Abstract
Cutaneous melanoma and uveal melanoma both derive from melanocytes but show remarkable differences in tumorigenesis, mode of metastatic spread, genetic alterations, and therapeutic response. In this review we discuss the differences and similarities along with the genetic research techniques available and the contribution to our current understanding of melanoma. The several chromosomal aberrations already identified prove to be very strong predictors of decreased survival in CM and UM patients. Especially in UM, where the overall risk of metastasis is high (45%), genetic research might aid clinicians in selecting high-risk patients for future systemic adjuvant therapies.
1. Introduction
Cutaneous melanoma (CM) has shown to be one of the life-threatening malignancies with the fastest rise in incidence over the last decades. The highest incidence of CM is observed in Australia (60–70 per 100.000 individuals). In Europe and the USA the incidence is lower (10–15 and 20–30 per 100.000 [1, 2], resp.). CM accounts for more than 90% of all melanomas [3], whereas uveal melanoma (UM) is only encountered in 5% [4]. Nevertheless, UM is the most frequently occurring intraocular malignancy (85%) in the western world. Although CM and UM both derive from melanocytes, these two distinct tumors show remarkable differences in tumorigenesis, mode of metastatic spread, genetic alterations, and therapeutic response [5, 6]. CM can occur anywhere on the body but is predominantly observed in sun-exposed body parts. This partly explains the high incidence of CM in the light-skinned residents of Australia and New-Zealand. UM can occur anywhere along the uveal tract but tend to occur more frequently in the choroid (80%) and the ciliary body (15%) (Figure 1). The incidence of UM appears to be relatively stable with around 7 new patients per 1 million individuals yearly in the western world. UV-light exposure has shown not to be of specific risk in UM. However recently, Schmidt et al. [7] demonstrated a positive interaction between UM and individuals with light colored eyes who sustained frequent UV-radiation. In addition, the tendency of iris melanomas to occur in the lower half of the iris has been explained by the increased sunlight exposure of this area [8]. Other known risk factors for CM and UM are fair skin type (CM and UM), familial occurrence of melanoma (CM) [9], number of melanocytic naevi (CM), light colored eyes (UM), and oculodermal melanocytosis (UM) [10, 11].
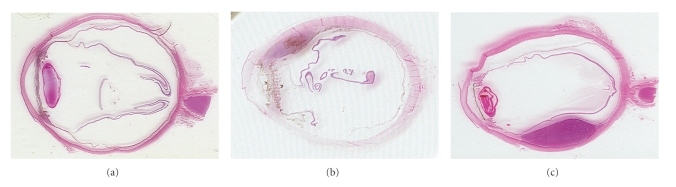
Figure 1.
Uveal melanoma located in: iris (a), ciliary body (b) and choroid (c).
2. Diagnostics
Clinical examination of suspicious lesions remains an important modality in diagnosing CM and UM. As for the diagnosis in CM, dermatologists rely mostly on clinical examination and reserve (excisional) biopsy for tumors of uncertain origin. Only UM of the iris can be diagnosed by external examination and is therefore detected in an early stage. For detection of UM of the choroid or ciliary body, a thorough ophthalmic examination including indirect ophthalmoscopy and ultrasonography of the retina has to be conducted. Tumor growth can lead to retinal detachment and result in extraocular extension of the tumor. At this point, defects in visual field or central vision may be present. Early symptoms of tumor growth, however, can be vague or absent to the patients' notion.
The overall survival is known to be dependent of the tumor thickness (CM) and largest tumor diameter (UM) at time of diagnosis. Therefore, clinicians still concentrate on early detection of CM and UM. This resulted in an average tumor thickness of 0.76 mm in CM at time of diagnosis nowadays. This was shown to relate to an overall 10-year survival of 90% [12] of these small lesions. Similarly, UMs with a diameter of under 4 mm relate to a 5-year survival of 84%. The 5-year survival rate for medium-sized UM (4–8 mm in diameter) is 68%, and 47% with large size UM (over 8 mm in diameter) [13]. The survival of CM and UM patients with metastatic disease is however equally bad with a dismal mean of 2–7 months [14–16].
