Abstract
Autophagy, the process by which proteins and organelles are sequestered in autophagosomal vesicles and delivered to the lysosome/vacuole for degradation, provides a primary route for turnover of stable and defective cellular proteins. Defects in this system are linked with numerous human diseases. While conserved protein kinase, lipid kinase, and ubiquitin-like (UBL) protein conjugation sub-networks controlling autophagosome formation and cargo recruitment have been defined, our understanding of the global organization of this system is limited. Here, we report a proteomic analysis of the autophagy interaction network (AIN) in human cells under conditions of ongoing (basal) autophagy, revealing a network of 751 interactions among 4 09 candidate interacting proteins with extensive connectivity among sub-networks. Many new AIN components have roles in vesicle trafficking, protein or lipid phosphorylation, and protein ubiquitination, and affect autophagosome number or flux when depleted by RNAi. The six ATG8 orthologs in humans (MAP1LC3/GABARAP proteins) interact with a cohort of 67 proteins, with extensive binding partner overlap between family members, and frequent involvement of a conserved surface on ATG8 proteins known to interact with LC3-interacting regions (LIR) in partner proteins. These studies provide a global view of the mammalian autophagy interaction landscape and a resource for mechanistic analysis of this critical protein homeostasis pathway.
Protein homeostasis in eukaryotes is controlled by proteasomal turnover of unstable proteins via the ubiquitin system and lysosomal turnover of the majority of stable proteins through macroautophagy (referred to hereafter as autophagy)1–3. The autophagy system orchestrates the engulfment of cytoplasmic proteins into a double-lipid bilayer – the autophagosome – and coordinates fusion of the autophagosome with the lysosome/vacuole, where degradation occurs. Autophagy is activated in response to low nutrient availability in order to provide building blocks for protein synthesis but can also be used to selectively degrade organelles (e.g mitochondria), misfolded and/or aggregated proteins, and infectious agents1. Defects in autophagy have also been linked to cancer and removal of apoptotic cell corposes1,2.
Autophagy is controlled by pathways that interpret the status of cellular energy (AMP-dependent protein kinase, AMPK), nutrients/amino acids (target of rapamycin, TOR), and growth factors such as insulin. Ground-breaking studies in yeast have revealed four conserved signaling modules encoded by autophagy (ATG) genes that control major steps in the process (Supplementary Fig. S1b–d)1–3. The Atg1p kinase complex (Atg1p-Atg13p-Atg17p)4 and its mammalian counterpart, the unc-51 like kinase (ULK1) complex, control early steps in autophagosome formation, and are regulated by nutrient availability via mammalian TOR (mTOR)5. The vacuolar protein sorting Vps34p-Vps30p complex, and its mammalian PIK3C3-BECN1 (Beclin) counterpart, control production of phosphoinositide signals that facilitate assembly of the incipient autophagosome6. A ubiquitin-like protein (UBL) conjugation cascade - composed of the E1 enzyme Atg7p, two E2 enzymes (Atg10p and Atg3p), and two UBLs (Atg8p and Atg12p) - is required for autophagosome maturation and cargo recruitment1–3. Atg12p is conjugated to a lysine residue in Atg5p via the Atg7p-Atg10p cascade, ultimately forming an oligomeric Atg12p--Atg5p-Atg16p complex that promotes conjugation of Atg8p's C-terminal Gly residue to phosphatidylethanolamine (PE) via Atg3p7 (were -- refers to a covalent bond). The Atg5p complex may also promote incorporation of Atg8p--PE into autophagosomes, allowing Atg8p to promote autophagosome closure and cargo recruitment. Finally, a recycling system containing Atg9p, Atg2p, Atg18p, and Atg21p participates in transfer and recycling of components from the isolation membrane, the presumed source of lipids for production of the autophagosome, to the growing autophagosome8.
While modules within the autophagy system are relatively well characterized, much less is known concerning the overall organization of the pathway and to what extent the various functional elements communicate with each other. Moreover, the autophagy system is more complex in mammalian cells, with several ATG proteins having multiple family members, bringing into question their individual roles in the process. For example, while Atg8p in yeast is represented by a single gene, the ATG8 family in humans contains 6 members (microtubule-associated protein-1 light chain 3A (MAP1LC3A), MAP1LC3B, MAP1LC3C, GABA(A) receptor-associated protein (GABARAP), GABARAPL1, and GABARAPL2). While MAP1LC3 proteins and GABARAP are known to be conjugated to PE and incorporated into autophagosomes, GABARAPL1 and L2 are largely unstudied and the biological underpinnings of the diversity of ATG8 proteins in mammals are unknown. Furthermore, it is likely that additional ATG proteins not found in yeast will participate in the process in vertebrate cells, such as the recently identified ATG101 protein5. Here, we report a systematic proteomic analysis of the human autophagy system coupled with a functional analysis of a subset of genes in the pathway, thereby providing a glimpse into the global architecture of the autophagy interaction network (AIN) and a resource for further mechanistic analysis of this pathway.
Proteomics of the autophagy system
Thirty-two human proteins linked to autophagy or vesicle trafficking were retrovirally expressed as Flag-HA-fusion proteins in 293T cells (Supplementary Fig. S1a, Table S1). Proteins in α-HA immune complexes were identified by mass spectrometry (LC-MS/MS). Total spectral counts for each of the 2553 unique proteins detected were processed using a modified version of the Comparative Proteomics Analysis Software Suite (CompPASS) to identify high-confidence candidate interaction proteins (HCIPs)9. CompPASS employs a database of interacting proteins (including data for all baits reported here and an additional 102 unrelated proteins)9 and three specificity metrics (WDN-Score, an associated p-value, and Z-score) to identify HCIPs (Methods and Supplementary Fig. S2a–d). In order to validate and further delineate the network, 33 HCIPs were chosen as secondary baits for reciprocal proteomic analyses based on interconnectivity with primary baits, the presence of functional domains or Gene Ontology (GO) terms linked to cellular processes associated with autophagy, and the apparent specificity of the interaction based on the WDN-Score (Supplementary Fig. S1a). The number of HCIPs did not correlate with bait abundance (Supplementary Fig. S2e), indicating that bait expression level does not unduly bias HCIP number, and that HCIPs can be identified for baits with very low expression levels. In total, we identified 409 non-redundant HCIPs making 751 interactions (Fig. 1, Supplementary Fig. S3), referred to here as the autophagy interaction network (AIN). Because the cells employed are actively undergoing autophagy under the growth conditions used, albeit with lower rates than found upon stimulation (Supplementary Fig. S6g), the AIN reported here is considered to represent the basal autophagy state.
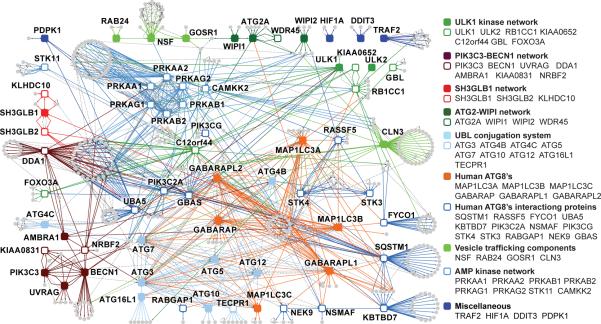
Figure 1. Overview of the autophagy interaction network (AIN).
HCIPs within the autophagy network are shown for 32 primary baits (solid squares) and 33 secondary baits (open squares). Sub-networks are color-coded. Interacting proteins are indicated by gray circles.
