce resistance to GNF-2 we performed two types of selections. In the first, Bcr-Abl transformed Ba/F3 cells were cultured in the presence of increasing concentrations of GNF-2 to allow cells to evolve drug resistance as previously described for the ATP-competitive inhibitor PD 16632613. In the second approach, Bcr-Abl was randomly mutated in E. coli and the mutant clones were expressed in Ba/F3 cells, which were then grown in the presence of inhibitor14. These screens resulted in the identification of a total of 306 mutants, 163 (12 sites) from the first and 143 (22 sites) from the second (Fig. S3). Strikingly, more than 80% of the resistant colonies contain Bcr-Abl mutations clustered in the myristate binding pocket or SH2 and SH3 domains (Fig. S3). This is in contrast to ATP-competitive inhibitors such as imatinib, PD166321, and AP2346413–15 for which the majority of resistance mutations cluster adjacent to the kinase catalytic site.
To validate the functional relevance of these mutations, we engineered individual mutant Bcr-Abl transformed Ba/F3 cells for nine of the most frequently isolated GNF-2 resistant mutations and for the T315I gatekeeper mutation. These selected mutations located in the SH3 domain (P112S), the SH3-SH2 domain linker (Y128D), the SH2 domain (Y139C), the SH2-kinase domain linker (S229P), the ATP-binding site (T315I), and adjacent to the myristate-binding site (C464Y, P465S, F497L, E505K, Y506L) were individually introduced into Bcr-Abl by site-directed mutagenesis. Of these, only the T315I substitution has previously been reported to confer resistance to imatinib16,17. GNF-2 displayed an IC50 against all ten mutants that was elevated by 5- to 50-fold relative to wild-type Bcr-Abl transformed Ba/F3 cells (Fig. 3b). The three most frequently recovered mutations (60% of total) were located in close proximity to the myristate binding site (C464Y, P465S, and E505K) and were demonstrated to confer complete resistance to GNF-2 up to a concentration of 10 µM (Fig. 3c).
In order to examine how the mutations affected the ability of GNF-2 to inhibit Bcr-Abl mediated signaling, we examined Bcr-Abl autophosphorylation and phosphorylation of a downstream substrate, STAT5, following inhibitor treatment (Fig. S4a). At a concentration of 10 µM, GNF-2 could inhibit Bcr-Abl and STAT5 phosphorylation of all mutants with the exception of the three myristate-site mutations (E505K, P465S and C646Y) and the gatekeeper T315I mutation.
Mutations in the myristate pocket interfere with GNF-2 binding
We tested the ability of mutant Bcr-Abl proteins, obtained from crude cell lysates, to bind to a GNF-2 affinity resin (Fig. S4b,c)7. These experiments revealed that only the mutations located in the myristate-binding site (C464Y, P465S, and E505K) ablated binding of Bcr-Abl to GNF-2 while all other mutations retained binding to GNF-2. Using an NMR-based titration, we confirmed that GNF-2 still binds to the gatekeeper mutant, T315I Abl (residues 229–500, not including helix I), albeit with a two-fold reduced affinity compared to wild-type (Fig. S5). These results suggest that the myristate-site mutations directly interfere with drug binding while the non-myristate site mutants function through a different mechanism, which may involve disfavoring the inhibited conformation induced upon binding of GNF-2.
GNF-5 and nilotinib combinations inhibit T315I Bcr-Abl
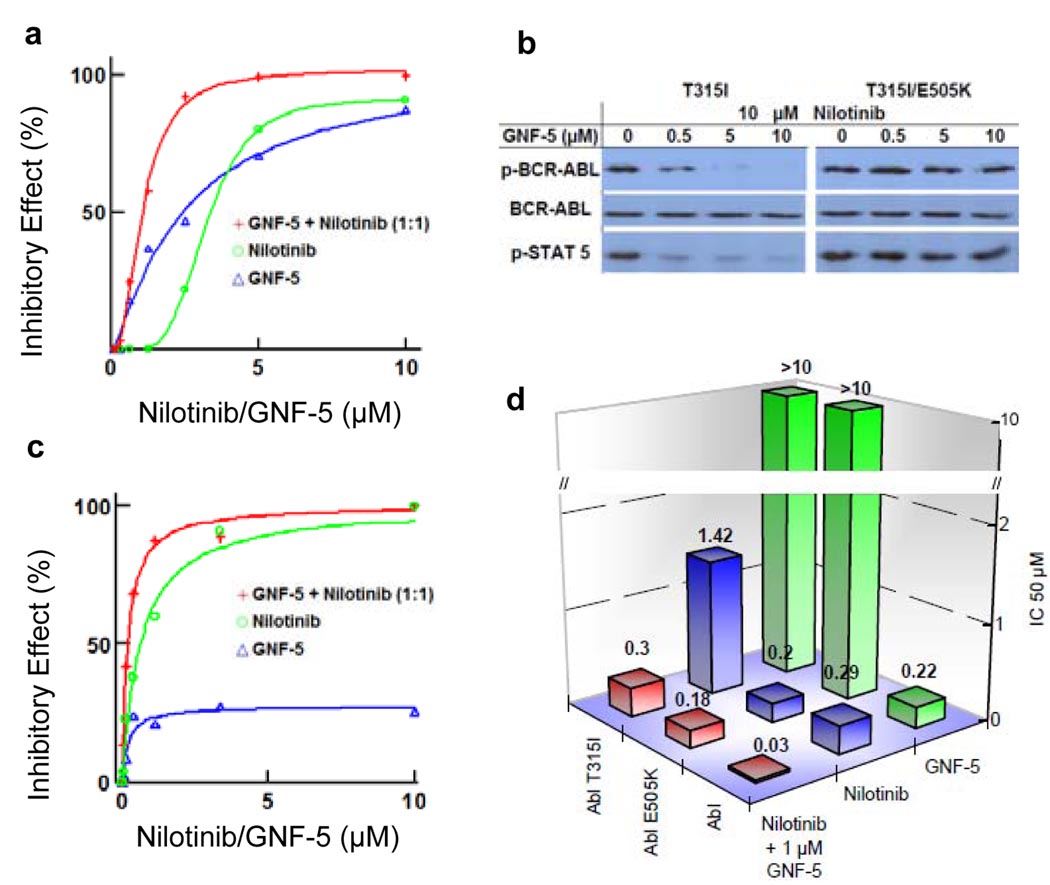
GNF-5, the N-hydroxyethyl carboxamide analog of GNF-2 possessing similar cellular Bcr-Abl inhibitory activity but having more favorable pharmacokinetic properties, was chosen for further in vitro and in vivo single and combination studies (Fig. S2b). Combinations of GNF-5 and nilotinib inhibited T315I Bcr-Abl-dependent cell growth with a calculated combination index (CI)18 of 0.6 indicating moderate synergy (Fig. 4a). For example, at a fixed GNF-5 concentration of 2 µM, nilotinib inhibits T315I Bcr-Abl-dependent proliferation with an IC50 of 0.8 ± 0.05 µM. Flow cytometry analysis showed that GNF-5 and nilotinib act additively to inhibit STAT5 phosphorylation which