3. Therapy
The most frequently used therapeutic option in CM is excision of the primary tumor and enucleation of the tumor containing eye in case of large UM. Most small and medium-sized UMs are currently managed by eye-saving treatments such as observation (small inactive tumors), episcleral brachytherapy or charged-particle radiotherapy, and several other variants of radiotherapy. In CM, radiotherapy is only used for palliative purposes as CM cells appear to be relatively radio-resistant. Adjuvant systemic therapy is mainly used in patients at high-risk of metastasis or in patients who already have developed metastasis. The response rates of chemotherapeutic agents in metastasized CM and UM are however as low as 7%–25% [17–20].
4. Metastasis
Both malignancies display a strong tendency to metastasize [3]. Although the mode of metastatic spread is different, CMs tend to metastasize by both hematogenous and lymphogenous route and local invasion. CMs are known to be able to give rise to metastases in skin (13%–38%), distant lymph nodes (5%–34%), distant subcutaneous tissues (32%), lung (18%–36%), liver (14%–20%), CNS (2%–20%), and/or bone (4%–17%) [21]. In UM, metastatic spread is almost exclusively by hematogenous route to remote organs of which the liver is involved in almost all cases (90%) [15, 22]. The reason why UM is not involved in metastatic spread by lymphogenous route is thought to be a direct result of the absence of draining lymphatics of the eye [6, 23]. It is however still unknown why the liver is especially affected by metastases although there are reports about sporadic metastases in lung (24%) and bone (16%) [24–27].
Eventually 45% of UM patients die of metastasis regardless of enucleation or radiotherapy [16]. This has led to theories about the early presence of micrometastasis in the disease process, which remain dormant for years before they give rise to clinically detectable macrometastasis [28]. The exact duration of this proposed state of dormancy and cues for metastatic development remains uncertain. Shields et al. [29, 30] reported tumors with a size of just 1.0 mm to be capable of metastasizing, hence the need for highly specific and sensitive prognostic markers to predict which patient is at risk of developing metastasis. In the quest for significant prognostic markers in UM, already several have been identified. Age (over 60 years), largest basal tumor diameter (over 18 mm), tumor cell type (epitheloid cellularity), and closed vascular patterns correlate with early metastatic disease and shorter survival [31–33]. In CM, tumor thickness (increasing Breslow thicknesses), level of invasion, age (old age), gender (males), anatomic site of primary tumor (head/neck or trunk), number of metastatic lymph nodes, and ulceration on histopathological research appeared to be independent significant prognostic factors of early metastasis [21]. These factors are summarized in a staging system known as the TNM-staging system. This system relies on tumor stage at time of diagnosis which has shown to be the most important prognostic factor in CM and is now widely used for prognostic purposes and clinical decision making [34].
5. Tumor Research Methods
Genetic analysis of tumor material, either from excised CM or from enucleated eyes, has led to the identification of genetic prognostic markers for both types of melanoma.
In the past years several cytogenetic and molecular genetic techniques have been used to investigate the genomic background of melanomas. With conventional karyotyping, we and others were able to identify chromosomal gains, losses, and translocations in UM (Table 1). Comparative Genomic Hybridization (CGH) allows a complete copy number analysis of the entire genome by comparative hybridization of differentially labelled genomic sample and reference DNA to normal human metaphase spreads. Both these techniques have a low resolution of 5–20 MB. Fluorescence in situ Hybridisation (FISH) provides a higher test resolution and even clonal gains and losses present in only a low percentage of tumor cells can be detected [35, 36]. Furthermore, FISH has high test specificity, and although time consuming, still it is a frequently used technique in tumor research and diagnostics. Also paraffin-embedded tissue sections can be assayed by FISH. A drawback to this technique, however, is that only a small number of loci can be analyzed in one single experiment. Molecular genetic techniques such as multiplex ligation-dependent probe amplification (MLPA) and microsatellite instability analysis (MSI) require input of isolated DNA and enable analysis of multiple loci in one experiment with a high resolution.
Table 1.