Overview of the AIN
Hierarchical clustering of the AIN revealed the presence of multi-protein complexes as well as baits that appeared to interact with a number of partners in a distributive manner (Supplementary Fig. S3). To visualize the interconnectivity of the AIN, we combined interaction maps for 10 functional sub-networks (Fig. 1). As expected, the UBL conjugation machinery sub-network displayed extensive connectivity with the ATG8 sub-network. However, we identified 22 interactions that connected individual proteins or established protein complexes with components in distinct sub-networks. This connectivity is further indicated by the identification of 34 proteins that are found in association with 3 or more bait proteins with WDN-scores>3.0, representing candidate nodes in the network (Supplementary Fig. 5d).
Using BIOGRID, MINT, and STRING protein interaction databases, we identified 84 known protein complexes (KPCs), representing 40 known bait-KPC interactions in the raw AIN (Supplementary Fig. S4b). Using tools in CompPASS that collapse known complexes into single nodes, we identified 497 total candidate associations, of which 429 were not present in these databases (Supplementary Fig. 4a,b). Thus, our analysis greatly expands the number of candidate autophagy network proteins.
Twenty-one percent of the HCIPs in the AIN were enriched in Gene Ontology process descriptors encompassing vesicle transport, proteolysis, signal transduction, and phosphorylation (p ~ 10−5 −10−9) (Supplementary Fig. S5a), accounting for 26% of GO process terms found throughout the AIN HCIPs. Fifty-seven proteins in the AIN lacked GO process descriptors. However, 6 of these are known autophagy proteins (ATG2A, ATG2B, ATG14, RUBICON, AMBRA1, NBR1), 3 could be linked to the ubiquitin system, and 14 could be linked to vesicle or membrane function based on GO component descriptors, functional domains, or previous studies on yeast orthologs (Supplementary Table S7). A variety of functional domains were also enriched (p~0.05−10−7), including protein and lipid kinases, G-protein regulatory motifs (TBC), ubiquitin ligase motifs (BTB), and WD40 repeats (Supplementary Fig. S5b,c).
Validation of sub-networks within the AIN
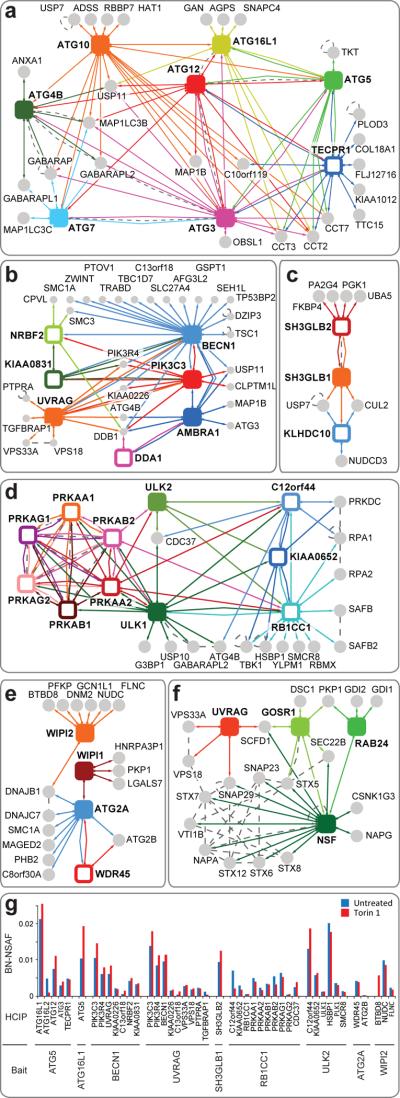
The connectivity within conjugation, ULK1 protein kinase, PIK3C3/BECN1 kinase, ATG2, SH3GLB1 and NSF sub-networks under basal conditions is shown in Fig. 2a–f (maps for individual complexes in Supplementary Fig. S6e). Of 27 interactions seen between 19 budding yeast Atg proteins found in BIOGRID and MINT, we found 23 corresponding human interactions (Supplementary Fig. S7). The UBL conjugation system is emblematic of the quality of the interaction data across the larger network. First, the E1 (ATG7) and the ATG8 hydrolase (ATG4B) associated with 6 and 5 ATG8 orthologs, respectively, as well as with ATG12 (Fig. 2a, 3a). As in yeast3, ATG5 associated with ATG12, ATG10, ATG16L1, ATG3, and a subset of ATG8 orthologs (Fig. 2a, Supplementary Fig. S7a), supporting the role of this complex as an “E3-like” activity for ATG8--PE production7. In total, 12 of 27 interactions in BIOGRID for the human UBL conjugation system were identified by our proteomics analysis (Supplementary Fig. S6a). The absence of complete representation may reflect the fact that 23 of 27 interactions reported in BIOGRID were from unvalidated high-throughput approaches (Supplementary Fig. S6b). For the UBL conjugation system overall, there were 71 previously unidentified HCIPs, and of the 44 potential reciprocal interactions tested by LC-MS/MS, 47% were validated (Supplementary Fig. S6a). For example, a previously uncharacterized tectonin β-propeller repeat protein TECPR1 associated reciprocally with ATG12, ATG3, and ATG5 (Fig. 2a and Supplementary Fig. S6f), suggesting its involvement in the ATG8 lipidation pathway. The WD40-repeat protein ATG16L1 and TECPR1 also associated with the CCT chaperonin complex implicated in the folding of WD40 proteins10.
Figure 2. Autophagy sub-network maps.
Common interacting proteins with sub-threshold WDN-scores were included if HCIP criteria were fulfilled in ≥ 1 IP-MS/MS experiment. Solid squares, primary baits; open squares, secondary baits; grey circles, HCIPs; dotted lines, interactions found in BIOGRID or MINT databases. a, UBL transfer cascade network, b, PIK3C3-BECN1 network, c, SH3GLB1 network, d, ULK1-AMPK network, e, ATG2 network, and f, NSF network. g, Effect of autophagy activation by Torin1 on bait-HCIP association. BN-NSAF, bait normalized-normalized spectral abundance factor.
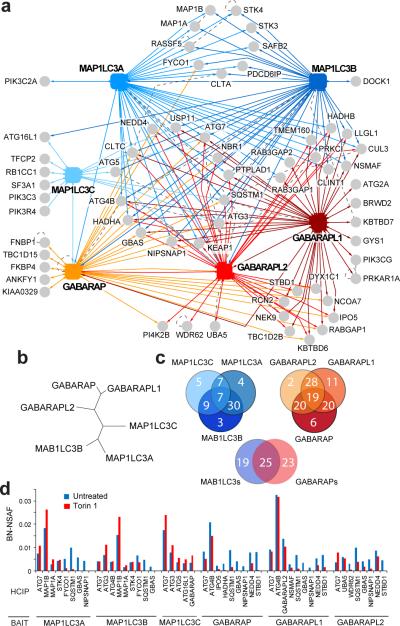
Figure 3. The ATG8 sub-network.
a, Proteomic analysis of MAP1LC3 and GABARAP isoforms (solid squares). b, Human ATG8 protein phylogenetic tree. c, Overlap of interacting proteins found between and among MAP1LC3 and GABARAP subfamilies by LC-MS/MS. d, Effect of autophagy activation by Torin1 on HCIP-ATG8 association. BN-NSAF, bait normalized-normalized spectral abundance factor.
