Abstract
Compared to traditional single-animal imaging methods, multiple-mouse MRI has been shown to dramatically improve imaging throughput and reduce the potentially prohibitive cost for instrument access. To date, up to a single radiofrequency coil has been dedicated to each animal being simultaneously scanned, thus limiting the sensitivity, flexibility, and ultimate throughput. The purpose of this study was to investigate the feasibility of multiple-mouse MRI with a phased-array coil dedicated to each animal. A dual-mouse imaging system, consisting of a pair of two-element phased-array coils, was developed and used to achieve acceleration factors greater than the number of animals scanned at once. By simultaneously scanning two mice with a retrospectively gated cardiac cine MRI sequence, a three-fold acceleration was achieved with SNR in the heart that is equivalent to that achieved with an unaccelerated scan using a commercial mouse birdcage coil.
Keywords: multiple-mouse MRI, imaging efficiency, parallel imaging, cardiac cine
INTRODUCTION
Relatively long acquisition times have prompted a great deal of research to improve the efficiency by which MRI data is acquired and images are formed. In a clinical setting, patient comfort must be balanced with the quantity and quality of data that is necessary to diagnose, stage, or assess the response of disease to medical intervention. Preclinically, efficiency improves our ability to scan a population of animals quickly and with minimal constraints on study design. Both the pulse sequence and detector geometry have a critical effect on the signal-to-noise (SNR) and overall acquisition time that can be achieved. Arrays of surface coils have been shown to improve efficiency over the use of individual volume coils by reducing the need for signal averaging (1) and allowing the integration of parallel imaging (PI) techniques (2–6) that add another degree of freedom for pulse sequence optimization.
PI techniques enable reconstruction of complete images from a reduced set of spatial phase-encoding (PE) observations. In the PI reconstruction process, missing data is estimated by using information about coil sensitivities that is gained either through a prescan or acquisition of additional autocalibrating signal (ACS) lines. However, image SNR is reduced by at least the square root of the reduction factor, (R) (2). The SNR advantage inherent to the use of phased arrays often mitigates this loss and makes PI a desirable means for reducing acquisition time.
Other techniques have been used to improve efficiency in small animal imaging. Instead of minimizing the scan time of a single animal, multiple animals can be simultaneously imaged within a single MRI scanner. Various methods have been suggested to accomplish this (7–12). Approaches involving arrays of volume coils, in which multiple transceive volume coils are strategically combined within a single gradient coil, have achieved image quality and acquisition times that are consistent with what is achieved when scanning one animal at a time (13,14). This technique can potentially increase animal throughput, decrease the cost associated with instrument access times, allow greater scanner accessibility, and reduce group variance of experimental subjects with pathologies that progress over time. Gradient-intensive sequences however, must be carefully optimized to avoid thermal overload when gradients that are large enough to accommodate several volume coils are used to encode a field of view (FOV) appropriate for a single mouse (15). Additionally, the increase in throughput resulting from arrays of volume coils will always be below the number of animals simultaneously scanned (N) because of the additional overhead associated with the preparation and adjustment of multiple animals.
Arrays of surface coils (16,17) could be used to increase sensitivity in multianimal imaging applications and allow the integration of PI techniques (18,19) to improve the efficiency and flexibility of small animal MRI. PI reduction factors could increase throughput beyond the number of animals that can be scanned at once. The ability to acquire accelerated images would reduce the total number of phase encode repetitions needed to encode image data, thus reducing gradient duty cycle and the thermal load placed on the gradient coils.
Simultaneous PI-encoding of multiple, separate regions of interest containing physically distinct specimens requires consideration of potential sources of image degradation that are not present when scanning a single subject. Because it is impossible to synchronize the cardiac and respiratory cycles among multiple animals, gated data acquisition can only be achieved relative to one animal at a time. Uncorrected motion from other specimens may cause artifacts in all volumes that project along the PE axis and complicate unfolding of data that is undersampled in the PI acquisition. In this regard, cardiac cine imaging is perhaps the most challenging application for PI-accelerated multi-animal imaging using multiple phased arrays. Retrospective gating strategies (14) enable data to be sorted for each animal in turn, but data that is synchronized for one sample may be contaminated by unsynchronized motion artifacts from other subjects. Questions regarding the propagation of motion artifact from one animal to the other are critically important and must be investigated.
The purpose of this study was to investigate the feasibility of multiple-mouse MRI with multiple arrays of receive coils (MARCs) for improving the sensitivity and animal throughput for investigations involving small animal imaging at high fields. Specifically, we demonstrate multiple-animal imaging with MARCs for PI-accelerated, retrospectively-gated (14) cine imaging of the murine cardiac cycle as appropriate for assessing models of cardiovascular disease or chemotherapy-related cardiac dysfunction. Two mice were simultaneously scanned using a pair of two-element phased-array coils with PI reduction factors ranging from one to four. Image quality and scan times for accelerated multiple-animal cardiac cine images are compared with corresponding unaccelerated acquisitions. We demonstrate that multiple separate phased arrays can be used to improve efficiency by a factor greater than the number of animals simultaneously scanned (> N), demonstrating a new versatile technique for efficient small-animal MRI.
MATERIALS AND METHODS
All imaging was performed on a four-channel, 7.0 T Biospec MRI system with a 30 cm bore (Bruker Biospin MRI, Billerica, MA) and all animal procedures were approved by our Institutional Animal Care and Use Committee, which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International.
Imaging Hardware
Two identical two-element surface coil arrays were manufactured using a ProtoMat C100/HF circuit prototyping system (LPKF Laser & Electronics, Wilsonville, OR) for imaging two mice simultaneously. The loop geometries were designed to cover a 3 × 3 cm coronal FOV near the plane of the array. Tuning and matching were achieved through a combination of fixed capacitors (American Technical Ceramics, Huntington Station, NY) and voltage-controlled varactor diodes (BB833E6327; Infineon Technologies, Milpitas, CA) that could be adjusted from the imaging console.