could be rescued by addition of IL-3 to the medium (Fig. S6a). Nilotinib and GNF-5 also exhibited cooperativity for inhibition of wild-type Bcr-Abl transformed Ba/F3 cells with a calculated combination index of 0.6 (Fig. S6b). We confirmed that the cooperativity observed between GNF-5 and nilotinib is directly mediated by inhibition of Bcr-Abl based upon the ability of a double mutation of T315I in the ATP-site and E505K in the myristate site to confer complete resistance to the combination of both inhibitors in phosphorylation (Fig 4b) and proliferation assays (Fig S6c & d). GNF-5 and nilotinib also acted cooperatively against p190 Bcr-Abl, a variant commonly found in acute lymphocytic leukemia which typically responds only transiently to imatinib therapy19, with a calculated combination index of 0.5 (Fig. S7).
Figure 4. Cellular and enzymatic inhibition of wild-type and mutants by combination treatments.
a, Effects of GNF-5, nilotinib and varying concentrations of GNF-5 in combination with nilotinib (0.3–10 µM) on the proliferation of T315I Bcr-Abl Ba/F3 cells. The combination curve (red, as well in Fig 4c) contains twice the total drug concentration of the single agent curves due to both drugs being present. b, Inhibition of Bcr-Abl autophosphorylation was determined by Bcr-Abl immunoprecipitation, followed by a immunoblot for phospho-Tyr (Y412)1, phospho-STAT5 (Y694) and total Bcr-Abl (antibody K-12) from cell lystates obtained after treatment of T315I Bcr-Abl expressing Ba/F3 with 10 µM of nilotinib and increasing concentrations of GNF-5 (0, 0.5, 5 and 10 µM) for 90 min. c, Percent inhibition of T315I Abl kinase by nilotinib and GNF-5 or the combination. d, IC50 for inhibition of wild-type, E505K and T315I Abl kinase activity by GNF-5, nilotinib or the combination at an ATP concentration of 20 µM.
Biochemical characterization of GNF-5 in combination with imatinib and nilotinib
To determine whether the additive interaction between GNF-5 and the ATP-competitive inhibitors imatinib and nilotinib observed in cellular assays could be confirmed at the protein level, we performed steady-state kinetic analyses of Abl kinase using a pyruvate kinase-lactate dehydrogenase detection system 20. We first tested the inhibitory activity of imatinib, nilotinib and GNF-5 in bacterially expressed wild-type, T315I, and E505K Abl kinases (Fig. S8). Inhibition of wild-type Abl was observed for all three inhibitors with GNF-5 exhibiting an IC50 = 0.22 ± 0.01 µM, imatinib IC50 = 0.24 ± 0.03 µM and nilotinib IC50 = 0.29 ± 0.06 µM using an ATP concentration of 20 µM, which is close to the apparent Km under our assay conditions. Although the activity of recombinant Abl was previously reported to be insensitive to GNF-27, we subsequently discovered that this was due to the presence of Brij-35, a common additive to kinase assay buffers, which masked inhibition by GNF-221. The myristate site mutant E505K was inhibited by imatinib with an IC50 = 0.22 ± 0.03 µM and nilotinib with an IC50 = 0.20 ± 0.01 µM, but not by GNF-5 (IC50 > 10 µM). The T315I mutant was not inhibited by imatinib or GNF-5, but nilotinib did exhibit weak activity against this mutant with an IC50 = 1.42 ± 0.3 µM. GNF-5 was confirmed to inhibit wild-type Abl in a non-ATP competitive fashion (Fig. S9).
We next examined whether combinations of GNF-5 and nilotinib resulted in additive inhibition of wild-type, T315I, or E505K recombinant Abl proteins. Positive cooperativity was observed for combinations of GNF-5 and nilotinib on the wild-type and T315I enzymes with calculated combination indices of 0.53 and 0.61, respectively (Fig. 4c,d and Fig S10). For example at a fixed GNF-5 concentration of 1 µM and an ATP concentration of 20 µM, the IC50 values of nilotinib against T315I and wild-type enzyme were reduced by 4.7 and 9.6-fold respectively compared to those calculated in the absence of GNF-5. As expected, no additivity was observed with the E505K myristate site mutant. These results demonstrate that allosteric communication between the myristate and ATP-site can be observed in biochemical kinase assays.