Overview of techniques used in (molecular) cytogenetics.
| Method | Resolution | Provides genome wide testing? | Detection balanced anomalies? | Detection unbalanced anomalies? |
|---|---|---|---|---|
| Karyotype | ~5–10 Mb | + | + | + |
| G-banding | ||||
| FISH | ~100 kb | − | + | + |
| SKY | ~1-2 Mb | + | + | + |
| MSI | <1 kb | − | − | + |
| CGH | ~5–20 Mb | + | − | + |
| MLPA | ~1–40 kb | − | − | + |
| SNP/ CGH array | >100 kb | + | − | + |
MLPA is a polymerase chain reaction- (PCR-) based technique which functions through the simultaneous hybridization of multiple (up to 50) probes to tumor DNA. Each probe with unique length is only amplified when ligated to its unique probe-counterpart. This provides high specificity of hybridized probes. The final amount of DNA, after several PCR-cycles, is dependent of its initial quantity and eventually copy number changes can be quantitated by relative quantification (RQ). MLPA has proven to be a suitable test for detection of chromosomal anomalies in tumor material [37].
For loss of heterozygosity (LOH) analysis, MSI is frequently used. With this technique, specific markers are required which are allowed to hybridize to the so-called microsatellites within genomic intronic DNA. These microsatellites are tandem repeats of simple polymorphic sequences that are randomly distributed and allow detection of the presence or absence of two different alleles. A drawback to this technique is that only a limited number of markers can be analyzed in a single experiment.
Microarray-based CGH, single-nucleotide polymorphism (SNP) arrays, and gene expression analysis are among the most frequently applied array-based techniques nowadays. All these techniques are based on series of DNA segments (oligonucleotides or bacterial artificial chromosomes; BACs) orderly arranged on a chip, to which fluorescently labeled DNA or RNA can be hybridized. This enables the analysis of copy number status or gene expression of one entire genome very rapidly. Nowadays, there are chips available which enable analysis of structural variation at high level of detail with up to 1.2 million markers. The use of SNP arrays can also provide evaluation of loss of heterozygosity or isodisomy of parts of the genome.
Drawbacks to array technology are its cost, which is about tenfold compared to FISH, and the inability to detect balanced anomalies and genomic abnormalities in frequencies below 10% of analyzed nuclei. Table 1 provides an overview of the differences in resolution and detection limits among the cytogenetic and array-based techniques. The different research techniques previously mentioned certainly contributed to our understanding of melanoma by identification of chromosomes and genes involved in the disease. In the following section we will discuss the most important chromosomal and genetic alterations UM and CM.
6. Chromosomal Aberrations in UM
6.1. Chromosome 3
The most frequently encountered chromosomal aberration in UM is loss of one of the two copies of chromosome 3 (monosomy 3 or −3). Monosomy 3 is observed in approximately 50% of cases [38–41] and appears rather specific for UM as this chromosomal anomaly is rarely encountered in CM or other cancer types [42] (Figure 2). Several groups have already shown that there is a strong correlation between monosomy 3 and the development of metastatic disease [43–46]. In addition, monosomy 3 strongly relates to several clinical and histopathological parameters such as epithelioid cytology, closed vascular patterns, large tumor diameter, and ciliary body involvement [41, 44, 47, 48]. Also, monosomy 3 is thought to represent an early event in tumorigenesis because the alteration is frequently seen in combination with all other known chromosomal abnormalities [49]. In 5%–10% of cases one copy of chromosome 3 is lost and the remaining copy is duplicated. This isodisomic state of chromosome 3 appears to be prognostically equivalent to monosomy 3 [50]. Rarely, partial deletions of chromosome 3 are found [26], and although this has hampered fine mapping studies, a common region of allelic loss on 3p25 and on 3q24–q26 could be defined [50, 51]. Most likely these regions harbor putative tumor suppressor genes but no specific genes have yet been identified.
Figure 2.
Chromosomal aberrations in cutaneous melanoma (CM) and uveal melanoma (UM): Based on all cases in the Mitelman Database of Chromosome Aberrations in cancer by Höglund [42].