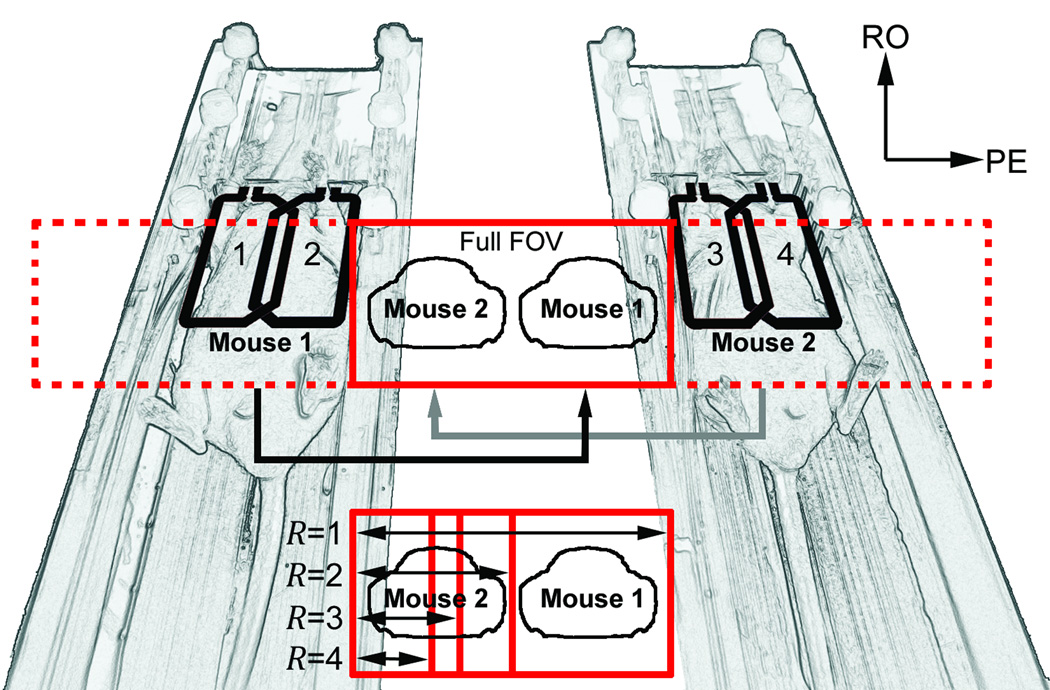
Coupling between adjacent elements was minimized by appropriate geometric loop overlapping to reduce mutual inductance (1) and by incorporating lattice-style baluns (20) that minimized cable coupling. Decoupling from the transmit coil was accomplished by both actively forward biasing the tuning varactors during the transmit phase (which dramatically shifted the resonant frequency of the receive coils) and by geometrically decoupling from the 15.4 cm inner diameter (ID) quadrature transmit birdcage coil (1P T9561; Bruker Biospin MRI) that was operated in linear mode. Likewise, the transmit coil was actively detuned during the receive portion of the imaging sequence. Interarray coupling was minimized by increasing the distance between the arrays. Coupling was determined by measuring scattering parameters with a network analyzer (3295A; Agilent Technologies, Palo Alto, CA). Figure 1 illustrates the 4-element array for dual-mouse imaging.
Figure 1.
Relationship between the two-element phased-array coil geometries and the aliasing strategies used for dual-mouse image acquisition. The full FOV (R = 1) contains the unaccelerated aliased images, making the animals appear as if they are adjacent to one another. The reduced FOVs prescribed to achieve up to four-fold accelerations are shown. Note that the FOV for R = 2 is identical to that which would be used to scan each of the two mice serially in a single-animal configuration.
Accelerated Imaging Scheme
A linear arrangement of coils and animals allows the prescription of single-animal parameters for 2D multiple-animal MRI (i.e. the R = 2 case shown in Figure 1). By strategically defining the FOV over a single animal, encoding by aliasing along the PE dimension (13) can yield centered images from all mice without additional postprocessing (21). By further reducing the PE FOV, more significant image overlap will occur, which can be later unfolded by using appropriate PI reconstruction strategies (Figure 2).
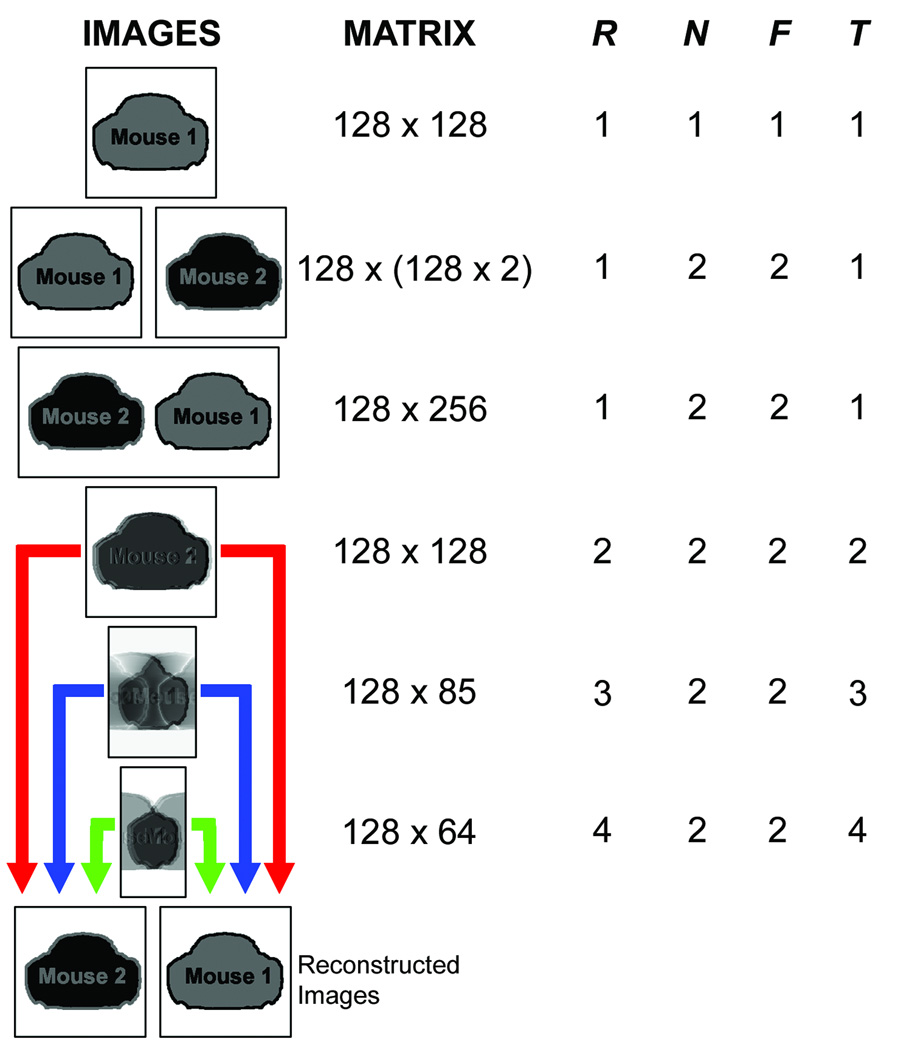
Figure 2.