GNF-5 binding alters the conformation of the ATP-binding site
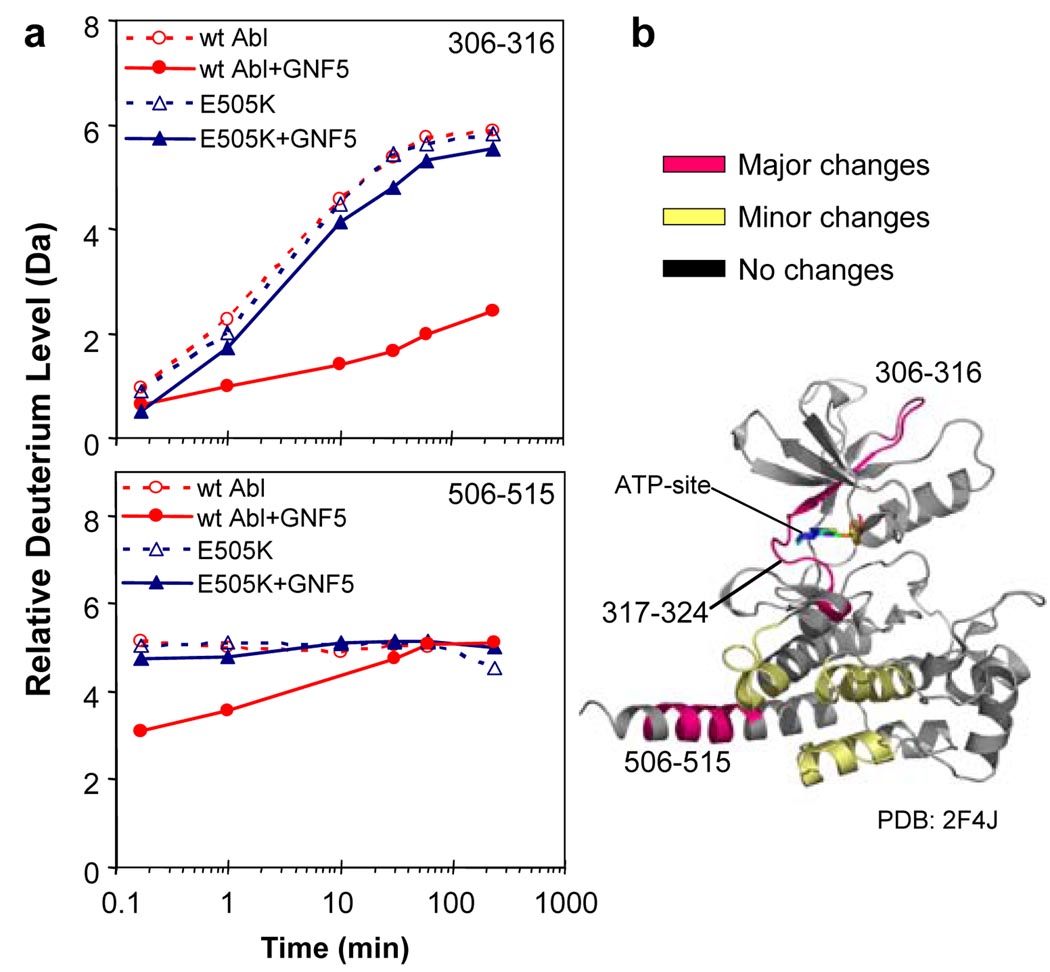
In order to examine how the binding of GNF-5 to the myristate binding site might influence the conformational dynamics of the ATP-binding site and other regions of Abl, we performed hydrogen exchange mass spectrometry (HX MS). HX MS allows the dynamics of a protein to be investigated by measuring the exchange of backbone amide hydrogens with the bulk solvent22. Unbound Abl and the GNF-5:Abl complex were independently exposed to D2O for periods ranging from 10 seconds to 4 hours as described previously23. Changes in deuterium incorporation in the presence of GNF-5 were observed in several peptides (Fig. 5) surrounding the myristate binding cleft. In addition, changes in peptides near the ATP-binding site (e.g., residues 306–316, 317–324) were also seen, implying that binding of GNF-5 binding affected the ATP-site. Hydrogen exchange in these peptides was not altered in a control experiment with GNF-5 and the non-binding Abl E505K myristate mutant, indicating that the binding of GNF-5 is what is responsible for the altered conformation and hydrogen exchange in the peptides near the ATP-site. These results demonstrate that ligation of the myristate-binding site can cause dynamic perturbations to residues in the ATP-binding site and provides a mechanism by which synergistic interactions between these two sites could occur.
Figure 5. Hydrogen exchange mass spectrometry upon binding of GNF-5 to Abl.
a, Deuterium uptake curves for the peptides 306–316 and 506–515. The y-axis maximum corresponds to the theoretical maximum amount of deuterium that could be incorporated into this peptide. No alterations in deuterium incorporation were seen with the E505K mutant whereas exchange was affected by GNF-5 binding to the wild-type protein. b, Location of the peptides that showed differences in deuterium incorporation mapped on the crystal structure of active Abl (PDB: 2F4J). This crystal structure was chosen because in the absence of myristoylation (as for the protein used here), Abl protein is believed to be in this conformation23. Major changes (colored magenta) were defined as a difference between exchange curves of 1.0 Da or more. Minor changes (colored yellow) were 0.4–1.0 Da. No changes (colored grey) were differences of 0.0–0.4 Da.
GNF-5 and nilotinib combinations inhibit T315I Bcr-Abl in vivo
The murine pharmacokinetic parameters of GNF-5 were measured and determined to be suitable for using the compound in vivo (Fig. S11). GNF-5 was demonstrated to be efficacious in vivo at well tolerated doses in a murine xenograft model of p210 Bcr-Abl Ba/F3 induced leukemia, but relapses were observed (Fig 6a and Fig S12). The in vivo target modulation was further confirmed by examining phosphorylation of STAT5 (Fig. S13).