We also identified additional components for other sub-networks. Multiple subunits of the ULK1 and ULK2 kinases were identified, including the recently reported regulatory subunits RB1CC1 (with sequence similarity to Atg11p), KIAA0652/ATG13, and C12orf44/ATG101 (Fig. 2d; Supplementary Fig. S6f)5. Moreover, ULK1 and ULK2 associated with catalytic and regulatory subunits of AMPK (Fig. 2d), which are genetically linked to autophagy in yeast and D. melanogaster11,12. Likewise, KIAA0831/ATG14 and KIAA0226/RUBICON, were found to interact with BECN1 and PIK3C3 (Fig. 2b), as recently reported13–15. Cumulatively, 213 of 243 HCIPs in these 6 sub-networks are not present in BIOGRID or MINT databases. Moreover, 50% of the interactions tested (47/94) by LC-MS/MS were found reciprocally (Supplementary Fig. S6c).
Inhibition of mTOR promotes autophagosome formation, but it is unclear to what extent core interactions within the AIN are reorganized upon mTOR inhibition. In 293T cells employed here, inhibition of mTOR using the small-molecule inhibitor Torin1 (200 nM) led to an increase in the extent of MAP1LC3—PE (LC3-II) above the level found in untreated cells (basal conditions) (Supplemental Figure S6g), signaling up-regulation of the pathway. We performed proteomics on 9 subunits of the conjugation, protein kinase, membrane recycling, and membrane curvature complexes with and without a 6 h incubation of cells with Torin1. In order to quantify changes in sub-network assembly, we employed a previously described label-free semi-quantitative LC-MS/MS approach (see Detailed Methods)16. Among the 44 HCIPs that passed the strict threshold necessary for quantification, 39 maintained interactions in the presence of Torin1 or increased somewhat (20–50%) (Fig. 2g). For example, enhanced association was seen between components within ATG12—ATG5-ATG16L1 and PIK3C3-BECN1-UVRAG complexes (Fig. 2g). In contrast, association between ATG2A, ATG2B, and WDR45 was unaltered by mTOR inhibition (Fig. 2g). While further studies using more quantitative approaches are required to elucidate how activation of the pathway globally affects temporal assembly and disassembly of the AIN, these data suggest the absence of large-scale changes in core conjugation, lipid kinase, and recycling complexes upon mTOR inhibition and imply the possible use of post-translational modification to induce pathway activation.
The ATG8 autophagy receptor sub-network
Proteins of the UBL-containing ATG8 family are central coordinators of autophagosome assembly, maturation, and lysosomal fusion1–3. Upon C-terminal lipidation with PE by an ATG7-ATG3 activation and transfer cascade17, ATG8 it is incorporated into the autophagosomal membrane where it promotes recruitment of cargo19,20, and possibly regulatory factors. Recent work has revealed that cargo recruitment often involves the use of a conserved surface on ATG8 we refer to as the LIR docking site (LDS) that interacts with a conserved hydrophobic W/YXXL motif in cargo binding proteins referred to as the LC3-interaction region (LIR)18 (Supplemental Figure S11c). Mutations in LDS or LIR motifs reduce binding of LIR containing cargo adaptors such as NBR1, p62/SQSTM1 (Sequestosome), and NIX to ATG8 proteins, and disrupt transfer of cargo to the lysosome18–21. NBR1 and SQSTM1 contain ubiquitin binding domains in addition to the LIR motif and have been implicated in recruitment of ubiquitinated cargo, while NIX recruits mitochondria21. Among the 6 ATG8 orthologs in mammals, MAP1LC3A, B, and C and GABARAP are known to be conjugated to PE, incorporated into autophagosomes, and interact with known cargo adaptors1–3,18–21. Much less is known about the functions of GABARAPL1 (ATG8L) and GABARAPL2 (GATE-16), although the latter has been implicated in intra-Golgi vesicle transport3. Given that the high frequency of LIR motifs across the proteome makes the use of predictive approaches in the identification of ATG8 interacting proteins challenging, more direct approaches are required to identify regulatory and cargo adaptor proteins that function together with ATG8.
Through an analysis of the 6 human ATG8 proteins, we identified 67 HCIPs (Fig. 3a,b, Supplementary Fig. S8a, Table S2), significantly expanding the number of candidate ATG8 interacting proteins. Included among these are known targets such as SQSTM1 and NBR118,19, as well as components of the conjugation apparatus (Fig. 3a). Fifty-two percent of the GO process terms in this network are linked with vesicle transport, GTPase signaling, protein/amino acid modification, protein localization and transport, proteolysis, ubiquitin, and phosphorylation (p~10−4 − 10−15) (Supplementary Fig. S5a), and ATG8 interacting proteins are enriched in lipid kinase, WD40, and GTPase regulatory domains (TBC, NIPSNAP), among others (p~10−2−10−5) (Supplementary Fig. S8b). Based solely on LC-MS/MS data, we found that approximately one-third of interacting proteins were specific for the MAP1LC3 and GABARAP sub-families, respectively, while one-third associated with both subfamilies (Fig. 3c). As described below, this may reflect the relative abundance and distribution of the various interacting proteins with ATG8 orthologs in vivo, rather than absolute specificity for a particular ATG8 ortholog.
We also examined the effect of pathway activation via mTOR inhibition on the ATG8 sub-network (Fig. 3d). ATG7 and ATG4B maintained association with ATG8 family members 6 h after Torin1 treatment, while MAP1B displayed increased association with MAP1LC3A and B. In contrast, several proteins including SQSTM1, GBAS/NIPSNAP2, NIPSNAP1, and NEDD4 displayed strongly reduced association with multiple ATG8 orthologs (Fig. 3d). SQSTM1 is degraded upon autophagic delivery to the lysosome, potentially explaining the loss of SQSTM1 in these experiments and raising the possibility that other proteins with this behavior are targeted for degradation in a similar manner.
In vitro ATG8 sub-network validation
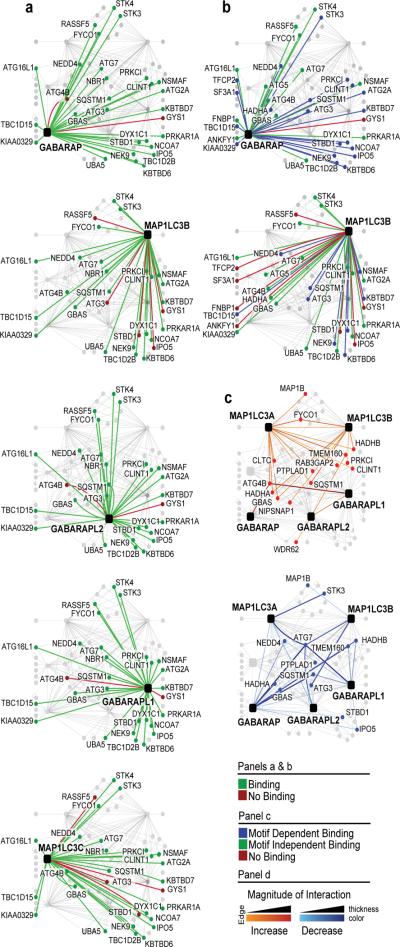
Reciprocal proteomic analysis revealed 40% validation (26/65) of interactions in the ATG8 network (Supplementary Fig. S8a). However, because the small size of ATG8 proteins (~14 kDa) limits the sensitivity in their detection by LC-MS/MS, we directly examined the interaction of GST-ATG8 proteins with 29 ATG8-interacting proteins synthesized in vitro. In total, 130 of the 145 interactions tested (90%) were confirmed (Fig. 4a, Supplementary Fig. S10a–c). Likewise, 85% of 115 interactions tested with 23 MYC-tagged ATG8-interacting proteins expressed in 293T cells were confirmed (Supplementary Fig. S9a–d). Of the proteins tested that were found only in association with the MAP1LC3 sub-network by proteomics, all could associate with GABARAP proteins in vitro (Fig. 4a). Similarly, of the 15 proteins tested that were found only in association with the GABARAP sub-network by proteomics, 12 and 14 were able to associate with MAP1LC3B and MAP1LC3C, respectively (Fig. 4a). Thus, specificity of ATG8 interacting proteins for MAP1LC3 and GABARAP sub-family members (Fig. 3d) may be more relaxed than appreciated by direct proteomic analysis in vivo, or alternatively, specificity may be controlled by cellular factors not present in the in vitro setting.