Grid describing various imaging options and calculated throughput parameters for dual-mouse imaging. The first three rows demonstrate unaccelerated imaging of a single mouse (first row), two mice serially (second row), and two mice at once (third row). All of these imaging strategies result in throughput, T = 1, where T=R×N/F, R is the traditional PI reduction factor, N is the number of simultaneously-scanned mice, and F is the factor by which the single-animal FOV must extended to cover all animals without acceleration. The next three rows indicate accelerated (R = 2, R = 3, and R = 4) dual mouse (N = 2) imaging made possible by using multiple arrays of receive coils. Note that the accelerated imaging schemes will ultimately result in separate reconstructed images for each mouse.
The definition of acceleration must be reconsidered when parallel imaging is used to encode multiple volumes. For traditional PI, R is the ratio of PE steps required for a fully-encoded, unaccelerated image to the number of PE steps that are acquired to encode the accelerated image. For multiple-animal MRI, multiple, separate FOVs are reconstructed from undersampled data that may or may not be prescribed over a single imaging volume. Unaccelerated imaging can be defined based on the number of PE steps required to fully-encode each animal scanned, in series, as illustrated in the first two rows of Figure 2. Multiple animals that are scanned at once but that require encoding of multiples of the desired image matrix size (Figure 2, row 3) also do not represent acceleration. The total increase in throughput, T, must account for the number of animals that are scanned and the factor by which the matrix would be extended to encode the entire image space, in addition to any accelerations due to parallel imaging:
| (1) |
Here, R is the traditionally defined PI reduction factor from the full multiple-animal FOV, N is the number of animals scanned at once, and F is the matrix extension factor (i.e. the ratio of the total number of PE lines required to cover all animals to the number of PE lines required for imaging a single animal). In this study, various reduction factors (R = 1, R = 2, R = 3, and R = 4) were considered for dual-mouse (N = 2) cardiac cine MRI with MARCs to investigate the potential to achieve an increase in throughput that is greater than the number of mice simultaneously scanned. For this study, F = 2 since the unaccelerated (R = 1) case (Figure 2, row 3) encodes a matrix that is twice as large as what would be used to encode a single animal (Figure 2, row 1). Figure 1 illustrates various acceleration schemes while Figure 2 provides acceleration examples and resulting throughput calculation parameters. Both axial and coronal images can be accelerated using these techniques because of the adjacent alignment of those anatomical views with this imaging configuration, whereas sagittal slices will require multiple slice packages to efficiently image both animals.
Phantom Imaging
An increase in SNR associated with the use of phased-array coils allows the flexibility to trade SNR for imaging speed by using PI. Phantom data was acquired to compare SNR of the phased-array coils with the commercially available birdcage coil that is used for routine mouse imaging. To accomplish this, phantoms filled with 0.1% Magnevist (Berlex Imaging, Montville, NJ) and distilled water were scanned with a fast low-angle shot (FLASH) sequence (in-plane resolution = 23 µm, TE/TR = 2.4/100 ms, flip angle (FA) = 16°, number of repetitions (NR) = 2, slice thickness (ST) = 1.0 mm) using both a 35 mm ID commercial mouse birdcage coil (1P T8102, Bruker Biospin MRI; QU/QL= 119/78 as measured with a 20 mL saline phantom) and the dual-mouse phased arrays. Signal was determined from the average of the two acquired images, whereas noise was determined from the difference image. SNR was calculated as square root of two times the ratio of the mean signal intensity divided by the standard deviation of the noise within an ROI 4 mm from the coil surface (corresponding to the location of the mouse heart). The average SNR within the ROI was calculated for the phased-array coil and the mouse birdcage coil.
To determine the SNR penalty paid as a function of PI reduction factor, an unaccelerated FLASH acquisition (R = 1, FOV = 3 × 6 cm, matrix = 128 × 256, TE/TR = 2.4/100 ms, FA = 16°, NR = 100, ST = 1.0 mm) of the two phantoms was repeated 100 times. The data was then downsampled to produce accelerated image sets (R = 2, R = 3, and R = 4) with four, eight, and twelve ACS lines respectively (Reff = 1.94, Reff = 2.75, and Reff = 3.37). Each of the 100-image sets was reconstructed with a multicolumn multiline interpolation (MCMLI) generalized autocalibrating partially parallel acquisition (GRAPPA) algorithm (4,22). SNR maps of each of the accelerated phantom images were generated on a pixel-by-pixel basis by dividing the mean signal value by the standard deviation calculated along the time series.
Propagation of Motion Artifacts through Coupling
To date, the majority of multiple-animal imaging has been performed using arrays of shielded transceive volume coils that could be geometrically-decoupled in a straightforward manner. By using multiple arrays of surface coils in conjunction with a single transmit body coil, phased-array coil decoupling techniques are used to reduce, but not eliminate, coupling between array elements and volumes of interest. Simulation was used to estimate the impact of motion on multiple-animal PI-accelerated acquisitions using surface coils.