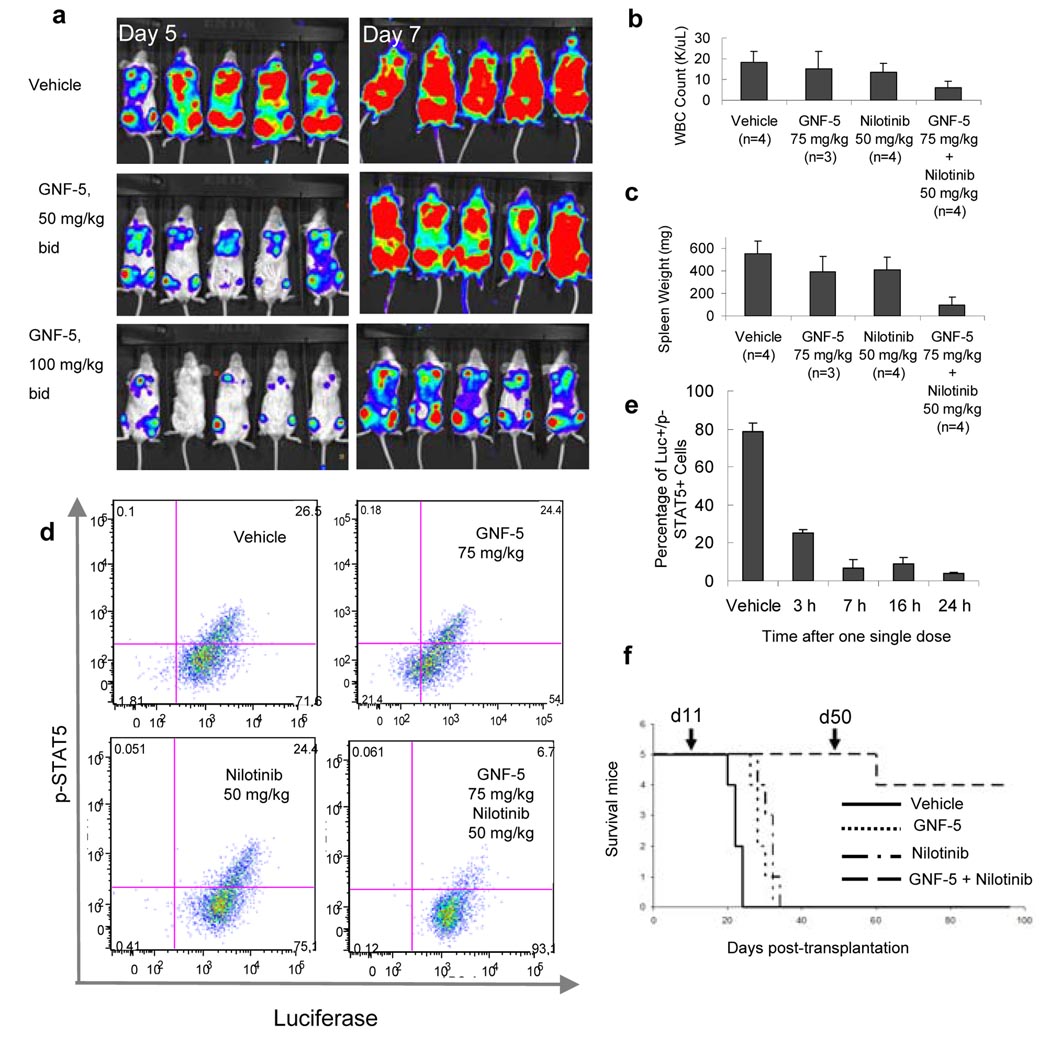
Figure 6. In vivo efficacy studies with GNF-5 on wild-type and T315I Bcr-Abl dependent proliferation in xenograft and bone marrow transplantation models.
a, Images of whole body luminescence of wild-type Bcr-Abl and luciferase expressing Ba/F3 cells on days five and seven after treatment with vehicle, GNF-5 50 mg/kg and 100 mg/kg b.i.d. b, Average white blood cell counts for vehicle, nilotinib, GNF-5, or combination treatments in the T315I Bcr-Abl bone marrow transplantation efficacy study. c, Spleen weight for vehicle, nilotinib, GNF-5, or combination treatments in the T315I Bcr-Abl bone marrow transplantation efficacy study. d, Quantification of p-STAT5-positive cells by flow cytometry in the different treatment groups after repeated doses. e, Time course inhibition of STAT5 phosphorylation after a single dose of GNF-5 and nilotinib combination. f, Kaplan-Meier plot showing survival of mice (n = 5 mice per group) transplanted with T315I Bcr-Abl transduced bone marrow and treated with vehicle (solid line), 75 mg/kg b.i.d. GNF-5 (dotted line), 50 mg/kg b.i.d. nilotinib (dots and dashes), or a combination of 75 mg/kg b.i.d. GNF-5 plus 50 mg/kg b.i.d. nilotinib (dashed line). Compound dosing was initiated on day 11 post-transplantation and discontinued on day 50 (indicated by arrows). Error bars are s.e.m.