Figure 4. Specificity within the ATG8 sub-network.
a, In vitro validation. S-tagged proteins were tested for GST-ATG8 binding (Supplementary Fig. S10). Green: binding. Red: no binding. b, LDS dependence. As in panel a using wild-type, GABARAPY49A/L50A or MAP1LC3BF52A/L53A proteins (Supplementary Fig. S11a,b). Blue edges: loss of binding. Green edges: binding maintained. Red edges: no binding with WT. c, ATG8ΔGly dependence. Wild-type and ATG8ΔGly proteins from 293T cells were subjected to LC-MS/MS. Blue edge: decreased binding. Red edge: increased binding. (Supplementary Fig. S12a, Table S3).
LIR-Dependence of the ATG8 sub-network
To examine the extent to which ATG8 binding proteins employ an LIR-LDS interface, 34 ATG8-interacting proteins were tested for in vitro binding to GST-tagged GABARAP or MAP1LC3B in which the LDS was mutated (Y49A/L50A for GABARAP or F52A/L53A for MAP1LC3B) (Fig. 4b, Supplementary Fig. S11c)19. An additional mutant (R70A in MAP1LC3B and R67A in GABARAP) near the LDS was also tested (Supplementary Fig. S11a,b)19. In total, 60% (20/33) of GABARAP-interacting proteins displayed a reduction (20–90%) in binding with the LDS-mutant when compared with wild-type GABARAP in parallel, while the R67A mutant had no effect on binding (Fig. 4b). Similarly, 38% (9/24) of MAP1LC3B-interacting proteins associated with GST-MAP1LC3B in an LDS-dependent manner (Fig 4c). Eight proteins displayed loss of binding with both MAP1LC3B and GABARAP LDS mutants (Fig. 4b). Thus, a substantial fraction of the ATG8 sub-network employs the LDS for assembly. In contrast with GABARAP, 18 proteins displayed reduced binding with the MAP1LC3BR70A mutant, indicating a specialized role for this residue (Supplementary Fig. S11b,d). These results imply LDS-independent and dependent mechanisms for ATG8 network assembly, and raise the possibility that a single ATG8 UBL fold may simultaneously associate with 2 or more interacting proteins to generate a signaling complex.
ATG8 lipidation and sub-network assembly
The ATG8 C-terminal glycine is required for conjugation to PE, facilitating incorporation into autophagosomes3. In order to determine the extent to which formation of the ATG8 sub-network depends upon the C-terminal glycine, we performed a comparative analysis of WT and ATG8ΔGly proteins (GABARAP, GABARAPL1, GABARAPL2, MAP1LC3A, and MAP1LC3B) (see Methods)16. Among the 28 HCIPs that passed the strict threshold required for quantification, 15 remained associated or increased association with ATG8ΔGly (Fig. 4c) (Supplementary Table S12). In contrast, 13 proteins displayed reduced binding with one or more ATG8ΔGly isoforms. Most dramatic was the near complete loss of association with both ATG7 and ATG3 (Fig. 4c, Supplementary Table S12), revealing the importance of the C-terminal glycine and possibly thioester formation for tight association with the conjugation apparatus. Interestingly, association with some proteins (e.g. SQSTM1) was maintained with MAP1LC3ΔGly proteins but was lost with GABARAPΔGly proteins (Fig. 4c), raising the possibility that the two ATG8 sub-families have distinct lipidation requirements for assembly with particular binding partners. These data suggest that a cohort of ATG8 sub-network proteins may assemble prior to ATG lipidation or independent of incorporation of ATG8--PE into the autophagosome.
RNAi analysis of the AIN
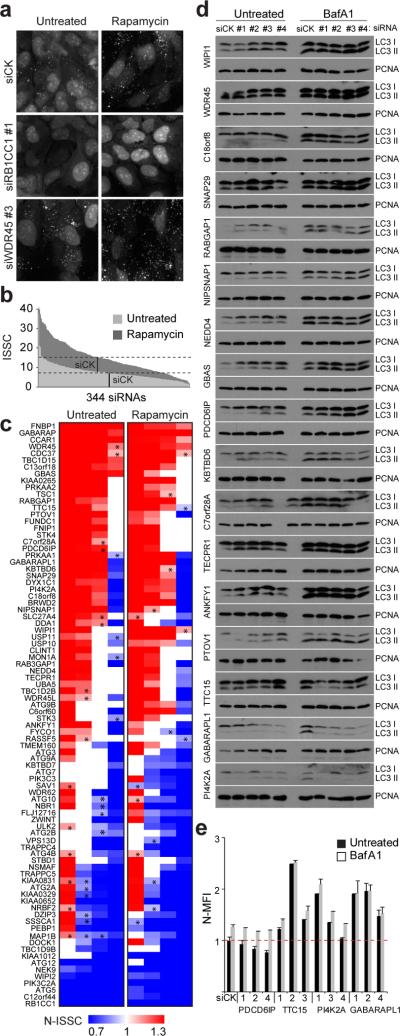
Autophagosomes can be marked with GFP-MAP1LC3 and GFP-GABARAP (Refs 1–3), and depletion of positive (e.g. PIK3C3) or negative (e.g. RUBICON) regulators decreases or increases the number of autophagosomes, respectively13. We initially examined whether depletion of 86 genes in the AIN using RNAi (4 siRNAs/gene in quadruplicate) altered basal or rapamycin-stimulated autophagosome production in U2OS cells expressing GFP-MAP1LC3B. To quantify autophagosome production, we employed an algorithm that determines cytoplasmic integrated spot signal per cell (ISSC) over 400 +/− 100 cells (Fig. 5a–c). The ISSC provides a measure of both the number and intensity of autophagosomes integrated across the cell cytoplasm (see Detailed Methods). Under these conditions, rapamycin induced a ~2.5-fold increase in ISSC in cells transfected with a control siRNA (siCK) (Fig. 5a, b, Supplementary Fig. S13b). A cross section of genes in the AIN were examined, including known autophagy proteins used as controls, but we focused primarily on proteins with multiple interactions within the network, and proteins whose domains or GO process identifiers potentially linked them to the autophagy process.
Figure 5. RNAi analysis of the autophagy interaction network.
a, GFP-MAP1LC3B foci after RNAi and/or Rapamycin in U2OS cells. b, Integrated spot signal/cell (ISSC) for 344 siRNAs targeting 86 genes (n = 4). Control siRNA (siCK) indicated by black vertical bars. c, Normalized ISSC (N-ISSC) for GFP-MAP1LC3B/U2OS with or without Rapamycin (6 h) (4 siRNAs/gene with each bar representing one of four siRNAs). Unless noted otherwise, p < 0.01 using Students T-test; *, p<0.05; white rectangles, p>0.05. d, α-LC3 blots of U2OS cell extracts after depletion of the indicated proteins in the presence or absence of BafA1 (100 nM, 3 h), and re-probed with α-PCNA. e, Normalized-mean fluorescence intensity (N-MFI) of GFP-MAP1LC3B cells after depletion with the indicated siRNAs in the presence or absence of BafA1. Error bars, Standard Deviation; n = 2.