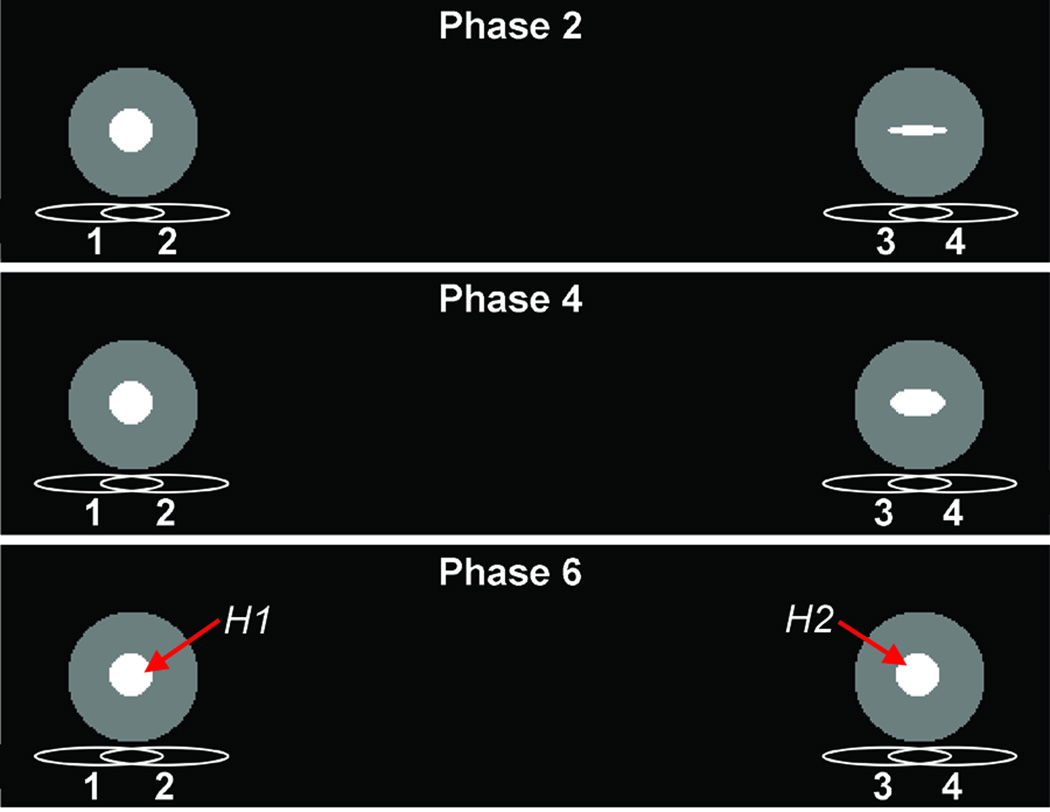
A numerical MATLAB (The MathWorks, Natick, MA) simulation was designed to assess the propagation of motion artifacts due to coupling between array elements. Coil sensitivity maps were calculated using a finite-element package (COMSOL Multiphysics; COMSOL AB, Stockholm, Sweden). The numerical phantom consisted of a series in time from two signal sources, each containing a circular region of interest (ROI) representing the heart and positioned according to the dual-mouse imaging geometry. One ROI was held constant, representing data that is synchronized with motion; the other was varied over 21 phases by expanding and contracting the circular object into variously shaped ellipses (Figure 3). A motion-free series of k-space data was generated by Fourier transform of the image series, and motion artifacts associated with non-synchronized data were induced in a synthetic acquisition by filling each line of k-space from a random phase of the image series. The synthetic raw data was then downsampled along the PE direction to simulate two-fold acceleration. This was repeated to produce 200 distinct data sets. A GRAPPA reconstruction was performed on each data set.
Figure 3.
Setup of numerical phantom simulation with respect to the phased-array coils. The gated phantom (left) is fixed with H1 in diastole while the ungated phantom (right) has H2 varied over 21 phases. Three representative phases are shown.
The effects of noise and coupled motion artifacts were independently calculated and combined into a figure of merit describing the artifact-to-noise (ANR). Artifact power within the gated ROI, as a function of interarray coupling, was determined by measuring the root mean square (RMS) of the difference between a perfectly decoupled image and the image containing coupled motion artifact. Intraarray elements were assumed to be modestly decoupled by 15 dB and the coupling between interarray elements was varied from 0 to −20 dB. This simulation was repeated for each of the data sets and then averaged. RMS noise levels corresponding to commonly-encountered SNR values were assumed.
Animal Preparation
Two female CD-1 mice were placed supine on a custom-built, dual-mouse sled distributing 0.5–2.5% isoflurane (IsoSol; VEDCO, St. Joseph, MO) in oxygen through nose cones. The nose cone positions were adjusted so that the mouse anatomies were similarly aligned. The two arrays were then each placed over the hearts of the mice and were secured to the sled via a set of thumb screws. The respiration rates of both mice were monitored throughout the study via a home-built, computer-based monitoring system that incorporated a low-cost, commercially available A/D conversion board (minilab 1008; Measurement Computing Corp., Norton, MA) with pressure transducers (PX138-001D5V; Omega Engineering, Inc., Stamford, CT). Respiration bellows with extended tubing were coupled to each mouse and were connected directly to the transducers. The respiratory rates of the two mice were maintained at roughly 24 to 30 breaths per minute.
Imaging Protocol
To align the mice along the field axis, sagittal FLASH images were acquired of a single animal. The transmit coil and each of the four array elements were sequentially tuned and matched. Automatic adjustments over a single animal and coil element were used to determine the required center frequency, shim currents, and transmit pulse power levels. A three-plane FLASH acquisition of each mouse further localized the heart positions for subsequent slice prescription. A spoiled gradient echo pulse sequence, modified to acquire multi-channel cardiac navigator signals and k-space using an efficient asymmetric short-echo readout, was used for simultaneous cardiac imaging (23). For each PE increment, the sequence was repeated continuously for a sufficient number of repetitions to ensure adequate temporal sampling of the cardiac cycle, and in anticipation of data that was acquired during inspiration or expiration being excluded during retrospective reconstruction. Axial slices based on the localizer scans were prescribed to include the hearts of both mice via aliasing as illustrated in Figure 1. Unaccelerated images (FOV = 3.1 cm × 6.2 cm, matrix = 128 × 256, TE/TR = 3.4/7.0 ms, FA = 16°, NR = 200, ST = 1.0 mm) were first acquired. These were followed by otherwise identical acquisitions but with two- (FOV = 3.1 × 3.1 cm, matrix = 128 × 128), three- (FOV = 3.1 × 2.07 cm, matrix = 128 × 85), and four-fold (FOV = 3.1 × 1.55 cm, matrix = 128 × 64) accelerations via parallel imaging. Four, eight, and twelve additional ACS lines were acquired for GRAPPA reconstruction of the two-, three-, and four-fold accelerated images yielding overall R = 1.94, R = 2.75, and R = 3.37, respectively.
Image Reconstruction
For each animal, retrospectively-gated cine data must be reordered to fill k-space with data that corresponds to the correct cardiac phase prior to PI reconstruction. The image reordering was performed using IDL software (ITT Industries, Inc., Boulder, CO) as described previously (23). Cardiac motion curves were derived from the magnitude of the points corresponding to the center of k-space along the temporal dimension (24,25). Data points were binned into the appropriate k-space set, representing an individual cine frame, according to the motion curves. Cardiac data that was contaminated by respiratory motion was discarded. Final image SNR was enhanced by averaging the inherently oversampled data (14).
The MCMLI GRAPPA reconstruction was implemented in MATLAB and was used to sequentially reconstruct coil images from all cine cardiac phases of the two mice. However, final mouse images were created strictly based on images from the coils dedicated to the particular animal. For example, the images from coils one and two were used to reconstruct mouse one and the images from coils three and four were used to reconstruct mouse two via a sum of squares image combination.
RESULTS
The four-element, dual-mouse imaging configuration of Figure 1 was used to acquire phantom and retrospectively-gated cine images. Through the use of overlap decoupling and cable baluns, more than 30 dB of intraarray element decoupling was achieved for both phased arrays. Nearest interarray elements (coils two and three from Figure 1) achieved approximately 40 dB of isolation and nonadjacent elements roughly 30 dB of isolation.