GNF-5 and nilotinib combinations against T315I Bcr-Abl
To further evaluate the in vivo efficacy of GNF-5 on wild-type and T315I Bcr-Abl, we used a bone marrow transduction/transplantation (BMT) mouse model that more closely resembles human CML disease24. Initial experiments using p210 Bcr-Abl demonstrated that 50 mg/kg GNF-5 twice daily could normalize blood counts and spleen size (Fig. S14). We then addressed whether the combination of GNF-5 with nilotinib would result in efficacy in a T315I Bcr-Abl BMT model. Mice treated b.i.d 15 days after transplantation with either nilotinib (50 mg/kg) or GNF-5 (75 mg/kg) alone showed no significant response compared to the vehicle group, with 2–3 fold higher cell counts and spleens four-fold larger than those of healthy mice. In contrast, the combination normalized blood cell counts and spleen size without signs of toxicity suggesting an additive effect of the compounds in a combination treatment (Fig. 6 b,c). In order to establish an efficacy/pharmacodynamic response correlation, bone marrow cells from the different mouse groups were isolated at the end of the efficacy study, stained with anti-p-STAT5 and anti-luciferase specific antibodies, and analyzed by flow cytometry. The percentage of p-STAT5 positive Bcr-Abl expressing bone marrow cells was similar (approx. 25%) in the vehicle, GNF-5, and nilotinib treated groups. In the combination group, the percentage of p-STAT5 positive cells was about 6%, reflecting a correlation between the tumor growth inhibition and a block in Bcr-Abl signaling (Fig. 6d). To determine the extent of the inhibition, mice transplanted with T315I Bcr-Abl expressing bone marrow cells were treated with a single dose of the combination (50 mg/kg nilotinib plus 75 mg/kg GNF-5) or vehicle on day 21 post-transplantation, and the bone marrow cells were collected and analyzed at 3, 7, 16, and 24 hours post-dose. In the vehicle group, about 80% of the luciferase-positive cells had phosphorylated STAT5. Three hours after dosing, STAT5 phosphorylation was reduced from 80% to 25% and, from 7 to 24 h, the number of p-STAT5 positive cells remained below 10%, demonstrating a strong and sustained inhibition of Bcr-Abl-mediated signaling following administration of the GNF-5/nilotinib combination (Fig. 6e).
In a third experiment, we monitored the survival of the mice transplanted with T315I Bcr-Abl transduced bone marrow and treated with GNF-5, nilotinib or the combination of the two. Mice transplanted with T315I Bcr-Abl transduced bone marrow and treated with vehicle control died by day 24 after transplantation, with a median survival of 22 days (Fig. 6f). GNF-5 (75 mg/kg b.i.d.) extended survival (median 28 days) significantly compared to vehicle treated controls (P = 0.023). Mice treated with nilotinib alone (50 mg/kg b.i.d.) also survived longer (median 32 days) than those treated with vehicle (P = 0.023). The overall survival of mice treated with GNF-5 plus nilotinib was improved compared to those treated with either GNF-5 alone (P = 0.002) or nilotinib alone (P = 0.002), with all the mice surviving by day 50 post-transplantation, after which the treatment was discontinued. Forty-six days after the combination treatment was completed, 4 out of 5 mice survived without signs of disease. Cumulatively, these results suggest that a combination of an ATP-competitive with an allosteric inhibitor may be a therapeutically appropriate strategy to target the T315I Bcr-Abl mutation.
DISCUSSION
The successful development of efficacious inhibitors against kinases such as Bcr-Abl, c-Kit and EGFR that are activated by genetic alterations has stimulated a massive effort to develop new ATP-competitive inhibitors targeted to a variety of kinases25,26. One major problem for all clinically approved ATP-competitive kinase inhibitors is resistance that results from the selection of drug-resistant mutant forms of the kinase target. One hotspot for resistance occurs at the “gatekeeper” residue, as it has been observed for Bcr-Abl4, EGFR27, FLT-328, c-KIT, SRC, PDGFRβ and FGFR-129. One strategy to overcome resistance mutations is to design new ATP-competitive inhibitors that derive potency and selectivity from alternative binding modes which has been clinically validated by the development of dasatinib and nilotinib, that target the majority of Bcr-Abl mutations with the exception of T315I. An alternative strategy is to find non-ATP competitive inhibitors that can allosterically regulate kinase activity. Allosteric inhibitors have been developed for multiple kinases including mTor30, Mek31, Akt 32, IKK33, Chk134, and CAMKII35.
GNF-2 and its analogs represent a new kind of non-ATP competitive Abl kinase inhibitors. The NMR, X-ray crystallography, mutagenesis and hydrogen exchange experiments are all consistent with binding of GNF-2/5 to the myristate-binding site located near the C-terminus of the kinase domain. Based upon our accumulated data, we propose the following model for the inhibition mechanism of GNF-2 class compounds. Binding of GNF-2/5 to the myristate site appears to induce a bent conformation of the αI helix that facilitates stabilization of an inhibited conformation. As supported by the mutagenesis studies and kinase assays, functional inhibition by GNF-2/5 requires the involvement of the SH3 and SH2 domains. The hydrogen exchange mass spectrometry data demonstrate that GNF-5 binding to the myristate-binding site results in dynamic or conformational changes at the ATP-site. This suggests that binding of GNF-2/5 to the myristate-binding site causes structural reorganization, possibly communicated via conformational rearrangement of other parts of Abl, that disrupts the catalytic machinery located in the ATP-site. As we currently do not have a crystal structure with a construct that contains the SH3-SH2-kinase domain of Abl bound to GNF-2, we cannot draw firm conclusions regarding the precise conformation of the kinase induced by GNF-2/5.
We demonstrate that GNF-2/5 can act cooperatively with an ATP competitive inhibitor to inhibit both wild-type and T315I Bcr-Abl in biochemical and cellular assays consistent with crystallographic and by differential scanning calorimetry results (Fig. S15). However, one puzzling question was why GNF-2/5 lacks significant biochemical and cellular potency against the T315I “gatekeeper” Abl mutation. T315I Abl biochemical kinase activity can be reduced by 20% at concentrations of GNF-5 below 0.5 µM but further increases in GNF-5 concentration up to 10 µM do not result in superior inhibition (Fig. 4c). Mutation of the gatekeeper residue does not interfere substantially with the ability of GNF-2/5 to bind to Abl as determined by NMR and affinity chromatography, but it appears to disfavor the GNF-2/5-induced inhibited conformation. The hydrogen exchange mass spectrometry data show that binding of GNF-5 results in reduced exchange in a peptide that contains the gatekeeper residue. While we have not proven that this exchange phenomenon is functionally relevant to the inhibitory mechanism of GNF-2/5, it does suggest that the conformation and/or dynamics of the gatekeeper containing segment may be coupled with the ability of GNF-2/5 to inhibit Abl36.