As expected, depletion of ATG5, ATG12, RB1CC1, C12orf44/ATG101, and KIAA0652/ATG13 revealed a significant reduction in normalized ISSC (N-ISSC) under either basal or induced conditions (Fig. 5a, c). Depletion of 31 genes with 2 or more siRNAs resulted in a reduction in autophagosomes, and this was increased to 38 genes in the presence of rapamycin (Fig. 5c, Supplementary Table S6 and Fig. 13g,h). Among the positive regulators of autophagosome formation were the protein kinase NEK9, the lipid kinase PIK3C2A, and the GTPase regulator TBC1D9B, each scoring with 3 or 4 independent siRNAs. The increase in the number of genes scoring in the presence of rapamycin may reflect a more stringent requirement for these genes when the pathway is induced. In contrast, depletion of 30 genes with 3 or 4 siRNAs led to increased autophagosomes under uninduced conditions and 25 genes in the presence of rapamycin (Fig. 5c, Supplementary Table S6), including WDR45, the BECN1 interacting protein C13orf18, and GBAS (Fig. 5c). Analogous studies in the context of GFP-GABARAP produced broadly similar results, suggesting that a subset of the genes identified also control formation of GFP-GABARAP-labeled vesicles (Supplementary Fig. S13d–f). Overall, there was strong correspondence (75%) between the extent of depletion by quantitative PCR or protein abundance and N-ISSC (see Detailed Methods).
Increased autophagosome number could reflect either an increase in the production of autophagosomes (for example, depletion of a negative regulator of an early step in the pathway) or inhibition of later steps in the pathway such as fusion with the lysosome. To examine this question, we employed established autophagosomal flux assays22,23 that examine the steady-state level of MAP1LC3--PE (LC3-II) in the presence and absence of Bafilomycin A1 (BafA1, an inhibitor of autophagosomelysosome fusion) (see Methods). Proteins whose depletion leads to an increase in the steady-state abundance of LC3-II upon BafA1 treatment are considered to cause up-regulation of the pathway, whereas proteins whose depletion has no effect on steady-state LC3-II levels upon BafA1 addition are considered to be required for flux through the pathway22,23. We tested 17 genes whose depletion led to increased N-ISSC using RNAi (Fig. 5d). In fourteen cases, addition of BafA1 led to an increase in the steady-state abundance of LC3-II relative to PCNA used as a loading control, indicating that depletion of these genes upregulates the autophagy system. In contrast, LC3-II levels in cells depleted of TTC15, GABARAPL1, and PI4K2A were not further increased upon BafA1 treatment (Fig. 5d). Thus, these genes appear to block flux through the pathway. These results were confirmed using an independent flow-cytometry assay (Fig. 5e; Supplementary Fig. S13i) (see Methods). While the normalized mean fluorescence intensity (N-MFI) of GFP-MAP1LC3B cells depleted of PCDC6IP was increased upon BafA1 treatment, as expected based on LC3 immunoblotting, cells depleted of GABARAPL1, TTC15, and PIK4C2A displayed an increase in the MFI that did not increase further upon BafA1 treatment (Fig. 5e). Thus, genes identified in the AIN appear to function at distinct stages in the autophagy process based on these flux assays. Further studies are required to pinpoint steps in the process in which genes in the AIN function.
Discussion
As with any screening approach, the results reported here will require additional validation using more directed experiments. The majority of interactions identified here represent the network under ongoing (basal) autophagy in cultured cells, and it is likely that components within the network will be enriched or released upon blockade or activation of the pathway. Indeed, our proteomic analysis of 15 pathway components revealed both gain and loss of components upon stimulation of autophagy by mTOR inhibition. Additional studies are required to fully elucidate and quantify the dynamics and importance of reorganization of the network upon pathway activation by various autophagic stimuli, and to identify mechanisms of action of genes whose depletion alters autophagosome formation in our RNAi experiments. Nevertheless, several potential functional links are suggested by analysis of proteins identified within the existing network. One surprising feature of the network is the extent of connectivity between the various signaling modules. In total, 22 cross-module links were found and each of the major sub-networks devoted to signaling (ULK1-RB1CC1, PIK3C3) and vesicle assembly (ATG2A) make interactions with ATG8 family members (Figs. 1 and 6), suggesting that ATG8 proteins may serve to organize regulatory functions during autophagosome formation. In addition, we identified a large cohort of proteins that associate with ATG8 orthologs, which may facilitate an understanding of the diversity of the human ATG8 proteins. The substantial overlap in binding partners for MAP1LC3 and GABARAP sub-families (Fig. 3 and 4) suggest that they play at least partially redundant roles in autophagy. ATG8-associated proteins could function as receptors that facilitate the engulfment of organelles and cellular proteins into the autophagosome, as is envisioned for NIX, NBR1 and SQSTM119,21, or alternatively, could be regulators of the process. ATG8-associated proteins could also be targeted for degradation directly, as appears to be the case for SQSTM1 and NBR119. Like MAP1LC3 and GABARAP, we found that GABARAPL1 and L2 also associated with ATG7, ATG3, and ATG5, implying that these poorly understood ATG8 family members are also under control of the PE conjugation system.
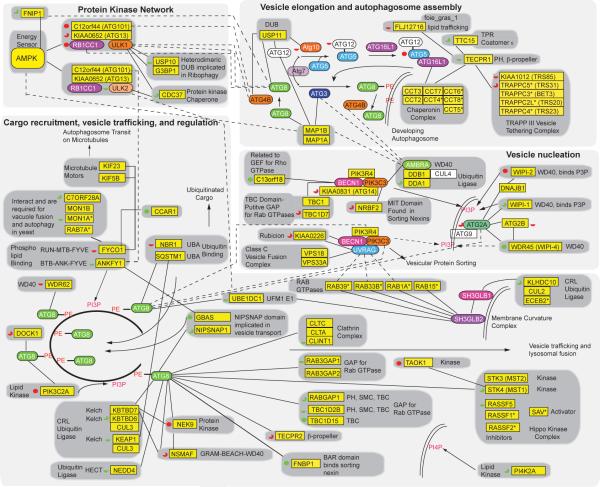
Figure 6. Functional integration of the Autophagy Interaction Network.
Proteins in yellow boxes are HCIPs and proteins labeled with an asterisk were sub-threshold (WDN-score <1.0) for HCIP identification. Dotted lines, cross-module interaction. Solid lines, this study. Dotted arrows, potential functional interactions. Red or green full circle, three-quarter circle, and half-circle represent a reduction or increase in autophagosomes with 4, 3, and 2 siRNAs, respectively, in GFP-MAP1LC3B expressing U2OS cells without rapamycin. Proteins in white boxes were not found by proteomics. The six ATG8 family members are represented by ATG8.