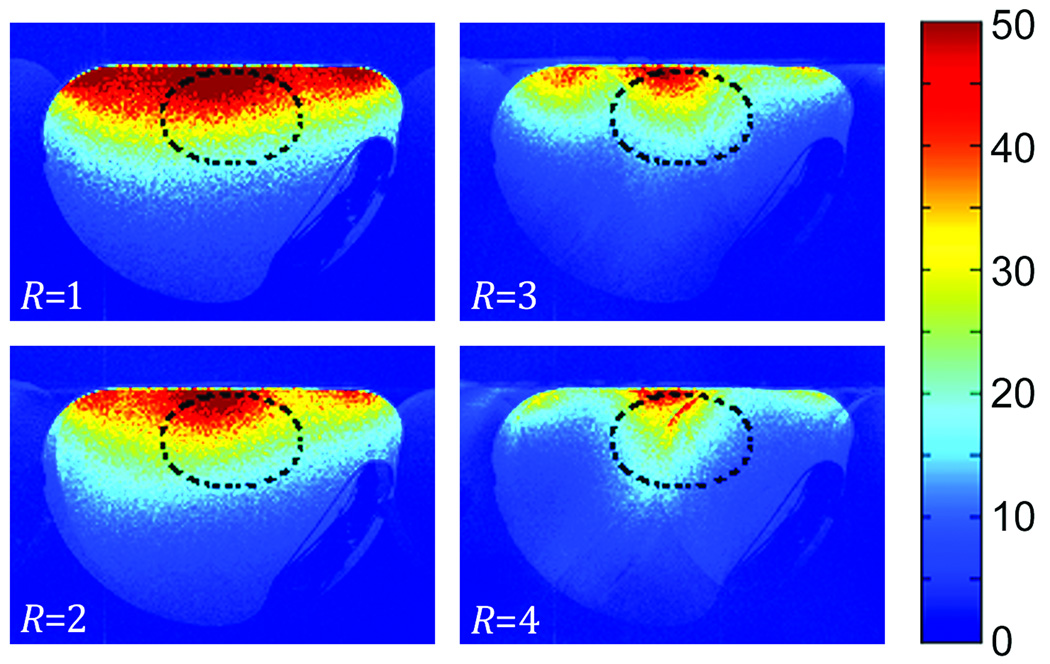
Figure 4 shows the resulting SNR maps from the unaccelerated and accelerated FLASH images from one of the two simultaneously-acquired phantoms. As expected, there is a noticeable reduction in SNR as R increases; however, the SNR in the ROI corresponding to the heart location remains comparatively high. The unaccelerated phantom images from the dual-mouse phased-arrays demonstrated a 60% increase in SNR compared with the commercial mouse birdcage coil over the illustrated heart ROI. Compared with the standard mouse birdcage coil, the R = 2 acceleration yielded a 32% SNR increase, while the R = 3 acceleration resulted in virtually equivalent SNR (1% higher). The largest acceleration considered (R = 4) resulted in images that were 12% lower in SNR than the commercial birdcage coil. This increase in SNR made available by the phased-array coils afforded the use of PI strategies to accelerate a mouse imaging study.
Figure 4.
SNR maps determined from GRAPPA-reconstructed images of the originally-acquired data set (R = 1) and subsequently downsampled data sets (R = 2, R = 3, and R = 4). Note that the SNR is highest in the location corresponding to the mouse heart as indicated by the oval ROI.
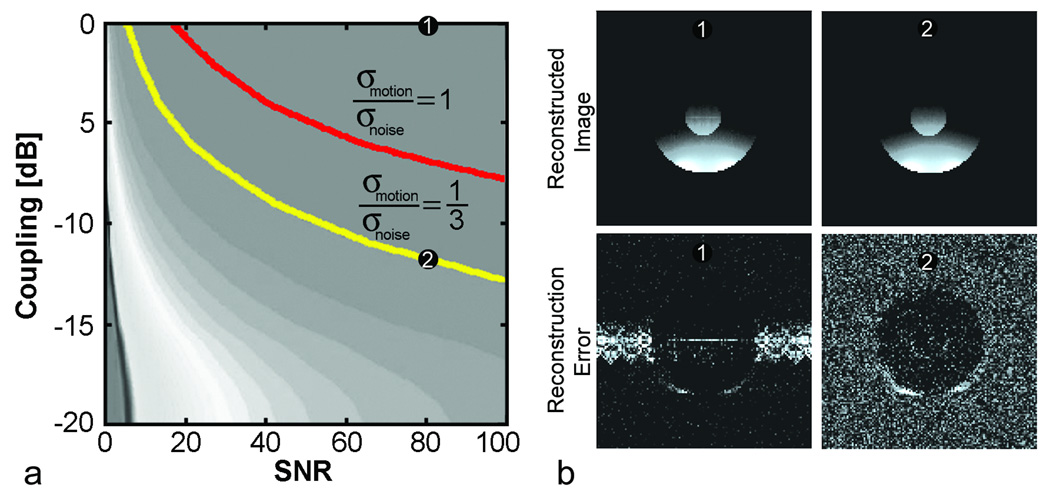
Simulations indicate that the prevalence of motion artifacts was strongly dependent on SNR and coupling between the phased-array coils. Figure 5 illustrates these relationships. Greater decoupling is required to suppress propagated motion artifacts that are more prevalent as image SNR increases. From these curves, it is possible to find the maximum coupling that is allowed for motion to interfere with adjacent images at a given artifact to noise. Contours representing an ANR of one and one-third are indicated. GRAPPA reconstructed images and the reconstructed images subtracted from ideal images (i.e. error images) are shown for two operating points. It can be seen that the motion artifact is prevalent in the highly-coupled error image whereas the motion artifact has disappeared into the noise floor when the arrays were decoupled by 12 dB.
Figure 5.
A) Propagated motion artifact-to-noise contours from an accelerated (R = 2) GRAPPA simulation. Two contours representing potential interarray decoupling requirements for the propagated motion artifact to be equal to and one third of the noise floor are labeled. B) Reconstructions of the static numerical phantom images and difference from the ideal reconstruction are shown, windowed to emphasize noise and artifact, for two example contour points (1 and 2).
The custom-built dual-animal imaging sled facilitated animal maintenance by providing unobstructed access to the animals and by integrating anesthesia distribution and channels for circulating warm water to maintain body temperature. Additionally, the adjustable nose cones allowed nearly identical animal and coil alignment along the field axis, making slice prescription to cover both animals straightforward.