While non-ATP competitive inhibitors will also be subject to inhibitor resistance through point mutation, we have demonstrated that the combined application of ATP and non-ATP competitive inhibitors reduces the number of resistant clones that emerge as a response to continued exposure to a single agent. Furthermore we have shown that the combined treatment of GNF-5 with nilotinib led to in vivo efficacy resulting in complete disease remissions in a T315I Bcr-Abl mutant murine bone-marrow transplantation model. These findings should encourage the search for non-ATP competitive inhibitors that can selectively target the large number of kinases that become deregulated in cancer and other diseases.
METHODS SUMMARY
The full methods section provides experimental procedures for: (1) NMR spectroscopy, (2) Abl crystallography, (3) wild-type and mutant Bcr-Abl Ba/F3 cellular proliferation assays, (4) selection for clones resistant to GNF-2 and imatinib, (5) Abl protein expression, and purification, (6) kinetic characterization of Abl inhibition, (7) hydrogen exchange experiments, (8) In vivo studies of GNF-5 and drug combinations. The supplementary material describes the experimental procedures for protein crystallization and synthetic chemistry,
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
(1), NMR spectroscopy
All NMR experiments were carried out at 296 K on a Bruker AV600 NMR spectrometer at a proton resonance frequency of 600 MHz as described previously37.
(2), Abl crystallography
Crystals were grown as described in Nagar et al8 using the conditions listed in Table S1. After soaking for 7 days at 4°C in an excess of GNF-2, data were collected from a single crystal at beamline PXII of the Swiss Light Source (Table S1). Statistics of the refinement and details of ligand-protein interactions in the final model are listed in Tables S2 and S3.
(3), Wild-type and mutant Bcr-Abl Ba/F3 cellular proliferation assays
Viability of wild type and mutant Bcr-Abl expressing Ba/F3 cells after a 48 hour treatment with various concentrations of single or combined agents was determined by AlamarBlue® (TREK Diagnostic Systems) reduction method. The combination index (CI) was calculated according to the method of Chou and Talalay18 using the Calcusyn software.
(4), Selection for clones resistant to GNF-2 and imatinib
Emergence of compound resistant Ba/F3.p210 clones was evaluated as previously described13. One 96-well plate was used for every compound concentration or combination, and the medium was renewed every 3–4 days. The plates were incubated for 21 days, and the number of wells with evident cell growth was recorded at days 9, 12 and 21.
(5), Abl protein expression and purification
The bacterial expression and purification of human c-Abl kinase (residues 46– 515, Abl 1a numbering38) was performed as previous described39. Phosphorylation (+80 or +160 Da) was observed in the intact protein spectra (Fig. S8). The location of each phosphorylation was determined by trypsin digestion followed by LC-MS/MS. For both wild-type Abl and Abl E505K, single phosphorylation corresponded to modification at Tyr412 and double phosphorylation involved Tyr412 and Tyr89. For Abl T315I, single phosphorylation was on Tyr89 and only a small quantity of the molecules contained phosphoryation at both Tyr89 and Tyr412.
(6), Kinetic characterization of Abl inhibition
The ATP/NADH-coupled assay system in a 96-well format was used to determine the initial velocity of Abl tyrosine kinase catalyzed peptide phosphorylation. The reaction mixture contained 20 mM Tris-HCl, (pH 8.0), 50 mM NaCl, 10 mM MgCl2, 2 mM PEP [2-(Phosphonooxy)- 2-propenoic acid, Sigma-Aldrich, cat. P-7002) and 20 µM Abl peptide substrate (EAIYAAPFAKKK, New England Biolabs, Cat No. P6051L), fixed or varied (to determine inhibitor kinetic parameters) concentration of inhibitor applied, 1/50 of the final reaction mixture volume of PK/LDH enzyme (pyruvate kinase/lactic dehydrogenase enzymes from rabbit muscle, Sigma-Aldrich, cat. P-0294), 160 µM NADH, 0.16 µM Abl, and ATP added last to start the reaction. Absorbance data were collected every 20s at 340 nm using a SpectraMax M5 Microplate Reader. The two-substrate kinase reaction was simplified to two one-substrate reactions to determine ATP kinetic parameters and inhibitor parameters separately. When determining ATP parameters, the inhibitor concentration was kept the constant. When determining inhibition parameters, the ATP concentration was kept the same at 20 µM. Steady-state initial velocity data were drawn from the slopes of the A340 curves and fit to the Michaelis-Menten equation to determine Vmax and Km values. Data were fit globally using GraphPad Prism (GraphPad Software) and Excel XLfit 4.0 to fit velocity equations for competitive and mixed inhibition.
(7), Hydrogen exchange experiments
Hydrogen exchange experiments were performed essentially as described in Iacob et al23. Abl protein (38 pmol) was incubated for 30 min at room temperature with 45 µM of GNF-5 at a protein:drug ratio 1:7 (4.38 µM GNF-5 at labeling solution). For an estimated EC50 of 0.169µM for GNF-5 (Fig. S9d), 97.41% of the protein was expected to be bound to GNF-5. Deuterium exchange was initiated by dilution of the protein-drug mixture 15-fold with 20 mM Tris, 100 mM NaCl (pD 8.3), D2O, 21 °C. The same procedure was applied for the Abl protein alone and for the E505K mutant. At each deuterium exchange time point (from 10 s to 4 hours) an aliquot from the exchange reaction was removed and labeling was quenched by adjusting the pH to 2.6 with an equal volume of quench buffer (50 mM potassium phosphate, pH 2.6, H2O). Quenched samples were immediately frozen on dry ice and stored at −80 °C until analysis. Each frozen sample was thawed rapidly to 0 °C and injected into a custom Waters nanoACQUITY UPLC system and analyzed as described previously40. The protein sample was digested online with pepsin and the resulting peptides were trapped and desalted for 3 min at 100 µL/min and then separated in 6 min by an 8%–40% acetonitrile:water gradient at 40 µL/min. The separation column was a 1.0×100.0 mm ACQUITY UPLC C18 BEH (Waters Corp., Milford, MA, USA) containing 1.7 µm particles and the back pressure averaged 8800 psi at 1 °C.