Multiple proteins in the ATG8 network (Fig. 6) contain related domains frequently implicated in vesicle function, including: 1) GBAS/NIPSNAP2 and NIPSNAP1 containing the NIPSNAP domain implicated in vesicle transport/fusion, 2) RABGAP, RAB3GAP1, RAB3GAP2, TBC1D2B and TBC1D15 which contain putative RAB GTPase activating functions, and 3) phosphatidylinositol 3-phosphate (PI3P)-binding FYVE domain-containing proteins FYCO1 and ANKFY1. FYCO1 was independently identified as a Rab7 effector and an ATG8 binding protein, and found to associate with the exterior of autophagosomes via its FYVE domain24, as we have also found (data not shown). Depletion of FYCO1 led to perinuclear accumulation of residual autophagosomes, and a role for FYCO1 in tethering autophagosomes to plus-end directed microtubule motor proteins was proposed to explain this phenotype24. We found that FYCO1 associates with 2 microtubule motor proteins – Kinesin (KIF) 5B and KIF23 (Fig. 6, Supplementary Fig. S6e). Interestingly, depletion of KIF5B leads to perinuclear accumulation of autophagosomes25, allowing us to propose that KIF5B links FYCO1-positive autophagosomes to microtubules to maintain cortical localization. We also identified C7orf28A and MON1B, as well as MON1A and RAB7A with sub-threshold WDN-scores, in association with FYCO1 (Fig. 6). MON1 is orthologous to Mon1p, which in yeast is required for vacuole fusion and autophagy, and interacts with Ccz1p, the RAB7 ortholog Ypt7p, and the vacuole cis-SNARE complex26. Although we were unable to find human orthologs of Ccz1p by conventional sequence searches, C7orf28A and Ccz1p share a distantly related domain of unknown function (DUF1712 in PFAM), suggesting that C7orf28A and Ccz1p are functional orthologs. C7orf28A depletion led to an increase in autophagosome number without blocking autophagosomal flux (Fig. 5d). Thus, FYCO1 appears to function as a platform for assembly of vesicle fusion and trafficking factors. Interestingly, a requirement for orthologs of Mon1p and Ccz1p in apoptotic cell phagocytosis in C. elegans was recently demonstrated27. ANKFY1 associates with both PI3P and RAB5 on endosomes and stimulates endosomal fusion28, and depletion of ANKFY1 also led to increased autophagosomal number without blocking flux (Fig. 5d). The interaction of ATG8 proteins with multiple RAB GTPase regulatory proteins suggests a role for ATG8 in controlling localized RAB activation and vesicle dynamics.
Protein kinases figure prominently in the AIN (Fig. 6). Consistent with recent studies, we found that both ULK1 and ULK2 associate with the RB1CC1-ATG13-C12ORF44/ATG101 regulatory complex (Fig. 2d), which is under mTOR control5. We also found that depletion of ULK2, like depletion of RB1CC1, leads to a reduction in autophagosome formation (Fig. 5c)29. In keeping with a common pathway for assembly, both ULK1 and ULK2 associated with the CDC37 subunit of the HSP90 chaperone complex implicated in stabilization of a substantial fraction of the kinome30. In yeast and Drosophila, AMPK is required for autophagy, but its target(s) in the pathway remain unknown. Interestingly, we found that both ULK1 and ULK2 associate with AMPK (Fig. 2), providing the first biochemical link between these kinases and the cells energy sensing system. Additionally, our proteomic analysis of the AMPK complex revealed several known and candidate AMPK targets, including FNIP1, which was also found in association with GABARAP (Supplementary Tables S2 and S3). Further studies are required to elucidate any regulatory links between AMPK, ATG8 family members, and the ULK1/2 complexes. Additional kinase complexes are linked with the cargo recruitment, vesicle trafficking, and regulation sub-network (Fig. 6), including NEK9 and components of the Hippo kinase complex - STK3/MST2 and STK4/MST1 - implicated in cell growth control. STK4 and NEK9 have opposing roles in autophagosome formation, as measured by our RNAi screen (Fig. 5), and appear to bind ATG8 through distinct surfaces (Fig. 4b).
New components of the ATG8 conjugation, vesicle elongation, and autophagosome assembly sub-network were identified, including the previously unstudied TECPR1 protein, which interacts with the ATG12--ATG5-ATG16 complex (Fig. 2a, 6), and a related protein TECPR2 which was found in association with ATG8 orthologs (Fig. 6). TECPR1 also associated with FLJ12716, TTC15, and components of the TRAPP vesicle-tethering complex, including KIAA1012/TRS85, as well as TRAPPC2L/TRS20, TRAPPC3/BET3, TRAPPC4/TRS23, and TRAPPC5/TRS31 with sub-threshold WDN-scores (Fig. 6, Supplementary Table S2). In yeast, Trs85p has been shown to associate specifically with a unique form of the TRAPP complex, TRAPPIII (containing Trs20p, Trs23p, Trs31p, Bet3p, Bet5p, and Trs33p), is required for assembly of Atg8p onto pre-autophagosomal structures, and serves as a guanine nucleotide exchange factor for the GTPase Ypt131. We found that depletion of TRAPPC5, KIAA1012/TRS85, and FLJ12716 (the human ortholog of the zebrafish foie gras (fgr) gene whose mutation leads to defects in lipid trafficking in hepatocytes32) led to a reduction in autophagosome formation (Fig. 5c). In contrast, depletion of TECPR1 and its associated protein TTC15 led to an increase in autophagosome number (Figs. 5,6). Interestingly, depletion of TTC15 also resulted in a defect in flux, while depletion of TECPR1 did not, suggesting that these proteins act at distinct steps in the process.
In yeast, Atg18p binds PI3P via its WD40 propeller, thereby directing the Atg18p-Atg2p complex to autophagosomal membranes. In mammalian cells, the Atg18p-related WIPI1 protein binds PI3P and associates with MAP1LC3-positive structures, but has not been reported to associate with either of the Atg2p orthologs (ATG2A and ATG2B)33. ATG2A was detected in WIPI1 immune complexes. However, while we did not detect WIPI1 in ATG2A complexes, a previously unstudied WD40 protein WDR45 (WIPI4) reciprocally bound ATG2A and also associated with ATG2B. WDR45 is phylogenetically related to both Atg18p and Atg21p (Supplementary Fig. S11e). Interestingly, ATG2A associated with ATG2B, suggesting that these two related proteins functionally interact (Fig. 2e). WIPI2, the putative ortholog of Atg21p, associated with ATG2A indirectly through DNAJB1 (Fig. 2e). Depletion of WDR45 or WIPI1 led to increased autophagosome number without blocking flux (Fig. 5d), while depletion of WIPI2 led to reduced autophagosome number (Fig. 5c), suggesting distinct roles for WIPI proteins in the autophagy process. It remains to be determined whether WDR45 (WIPI4) also binds PI3P, and whether it functions in a manner analogous to Atg18p in yeast, possibly in combination with WIPI1.
Finally, several components of the ubiquitin-proteasome system are present in the AIN, including AMBRA1/DCAF3, which we previously identified as a putative substrate adaptor for the DDB1-CUL4-DDA1 ubiquitin ligase34 (Fig. 2b, Supplementary Fig. S6). The association of CUL4-DDB1-DDA1AMBRA1 with the PIK3C3-BECN1 complex6 suggests a role for ubiquitin-dependent proteolysis in the function or regulation of this complex. Two additional cullin-related ubiquitin ligases (CRLs) are also found in the AIN – the CUL2-ELOB-ELOCKLHDC10 complex associated with the SH3GLB1 network (Fig. 2c), and the CUL3KBTBD6-KBTBD7 complex found associated with the GABARAP network (Fig. 3a, Supplementary Fig. S6). Moreover, the HECT E3 NEDD4 associates with multiple ATG8 proteins and has been previously linked with endosomal protein degradation35. KBTBD6/7 and NEDD4 associate with ATG8 in an LDS-independent manner and NEDD4 association decreases upon loss of the ATG8 C-terminal glycine, suggesting a role for lipidation in NEDD4 assembly. Enzymes that remove ubiquitin from proteins also figure prominently in the AIN. For example, the ubiquitin specific protease USP10 and its adaptor protein G3BP19 were found to associate with the ULK1 complex and depletion of USP10 led to an increase in autophagosome number (Fig. 5c). Yeast orthologs of USP10 and G3BP1 – Ubp3p and Bre5p – have been implicated in ribophagy36. Further studies are required to determine if ULK1 regulates USP10/G3BP1 function or vice versa.