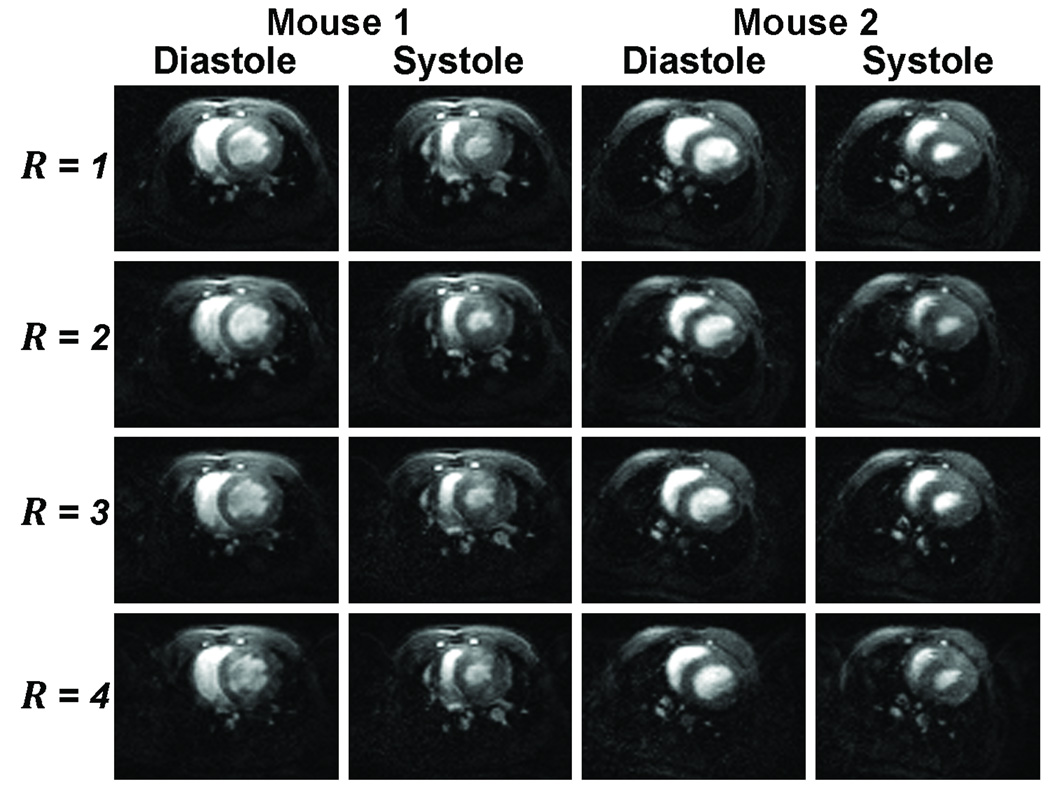
PI-accelerated retrospective cine images of a murine heart yielded excellent results for up to a PI reduction factor of four. Figure 6 illustrates representative ventricular diastole and systole phases of the two mice from the four accelerated cardiac cine acquisitions. As expected and as demonstrated by phantom imaging, image quality degrades with increased acceleration. By decreasing the required number of PE lines to reconstruct a full image (i.e. a reduction in the time spent encoding space), we were able to keep temporal sampling and thus temporal resolution unchanged, while substantially reducing the overall cine MRI acquisition time. The two-fold accelerated cardiac cine images required 1 minute 33 seconds per slice per animal while the three- and four-fold accelerated images required only 1 minute 6 seconds and 54 seconds per slice per animal respectively: an improvement over the 3 min per slice per animal required for unaccelerated imaging.
Figure 6.
Representative cardiac cine images of a single slice during ventricular systole and diastole from the four accelerated dual-mouse acquisitions.
DISCUSSION
This work demonstrates the feasibility of combining PI techniques with multiple-animal MRI strategies to improve animal throughput and extend the flexibility and sensitivity of small animal imaging studies including challenging applications such as cardiac cine MRI. The development of dual-array imaging hardware, multiple-animal acquisition strategies, and use of a GRAPPA reconstruction algorithm enabled the acceleration of a retrospectively-gated multiple-mouse cardiac cine MRI acquisition by two-, three-, and four-fold. Numerical simulations indicated that approximately 12 dB of interarray decoupling would be sufficient to eliminate the effects of propagated motion artifact from one VOI to the other. The array decoupling that was achieved was well beyond those determined via simulation and minimal error in the GRAPPA reconstruction was expected. Artifacts that are due to unsynchronized motion in other volumes of interest were not apparent in cardiac cine images.
Recently, a retrospectively-gated, multiple-mouse MRI study was successfully performed on three mice with an array of volume coils. Bishop, et al. report achieving nearly equivalent image quality with retrospective gating compared to prospective gating in an equal amount of time (14). In the current work, we dedicated an individual phased-array coil to each animal being simultaneously imaged to achieve an unaccelerated baseline SNR that was 60% greater than that produced by a commercial mouse volume coil. This increase in intrinsic SNR is achieved through detector design and is not sequence dependent. In this work, the increase in SNR afforded an acceleration factor of up to three-fold without any degradation in SNR over the mouse heart. A four-fold imaging acceleration was also demonstrated, although this resulted in a slight degradation (approximately 12%) in SNR compared to an unaccelerated acquisition through the volume coil. For many applications where excess SNR is available, such a reduction in image quality may be tolerated to achieve a substantial reduction in imaging time.
PI techniques for multiple separated VOIs can also be applied to certain extremity applications in human imaging. In particular, the accelerated imaging method described in this work can allow improved imaging speed when imaging two distinct, but similar FOVs that are separated by some distance such as with bilateral imaging of the breasts, legs, or other extremities. Acceleration improvements are possible by eliminating the separation space from the full FOV and rather treating the VOIs as independent. Significant scan time reductions could be realized to promote patient comfort and compliance, especially as the distance between VOIs increases.
The use of MARCs increases the flexibility of traditional multiple-animal MRI. For example, imaging sequences that provide fast image update rates (such as for cardiac cine or DCE-MRI) can potentially suffer from constraints imposed by the larger gradient coils required to accommodate multiple animals and the appropriate imaging hardware (14). Parallel imaging techniques can reduce this gradient load by either reducing the overall scan time or by lowering the gradient duty cycle of a fixed-duration scan by reducing the number of phase encode steps required to encode each image.