The mass spectra were obtained with a Waters QTOF Premier equipped with standard ESI source (Waters Corp., Milford, MA, USA) as described previously23. There was no correction made for back-exchange and all results are reported as relative deuterium level22. Mass spectra were processed with the software HX-Express41 where the deuteration levels were calculated by subtracting the centroid of the isotopic distribution for peptide ions of undeuterated protein from the centroid of the isotopic distribution for peptide ions from the deuterium labeled sample.
(8), In vivo studies of GNF-5 and drug combinations
GNF-5 pharmacokinetic parameters in mice. Male Balb/c mice were dosed with GNF-5 in PEG400/saline, 1:1 at 5 mg/kg intravenously or 20 mg/kg orally. The compound plasma concentration at any given time point was determined by Liquid Chromatography/Mass Spectrometry (LC/MS/MS). Pharmacokinetic parameters were calculated by non-compartmental regression analysis using Winnonlin 4.0 software (Pharsight, Mountain View, CA, USA).
In vivo efficacy in Ba/F3.p210 xenograft model. Female SCID beige mice, 6–8 weeks of age (n=5 for each GNF-5-treated or vehicle control group) were injected via tail vein with 1×106 Ba/F3 cells co-expressing Bcr-Abl p210 and luciferase. Three days post-injection, mice were orally dosed twice daily with 50 or 100 mg/kg GNF-5 for seven days. At days 5 and 7, bioluminescence was quantified using luciferin and an IVIS imaging system (Xenogen Corp., Alameda, CA).
GNF-5 pharmacodynamics in Ba/F3.p210 xenograft mice. Bone marrow samples were collected at 3 hours (n=3 mice per time point) following a single oral administration of GNF-5 at 50 and 100 mg/kg at day 7 post cell injection. Fixed and permeabilized bone marrow cells were stained with PE-conjugated anti-phospho-Y694 STAT5 antibody (pSTAT5, Becton Dickinson) and subjected to flow cytometry.
In vivo efficacy in bone marrow transduction/transplantation model. Bone marrow cells harvested from 6–8 weeks old 5-FU injected male Balb/c mice were transduced with a pMSCV Bcr-Abl wt or T315I Bcr-Abl retroviral construct and transplanted into irradiated recipient female Balb/c mice (6–8 weeks). A seven days long treatment with GNF-5, nilotinib or vehicle control started on days 7 (wt Bcr-Abl) or 15 (T315I) after transplantation (10 (wt Bcr-Abl) or 4 (T315I Bcr-Abl) mice per treatment group). Blood cell counts and spleen size were determined at treatment day 7. Bone marrow cells were isolated, fixed, permeabilized, stained with anti-p-STAT5 and anti-luciferase antibodies, and analyzed by flow cytometry. For survival studies, the treatment with vehicle, GNF-5, nilotinib, or the combination of both (n = 5 mice per group) was initiated 11 days after transplantation and prolonged until day 50 post-transplantation or until the mice had to be sacrificed due to becoming moribund. Overall survival and time to relapse were determined by the Kaplan-Meier method. Statistical significance was assessed using the Kaplan-Meier survival analysis, under the assumption of a normal distribution of normalized ratios with an estimate of variance (α = 0.05, two-sided).
Supplementary Material
Acknowledgements
We thank Chrystèle Henry and Gabriele Rummel for technical assistance and Ronak Beigi for her help with the bone marrow transplantation studies and Anastasia Velentza for performing the DSC experiments. We also thank John Kuriyan, Markus Seeliger, Caihong Yun, Michael Eck, Ellen Weisberg, Doriano Fabbro, Priscilla L. Yang, Giulio Superti-Furga, and Andrew Kung for helpful discussions. We would also like to acknowledge the support of staff at beamline PXII of the Swiss Light Source, Villigen, Switzerland, during X-ray data collection, ICCB-Longwood Screening facility at Harvard Medical School for cell proliferation and enzyme assay, and this is Barnet Institute contribution 935.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Accession code. Protein Data Bank: the coordinates and structure factors of the complete Abl-imatinib-GNF-2 complex crystal structure have been deposited under accession code 3K5V.
Author contributions F.J.A., J.Z., J.P., Y.C., G.L., M.A. and G.D. designed and performed cellular and biochemical experiments. J.Z. performed bacterial Abl expression and enzyme assays. W.J., N.V. and S.G designed and performed the NMR experiments. S.W.C.-J. designed and performed the crystallographic experiments. G.F. and A.S. produced the protein for the NMR and X-ray experiments. T.S., Q.D., B.O., A.W. and X.D. designed and synthesized the compounds. A.G.L., C.D., F.S., G.-R.G. and T.T. carried out the in vivo studies. Y.L. and B.B. contributed to the design of the compounds. R.E.I. and J.R.E. performed and designed the hydrogen exchange experiments. M.W. contributed to the design of the in vivo experiments. F.J.A., M.W., and P.M. provided critical input to the overall research direction. N.S.G. directed the research and wrote the paper with input from all co-authors.
Author information The authors declare competing financial interest.