These studies provide insight into the organization of the mammalian autophagy interaction landscape and will serve as a resource for further mechanistic analysis of this pathway so critical for protein homeostasis.
METHODS SUMMARY
Proteins (Supplementary Table S1) were expressed 293 (or 293T) cells and purified using α-HA prior to proteomic analysis using CompPASS9. Proteomic data in searchable and downloadable formats are available at: http://pathology.hms.harvard.edu/labs/harper/Welcome.html. For in vitro binding, GST-ATG8 proteins (2 μg) were incubated with 35S-methionine-labeled interacting protein (3 μl) in 1 ml of lysis buffer. Proteins were subjected to SDS-PAGE and autoradiography. To measure autophagosome formation, U2OS cells expressing GFP-MAP1LC3B or GFP-GABARAP were transfected in quadruplicate with 30 nM of the indicated siRNAs (Supplementary Table S5) using RNAiMax (Invitrogen). After 72 hr with or without a 6 h treatment with rapamycin (200 nM), nuclei were stained with DRAQ5 and GFP-positive foci in 400 +/− 100 cells quantified using an Opera high-throughput confocal microscope (Evotec) and an Acapella algorithm that measures integrated spot signal per cell cytoplasm, normalizing to a control siRNA (siCK) (Supplementary Table S6). Students T-test was used to determine statistical significance.
Detailed Methods
Plasmids and Cell Lines
Sequence verified ORF clones (Supplementary Table S1) in pDONR223 were recombined into either the Gateway destination vector MSCV-N-Flag-HA-IRES-PURO (LTR-driven expression) or pHAGE-N-Flag-HA (lentiviral vector) using λ recombinase9. After packaging in 293T cells, viruses were used to infect the indicated cell lines and selection accomplished using 1 μg/ml puromycin. The pHAGE-N-Flag-HA vector was employed in transient transfections (293T cells, Lipofectamine 2000 (Invitrogen)) for a subset of AIN proteins that were toxic when expressed constitutively from the LTR promoter (Supplementary Table S1).
Protein Purification
For standard purifications, cells from four 15-cm tissue culture dishes at ~80% confluence (~107 cells) were lysed in a total volume of 4 ml of lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% Nonidet P40, Roche complete EDTA-free protease inhibitor cocktail) for 1 hour with gentle rocking at 4°C. In some experiments, cells were incubated with 200 nM Torin137 (a gift from N. Gray, Dana Farber Cancer Institute) for 6 h prior to harvesting. Lysates were cleared using centrifugation (13,000rpm, 10min), the supernatant was filtered through 0.45 μm spin filters (Millipore) to further remove cell debris, and the resulting material subjected to immunoprecipitation with 60 μl of immobilized anti-HA (Sigma) resin (50% slurry) at 4°C with gentle inversion. Resin containing immune complexes was washed with 1 ml ice-cold lysis buffer 5 times followed by five 1 mL phosphate buffered saline washes. Proteins were eluted with three 50 μl incubations with 250 μg/ml HA-peptide (Sigma) in PBS for 30 minutes each at 22°C, and elutions were pooled for a final volume of 150 μl. Proteins in each elution were precipitated with 20% tri-chloroacetic acid (TCA) and the resulting pellet washed once with 10% TCA and 4 times with cold acetone.
Mass Spectrometry
Trichloroacetic acid (TCA)-precipitated proteins were re-suspended in 30 μl 100 mM ammonium bicarbonate pH 8.0 with 10% acetonitrile and sequencing grade trypsin (750 ng, Promega) and incubated at 37°C for 4 hours. Digested samples were then loaded onto stagetips and washed as described previously9. Peptides were eluted with 50% acetonitrile, 5% formic acid, dried, and re-suspended in 10 μl of 5% acetonitrile, 5% formic acid. For each LC-MS/MS run using an LTQ linear ion trap mass spectrometer (ThermoFinnigan), 4 μl was loaded onto an 18 cm × 125 μm (ID) C18 column and peptides eluted using a 50 minute 8%–26% acetonitrile gradient. Spectra were acquired using a Top-10 method. Each sample was shot twice in succession, followed by a wash with 70% acetonitrile, 30% isopropanol. The resulting spectra were searched using Sequest against a target-decoy database of human tryptic peptides. The resulting list of identifications for each was loaded into CompPASS for further processing and analysis9. All the proteomic data from this work can be obtained at: http://pathology.hms.harvard.edu/labs/harper/Welcome.html.
Validation of Protein Interactions
For reciprocal tagging MS validation, HCIPs were tagged, expressed, and purified using the same methodology as employed for the primary baits. For western analysis, the ORF for the HCIP of interest was recombined into pDEST-CMV-N-Myc using Gateway cloning methods and transfected into 293T cells using Lipofectamine 2000 (Invitrogen). After 48 hrs, whole cell extracts were incubated with the indicated GST-ATG8 protein (2 μg/10 μl resin) purified from bacteria. Washed beads were subjected to SDS-PAGE and immunoblotting using anti-MYC antibodies. For GST pulldown with in vitro expressed 35S-labeled proteins, ORFs of interest were recombined into pET-56-DEST (Novagen) as described above and co-transcribed/translated in the TNT T7 coupled reticulocyte lysate system (Promega). The indicated GST-ATG8 ortholog (or GST as a control) was expressed in E. coli (BL21) cells and purified using GSH-Sepharose. GST-fusion proteins (2 μg in 10 μl of resin) were incubated with 3 μl of in vitro translated and 35S-methionine-labeled interacting protein in a total volume of 1 ml of 25 mM Tris-HCl, 0.5% NP40, 150 mM NaCl, 6 mM EDTA (lysis buffer). After 1 h, the resin was washed 5 times with 1 ml of lysis buffer, bound proteins subjected to SDS-PAGE, Coomassie staining, and autoradiography.
High-throughput autophagosome assays
To develop a 384-well plate based microscopic assay that measures autophagosome formation, we created clonal U2OS cells that stably express either GFP-MAP1LC3B or GFP-GABARAP via a lentivirus. These cells were reverse transfected in quadruplicate with 30 nM of the indicated siRNAs (Supplementary Table S5) using RNAiMax (Invitrogen) and after 72 hr with or without a 6 h treatment with rapamycin (200 nM), nuclei were stained with DRAQ5 (Cell Signaling) and GFP-positive foci quantified using an Opera high-throughput confocal microscope (Evotec) in the Drosophila RNAi Screening Center (DRSC) at Harvard Medical School. As a control siRNA and for normalization purposes, we employed siCK (GAUCCGCAGCGACAUCAACCUGA) (Supplemental Table S5). This siRNA lacks complementarity with any mRNA in the human and mouse genome and gives an indistinguishable number of autophagosomes when compared with mock-transfected cells (data not shown). We imaged 5 planes at 10 positions in each well of the plate with a 40× lens and then performed image analysis on 400 +/− 100 cells using custom scripts in Acapella software (Evotec) that determine the maximum intensity projection, identify nucleus and cytoplasm, and individual spots, and then outputs the number of spots (GFP-MAP1LC3B or GFP-GABARAP foci) per cell cytoplasm and the integrated spot signal per cell cytoplasm. The results of quadruplicate assays were averaged prior to determination of the standard deviation and the normalized average spot signal per cell across multiple plates (Supplementary Table S6). Students T-test was used to determine statistical significance.