In this work, a GRAPPA PI strategy was employed. It is important to note that other PI techniques such as k-tSENSE (26), TSENSE (27), or TGRAPPA (28) can be used for studies involving temporal imaging series, such as cardiac cine imaging (29–33) or DCE-MRI (34–36). The performance trends that were observed during this work are not limited to temporal imaging series, however, or to the particular cine imaging protocol that was used to explore multiple-animal parallel imaging in the presence of motion. This flexibility will be critical during the translation of a greater range of imaging sequences from single-animal to multiple-animal imaging operation.
Higher channel counts will allow increases in both the number of animals simultaneously scanned and the number of array elements that can be dedicated to each animal. Due to the limited availability of receive channels, it was only possible to scan two mice with a pair of two-element phased array coils in this feasibility study. The ultimate gains in throughput, given an appreciable but finite number of receive channels; will depend largely on the target application. Recently, simultaneous in vivo imaging of up to 16 mice was demonstrated for phenotyping the mouse brain (37). In some circumstances, scanning fewer animals faster can improve throughput more effectively than arrays with a very large number of animals. Here, the total increase in data throughput in a dual-mouse cardiac scan using phased-arrays was three, with SNR that was equivalent to that achievable with standard imaging hardware. It is thus feasible to achieve multiple-animal imaging acceleration beyond the number of animals simultaneously scanned without a sacrifice in image quality. Multiple arrays of receive coils improve sensitivity and enable accelerated imaging techniques that achieve increases in throughput while requiring fewer animals to be simultaneously scanned, reducing the complexity with regard to animal preparation and monitoring.
It is important to note that the complexity involved with multiple-animal procedures increases setup time compared to single-animal imaging. Thus, the throughput improvement for the entire imaging protocol is inherently less than that achieved for any individual scan alone. However, we are optimistic that future increases in both the number of animals scanned at once, N, and the number of phased-array elements dedicated to each of the N mice, will allow throughput improvements greater than N for an entire functional mouse protocol.
In conclusion, multiple-mouse MRI with multiple phased-arrays has been shown to further improve imaging efficiency by enabling the use of accelerated imaging techniques. These findings have the potential to substantially improve the efficiency and flexibility of small animal MRI while reducing the overall cost and complexity of imaging in biological investigations. As these techniques mature, they will further promote the use of imaging as a primary noninvasive quantitative tool for basic and preclinical biomedical research.
ACKNOWLEGEMENTS
This work was supported in part by the National Institutes of Health (P30-CA-16672 and U24-CA-126577) and internal research grants. We thank Jorge de la Cerda, Charles Kingsley, Dustin Ragan, Catherine Stambouzou, and Douglas Webb for their assistance.
REFERENCES
- 1.Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller OM. The NMR phased array. Magn Reson Med. 1990;16(2):192–225. doi: 10.1002/mrm.1910160203. [DOI] [PubMed] [Google Scholar]
- 2.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42(5):952–962. [PubMed] [Google Scholar]
- 3.Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): fast imaging with radiofrequency coil arrays. Magn Reson Med. 1997;38(4):591–603. doi: 10.1002/mrm.1910380414. [DOI] [PubMed] [Google Scholar]
- 4.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 5.Griswold MA, Jakob PM, Nittka M, Goldfarb JW, Haase A. Partially parallel imaging with localized sensitivities (PILS) Magn Reson Med. 2000;44(4):602–609. doi: 10.1002/1522-2594(200010)44:4<602::aid-mrm14>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Kyriakos WE, Panych LP, Kacher DF, Westin CF, Bao SM, Mulkern RV, Jolesz FA. Sensitivity profiles from an array of coils for encoding and reconstruction in parallel (SPACE RIP) Magn Reson Med. 2000;44(2):301–308. doi: 10.1002/1522-2594(200008)44:2<301::aid-mrm18>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 7.Schneider JE, Bose J, Bamforth SD, Gruber AD, Broadbent C, Clarke K, Neubauer S, Lengeling A, Bhattacharya S. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev Biol. 2004;4(1):16. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazovic J, Stojkovic DS, Collins CM, Yang QX, Vaughan JT, Smith MB. Hexagonal zero mode TEM coil: a single-channel coil design for imaging multiple small animals. Magn Reson Med. 2005;53(5):1150–1157. doi: 10.1002/mrm.20459. [DOI] [PubMed] [Google Scholar]
- 9.McConville P, Moody JB, Moffat BA. High-throughput magnetic resonance imaging in mice for phenotyping and therapeutic evaluation. Curr Opin Chem Biol. 2005;9(4):413–420. doi: 10.1016/j.cbpa.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Koutcher JA, Hu X, Xu S, Gade TP, Leeds N, Zhou XJ, Zagzag D, Holland EC. MRI of mouse models for gliomas shows similarities to humans and can be used to identify mice for preclinical trials. Neoplasia. 2002;4(6):480–485. doi: 10.1038/sj.neo.7900269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuda Y, Utsuzawa S, Kurimoto T, Haishi T, Yamazaki Y, Kose K, Anno I, Marutani M. Super-parallel MR microscope. Magn Reson Med. 2003;50(1):183–189. doi: 10.1002/mrm.10515. [DOI] [PubMed] [Google Scholar]
- 12.Xu S, Gade TP, Matei C, Zakian K, Alfieri AA, Hu X, Holland EC, Soghomonian S, Tjuvajev J, Ballon D, Koutcher JA. In vivo multiple-mouse imaging at 1.5 T. Magn Reson Med. 2003;49(3):551–557. doi: 10.1002/mrm.10397. [DOI] [PubMed] [Google Scholar]