REFERENCES
- 1.Weisberg E, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7(2):129. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Quintas-Cardama A, Kantarjian H, Cortes J. Flying under the radar: the new wave of BCR-ABL inhibitors. Nat Rev Drug Discov. 2007;6(10):834. doi: 10.1038/nrd2324. [DOI] [PubMed] [Google Scholar]
- 3.Shah NP, et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305(5682):399. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 4.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 5.Nagar B, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62(15):4236. [PubMed] [Google Scholar]
- 6.Bradeen HA, et al. Comparison of imatinib mesylate, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: high efficacy of drug combinations. Blood. 2006;108(7):2332. doi: 10.1182/blood-2006-02-004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adrian FJ, et al. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat Chem Biol. 2006;2(2):95. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- 8.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112(6):859. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 9.Hantschel O, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112(6):845. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]; Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385(6617):595. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 10.Jahnke W, Widmer H. Protein NMR in biomedical research. Cell Mol Life Sci. 2004;61(5):580. doi: 10.1007/s00018-003-3382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vajpai N, et al. Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of imatinib/nilotinib and dasatinib. J Biol Chem. 2008;283(26):18292. doi: 10.1074/jbc.M801337200. [DOI] [PubMed] [Google Scholar]
- 12.Ray A, et al. Identification of BCR-ABL point mutations conferring resistance to the Abl kinase inhibitor AMN107 (nilotinib) by a random mutagenesis study. Blood. 2007;109(11):5011. doi: 10.1182/blood-2006-01-015347. [DOI] [PubMed] [Google Scholar]
- 13.von Bubnoff N, et al. A cell-based screen for resistance of Bcr-Abl-positive leukemia identifies the mutation pattern for PD166326, an alternative Abl kinase inhibitor. Blood. 2005;105(4):1652. doi: 10.1182/blood-2004-06-2445. [DOI] [PubMed] [Google Scholar]
- 14.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112(6):831. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 15.Azam M, et al. Activity of dual SRC-ABL inhibitors highlights the role of BCR/ABL kinase dynamics in drug resistance. Proc Natl Acad Sci U S A. 2006;103(24):9244. doi: 10.1073/pnas.0600001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branford S, et al. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood. 2002;99(9):3472. doi: 10.1182/blood.v99.9.3472. [DOI] [PubMed] [Google Scholar]
- 17.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2(2):117. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 18.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 19.Mishra S, et al. Resistance to imatinib of bcr/abl p190 lymphoblastic leukemia cells. Cancer Res. 2006;66(10):5387. doi: 10.1158/0008-5472.CAN-05-3058. [DOI] [PubMed] [Google Scholar]
- 20.Kornberg A, Pricer WE., Jr Di- and triphosphopyridine nucleotide isocitric dehydrogenases in yeast. J Biol Chem. 1951;189(1):123. [PubMed] [Google Scholar]
- 21.Choi Y, et al. N-myristoylated c-Abl tyrosine kinase localizes to the endoplasmic reticulum upon binding to an allosteric inhibitor. J Biol Chem. 2009;284(42):29005. doi: 10.1074/jbc.M109.026633. [DOI] [PMC free article] [PubMed] [Google Scholar]; Choi Y, et al. N-myristoylated c-ABL tyrosine kinase localizes to the endoplasmic reticulum upon binding to an allosteric inhibitor. J Biol Chem. 2009 doi: 10.1074/jbc.M109.026633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25(1):158. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 23.Iacob RE, et al. Conformational disturbance in Abl kinase upon mutation and deregulation. Proc Natl Acad Sci U S A. 2009;106(5):1386. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Etten RA. Disease progression in a murine model of bcr/abl leukemogenesis. Leuk Lymphoma. 1993;11 Suppl 1:239. doi: 10.3109/10428199309047893. [DOI] [PubMed] [Google Scholar]
- 25.Joensuu H, et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med. 2001;344(14):1052. doi: 10.1056/NEJM200104053441404. [DOI] [PubMed] [Google Scholar]
- 26.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 28.Cools J, et al. Prediction of resistance to small molecule FLT3 inhibitors: implications for molecularly targeted therapy of acute leukemia. Cancer Res. 2004;64(18):6385. doi: 10.1158/0008-5472.CAN-04-2148. [DOI] [PubMed] [Google Scholar]
- 29.Blencke S, et al. Characterization of a conserved structural determinant controlling protein kinase sensitivity to selective inhibitors. Chem Biol. 2004;11(5):691. doi: 10.1016/j.chembiol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 30.Brown EJ, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369(6483):756. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 31.Dudley DT, et al. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1995;92(17):7686. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barnett SF, et al. Identification and characterization of pleckstrin-homology-domain- dependent and isoenzyme-specific Akt inhibitors. Biochem J. 2005;385(Pt 2):399. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke JR, et al. BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J Biol Chem. 2003;278(3):1450. doi: 10.1074/jbc.M209677200. [DOI] [PubMed] [Google Scholar]
- 34.Converso A, et al. Development of thioquinazolinones, allosteric Chk1 kinase inhibitors. Bioorg Med Chem Lett. 2008 doi: 10.1016/j.bmcl.2008.12.076. [DOI] [PubMed] [Google Scholar]
- 35.Tokumitsu H, et al. KN-62, 1-[N,O-bis(5-isoquinolinesulfonyl)-N-methyl-Ltyrosyl]- 4-phenylpiperazi ne, a specific inhibitor of Ca2+/calmodulin-dependent protein kinase II. J Biol Chem. 1990;265(8):4315. [PubMed] [Google Scholar]
- 36.Lee TS, et al. Molecular basis explanation for imatinib resistance of BCR-ABL due to T315I and P-loop mutations from molecular dynamics simulations. Cancer. 2008;112(8):1744. doi: 10.1002/cncr.23355. [DOI] [PubMed] [Google Scholar]
- 37.Strauss A, et al. Efficient uniform isotope labeling of Abl kinase expressed in Baculovirus-infected insect cells. J Biomol NMR. 2005;31(4):343. doi: 10.1007/s10858-005-2451-3. [DOI] [PubMed] [Google Scholar]
- 38.Oppi C, Shore SK, Reddy EP. Nucleotide sequence of testis-derived c-abl cDNAs: implications for testis-specific transcription and abl oncogene activation. Proc Natl Acad Sci U S A. 1987;84(23):8200. doi: 10.1073/pnas.84.23.8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seeliger MA, et al. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14(12):3135. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80(17):6815. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17(12):1700. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.