Autophagic flux was measured in live cells using a previously described flow cytometric analysis38 and by immunoblotting of LC322. For flow cytometry, U2OS cells expressing GFP-MAP1LC3B in 96-well format were reverse transfected with siRNAs (20 nM) in duplicate and after 72 h, cells were treated with Bafilomycin A1 (3h, 100 nM). A total of 10,000 cells were analyzed for GFP by flow cytometry and analyzed using FLOWJO. For LC3-II immunoblotting assays, cells were transfected with siRNAs and after 72 h, cells were either left untreated or treated with Bafilomycin A1 (3h, 100 nM) prior to lysis in 50 mM Tris (pH 7.5) buffer containing 150 mM NaCl, 0.1% SDS, 1 % Nonidet-P40, and 0.5% deoxycholate. Blots were probed with anti-LC3 (Cell Signaling Technologies) and re-probed with anti-PCNA (Santa Cruz Biotechnology) as a loading control.
Quantitative RT-PCR
U2OS cells were reverse transfected in triplicate with RNAiMAX (Invitrogen) and 30 nM siRNA oligo (Supplementary Table S5). Total RNA was extracted from each sample 72 h after transfection using Trizol reagent (Invitrogen). cDNA was then prepared form ~1μg of total RNA using Superscript II First-Strand Synthesis kit (Invitrogen). Quantitative RT-PCR was performed using LightCycler 480 SYBR Green I Master RT-PCR kit (Roche). Primer pairs used for RT-PCR experiments are provided in Supplementary Table S5. Signals were normalized to GAPDH and to control-transfected cells. Values represent relative mRNA abundance. Using a depletion cut-off set to 60% reduction in mRNA levels, the correlation between depletion and effects on autophagosome number is 75% (67 out of 96). For data obtained by immunoblotting, there was a 79% correlation (19 out of 24).
CompPASS and Bioinformatics
Data Processing and Initial Analysis
Mass spectral data was processed using CompPASS, as previously described9 with modifications. Sequest summary files were processed into a high threshold dataset based on a 2% protein false-positive rate as described earlier.9 These processed data sets were merged for each duplicate run and used to populate a “stats table” consisting of each dataset for the AIN as well as 102 unrelated proteins (Dubs and their selected HCIPs;https://harper.hms.harvard.edu/CompPASS_Dubs.html). The DN- and Z-scores were calculated from total spectral counts (TSCs) for each protein found in association with each bait. Because CompPASS was originally designed for analysis of mostly non-reciprocal datasets9, we devised a new weighted DN-score (WDN-score) (Supplementary Fig. S2), which aids in the identification of HCIPs that are associated with multiple baits in a network. Proteins identified in each LC-MS/MS experiment with a WDN-scores ≥ 1 and a p-value ≤ 4.9 × 10−6 are considered HCIPs.
Additional Bioinformatic Analysis
Interactions used for generating protein networks were from the STRING database, found at http://string.embl.de/, the BioGRID database found at http://www.thebiogrid.org/downloads.php, and the MINT database found at http://mint.bio.uniroma2.it/mint/download.do. When necessary, Ensembl nomenclature was converted into NCBI Gene Symbols using Ensembl's BioMart (http://www.ensembl.org/biomart/index.html). Output files from CompPASS for network analysis are in the “sif” format, which is compatible with Cytoscape (http://www.cytoscape.org) and additional files containing both node and edge attributes are generated. Attribute files were used in Cytoscape to assign values for nodes and edges, as indicated. Gene Ontology (GO) process terms were analyzed in CompPASS, essentially as described9.
Pfam analysis was performed in house using HMMER software and the Pfam_ls set of Hidden Markov matrices from ftp://ftp.sanger.ac.uk/pub/databases/Pfam/. All clustering was done using Multiple Experiment Viewer (www.tm4.org/mev.html) and protein interaction maps were generated using Cytoscape (http://www.cytoscape.org/).
Comparison of HCIP abundance
In order to compare the abundance of HCIPs found in the wild type and mutant ATG8 protein IP-MS/MS experiments, and in experiments examining the effect of Torin1 on sub-networks, we used the normalized spectral abundance factor (NSAF) approach previously applied to determine the abundance of proteins found in IP-MS/MS datasets16. For each interactor in each IP-MS/MS experiments, the NSAF was calculated and then difference in NSAF values for that protein in wild type control and mutant experiment was determined (see Supplemental Methods for detailed formulas). In order to plot the data using the log2 values of this difference while maintaining the proper sign of the value (positive for increase and negative for decrease), the conventional NSAF was multiplied by 100,000 so that each value was ≥ 1 before taking the log2 of the difference.
Supplementary Material
Acknowledgements
We thank I. Dikic for discussions, D. Bowman and J. Ringeling (Millennium Pharmaceuticals) for assistance with Acapela software, N. Perrimon, S. Mohr, and M. Ocana (Harvard Medical School) for access to the Opera microscope, and N. Gray (Dana Farber Cancer Institute) for Torin1. This work was supported by grants to J.W.H. from Millennium Pharmaceuticals, the National Institutes of Health, and the Paul F. Glenn Foundation on Aging. C.B. is a Humboldt Postdoctoral Fellow.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Conflict of Interest Statement J.W.H. is a consultant for Millennium Pharmaceuticals.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–67. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 4.Kabeya Y, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–53. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–82. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 8.Legakis JE, Yen WL, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–32. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- 9.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001;21:5742–52. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lippai M, et al. SNF4Agamma, the Drosophila AMPK gamma subunit is required for regulation of developmental and stress-induced autophagy. Autophagy. 2008;4:476–86. doi: 10.4161/auto.5719. [DOI] [PubMed] [Google Scholar]
- 13.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 14.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sardiu ME, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–9. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulman BA, Harper JW. Ubiquitin-like protein activation: the apex for diverse signaling pathways. Nature Reviews Molecular Cell Biology. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noda NN, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–8. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 19.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–69. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 24.Pankiv S, Endalkachew AA, Brech A, Bruun J-A, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cardoso CM, et al. Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS One. 2009;4:e4424. doi: 10.1371/journal.pone.0004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang CW, Stromhaug PE, Kauffman EJ, Weisman LS, Klionsky DJ. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol. 2003;163:973–85. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 2010;464:778–782. doi: 10.1038/nature08853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnatwinkel C, et al. The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2004;2:E261. doi: 10.1371/journal.pbio.0020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karnitz LM, Felts SJ. Cdc37 regulation of the kinome: when to hold 'em and when to fold 'em. Sci STKE. 2007;2007:pe22. doi: 10.1126/stke.3852007pe22. [DOI] [PubMed] [Google Scholar]
- 31.Lynch-Day MA, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1000063107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–72. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- 33.Jeffries TR, Dove SK, Michell RH, Parker PJ. PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49. Mol Biol Cell. 2004;15:2652–63. doi: 10.1091/mbc.E03-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–21. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Sakata T, et al. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol. 2004;14:2228–36. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 36.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–10. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 37.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–32. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shvets E, Elazar Z. Flow cytometric analysis of autophagy in living mammalian cells. Methods Enzymol. 2009;452:131–41. doi: 10.1016/S0076-6879(08)03609-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.