- 13.Bock NA, Konyer NB, Henkelman RM. Multiple-mouse MRI. Magn Reson Med. 2003;49(1):158–167. doi: 10.1002/mrm.10326. [DOI] [PubMed] [Google Scholar]
- 14.Bishop J, Feintuch A, Bock NA, Nieman B, Dazai J, Davidson L, Henkelman RM. Retrospective gating for mouse cardiac MRI. Magn Reson Med. 2006;55(3):472–477. doi: 10.1002/mrm.20794. [DOI] [PubMed] [Google Scholar]
- 15.Ramirez MS, Ragan DK, Kundra V, Bankson JA. Feasibility of multiple-mouse dynamic contrast-enhanced MRI. Magn Reson Med. 2007;58(3):610–615. doi: 10.1002/mrm.21348. [DOI] [PubMed] [Google Scholar]
- 16.Beuf O, Jaillon F, Saint-Jalmes H. Small-animal MRI: signal-to-noise ratio comparison at 7 and 1.5 T with multiple-animal acquisition strategies. Magma. 2006;19(4):202–208. doi: 10.1007/s10334-006-0048-9. [DOI] [PubMed] [Google Scholar]
- 17.Bernardo M, Kobayashi H, Metzger G, Koyama Y, Shaw C, Thomasson D, Choyke P. In-vivo multiple mouse MRI using parallel receive-only coils on a 3.0 T clinical scanner for molecular imaging research. Proceedings of the 14th Annual Meeting of ISMRM; Seattle, WA, USA.2003. [Google Scholar]
- 18.Bock NA, Nieman BJ, Bishop JB, Henkelman RM. In vivo multiple-mouse MRI at 7 Tesla. Magn Reson Med. 2005;54(5):1311–1316. doi: 10.1002/mrm.20683. [DOI] [PubMed] [Google Scholar]
- 19.Sutton BP, Ciobanu L, Zhang X, Webb A. Parallel imaging for NMR microscopy at 14.1 Tesla. Magn Reson Med. 2005;54(1):9–13. doi: 10.1002/mrm.20531. [DOI] [PubMed] [Google Scholar]
- 20.Vizmuller P. RF Design Guide: Systems, Circuits, and Equations. Boston: Artech House; 1995. [Google Scholar]
- 21.Ramirez MS, Bankson JA. A practical method for 2D multiple-animal MRI. J Magn Reson Imaging. 2007;26(4):1162–1166. doi: 10.1002/jmri.21112. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Wang J, Detre JA. Improved data reconstruction method for GRAPPA. Magn Reson Med. 2005;54(3):738–742. doi: 10.1002/mrm.20601. [DOI] [PubMed] [Google Scholar]
- 23.Esparza-Coss E, Ramirez MS, Bankson JA. Wireless self-gated multiple-mouse cardiac cine MRI. Magn Reson Med. 2008;59(5):1203–1206. doi: 10.1002/mrm.21562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spraggins TA. Wireless retrospective gating: application to cine cardiac imaging. Magnetic resonance imaging. 1990;8(6):675–681. doi: 10.1016/0730-725x(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 25.Kim WS, Mun CW, Kim DJ, Cho ZH. Extraction of cardiac and respiratory motion cycles by use of projection data and its applications to NMR imaging. Magn Reson Med. 1990;13(1):25–37. doi: 10.1002/mrm.1910130105. [DOI] [PubMed] [Google Scholar]
- 26.Tsao J, Boesiger P, Pruessmann KP. k-t BLAST and k-t SENSE: dynamic MRI with high frame rate exploiting spatiotemporal correlations. Magn Reson Med. 2003;50(5):1031–1042. doi: 10.1002/mrm.10611. [DOI] [PubMed] [Google Scholar]
- 27.Kellman P, Epstein FH, McVeigh ER. Adaptive sensitivity encoding incorporating temporal filtering (TSENSE) Magn Reson Med. 2001;45(5):846–852. doi: 10.1002/mrm.1113. [DOI] [PubMed] [Google Scholar]
- 28.Breuer FA, Kellman P, Griswold MA, Jakob PM. Dynamic autocalibrated parallel imaging using temporal GRAPPA (TGRAPPA) Magn Reson Med. 2005;53(4):981–985. doi: 10.1002/mrm.20430. [DOI] [PubMed] [Google Scholar]
- 29.Schneider JE, Lanz T, Barnes H, Medway D, Stork LA, Lygate CA, Smart S, Griswold MA, Neubauer S. Ultra-fast and accurate assessment of cardiac function in rats using accelerated MRI at 9.4 Tesla. Magn Reson Med. 2008;59(3):636–641. doi: 10.1002/mrm.21491. [DOI] [PubMed] [Google Scholar]
- 30.Kim D, Kellman P. Improved cine displacement-encoded MRI using balanced steady-state free precession and time-adaptive sensitivity encoding parallel imaging at 3 T. NMR Biomed. 2007;20(6):591–601. doi: 10.1002/nbm.1126. [DOI] [PubMed] [Google Scholar]
- 31.Wintersperger BJ, Bauner K, Reeder SB, Friedrich D, Dietrich O, Sprung KC, Picciolo M, Nikolaou K, Reiser MF, Schoenberg SO. Cardiac steady-state free precession CINE magnetic resonance imaging at 3.0 tesla: impact of parallel imaging acceleration on volumetric accuracy and signal parameters. Investigative radiology. 2006;41(2):141–147. doi: 10.1097/01.rli.0000192419.08733.37. [DOI] [PubMed] [Google Scholar]
- 32.Wintersperger BJ, Reeder SB, Nikolaou K, Dietrich O, Huber A, Greiser A, Lanz T, Reiser MF, Schoenberg SO. Cardiac CINE MR imaging with a 32-channel cardiac coil and parallel imaging: impact of acceleration factors on image quality and volumetric accuracy. J Magn Reson Imaging. 2006;23(2):222–227. doi: 10.1002/jmri.20484. [DOI] [PubMed] [Google Scholar]
- 33.Yamamuro M, Tadamura E, Kanao S, Okayama S, Okamoto J, Urayama S, Kimura T, Komeda M, Kita T, Togashi K. Cardiac functional analysis by free-breath real-time cine CMR with a spatiotemporal filtering method, TSENSE: comparison with breath-hold cine CMR. J Cardiovasc Magn Reson. 2006;8(6):801–807. doi: 10.1080/10976640600777686. [DOI] [PubMed] [Google Scholar]
- 34.Irwan R, Lubbers DD, van der Vleuten PA, Kappert P, Gotte MJ, Sijens PE. Parallel imaging for first-pass myocardial perfusion. Magnetic resonance imaging. 2007;25(5):678–683. doi: 10.1016/j.mri.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Kellman P, Derbyshire JA, Agyeman KO, McVeigh ER, Arai AE. Extended coverage first-pass perfusion imaging using slice-interleaved TSENSE. Magn Reson Med. 2004;51(1):200–204. doi: 10.1002/mrm.10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber S, Kronfeld A, Kunz RP, Fiebich M, Horstick G, Kreitner KF, Schreiber WG. Comparison of three accelerated pulse sequences for semiquantitative myocardial perfusion imaging using sensitivity encoding incorporating temporal filtering (TSENSE) J Magn Reson Imaging. 2007;26(3):569–579. doi: 10.1002/jmri.21047. [DOI] [PubMed] [Google Scholar]
- 37.Bishop JE, Spring S, Dazai J, Chugh BP, Portnoy S, Suddarth S, Morris GR, Henkelman RM. Multiple Mouse Imaging of 16 Live Mice. Proceedings of the 16th Annual Meeting of ISMRM; Toronto, Ontario, Canada. 2008. [Google Scholar]