Gene expression profiling on UM tumor material does show promising results. By this technique UMs were found to cluster naturally in two distinct molecular classes (class I or class II) based on classifier gene sets [52]. Both classes appear to have clinical prognostic relevance; patients with class I tumors rarely die of metastases, while patients with class II tumors have a high risk of death due to metastases [33, 53]. Onken et al. [54] reported an eight-year survival of 95% for patients with class I UM and 31% for patients with class II UM. Moreover, class II tumors display the previously identified poor prognostic factors: monosomy 3, epitheloid cytology, and closed vascular patterns. The strong significant relation between molecular class and survival indicates that array technology clearly outperforms clinical and histopathological parameters [53–56].
6.2. Chromosome 8
Gain of 8q (+8q) is found in around 40% of UM cases and proved to be an independent significant prognostic marker for decreased survival [43, 45]. It frequently occurs in combination with monosomy 3, either as +8q or as isodisomy 8q, and this combination also shows a strong relation with metastatic disease [43–45, 57]. Abnormalities of chromosomes 3 and 8 are more common in ciliary body-located UMs; whilst alterations of the long arm of chromosome 8 tend to relate to choroid-derived UMs [43, 46, 48, 49]. However, in the study by Kilic et al. [58], chromosome 8q abnormalities were shown to correlate with large tumor diameter but there was no significant relation found between gain of 8q and the metastatic phenotype by univariate analysis. Gain of 8q is also frequently observed in different copy numbers in different UMs, therefore this is speculated to be a late event following the initiation of monosomy 3. The common region of amplification was found to range from 8q24.1 to 8q24.3 [59, 60]. Although gain of chromosome 8q is observed in 25% of CMs, the simultaneous occurrence of monosomy 3 and gain of 8q, as in UM, is rarely observed in CM. Several oncogenes on chromosome 8q were hinted as possible factors in UM pathogenesis; among these genes are MYC (on 8q24), NBS1 (on 8q21), and DDEF1(on 8q24) [46, 61–64]. A potential metastasis suppressor gene located on 8p21, named LZTS1, has been pointed out by Onken et al. [23].
6.3. Other Chromosomal Aberrations in UM
Kilic et al. [65] showed loss of 1p36 in combination with monosomy 3 to be of prognostic significance: these aberrations occurring together display a stronger correlation with decreased survival than monosomy 3 or loss of 1p36 alone (−1p36 by itself is not of prognostic significance). One of the suggested tumor suppressor genes in the 1p36 region, APITD1, was found to be not of significance in patients survival [66]. The common deleted regions on chromosome 1 were found to range from 1p34.3 to 36.2 [48, 67].
Alterations of chromosome 6 are frequently encountered in both UM & CM (discussed later) but show less prognostic value compared to monosomy 3 or gain of 8q in UM [42, 46]. Of these alterations, gain of DNA-material on the short arm of chromosome 6 (+6p) is found in 25%–29% of UM and relates to spindle cell cytology and low risk for development of metastasis [33, 42, 49, 68, 69]. Hughes et al. [60] reported the shortest region of overlap on the p-arm on chromosome 6 to be restricted to 6p22.3–p25. The simultaneous occurrence of +6p and −3, however, is rarely observed. Loss of DNA material on the long arm of chromosome 6 (−6q), observed in 25%–38%, possibly represents another late event in tumorigenesis and correlates with worse prognosis [39, 42, 48, 69, 70]. The region of common deletion on the long arm was found to range from 6q16.1 to 22.3 [60].
Infrequently, abnormalities of the other chromosomes such as loss of 9p, loss of chromosome 10, loss of 11q23–q25, and gain of chromosomes 7 and 10 have been reported [39, 40, 44, 46, 47] but a possible role in tumorigenesis and/ or development of metastasis in UM has yet to be evaluated.
6.4. Genes
Much less is known about genes involved in the development and progression to metastasis in UM compared to CM. This is mainly the result of the lower incidence of UM and the small quantities of tumor sample available for research. While there are many different potential tumor genes identified in CM every year, UM lags behind. However, several candidate genes were proposed in UM recently, such as GNAQ, DDEF1, NBS1, HDM2, BCL-2, and CCND1. For most of these genes, a definite role in tumorigenesis or progression towards metastasis has to be validated.
G protein alpha subunit q (GNAQ) is the first gene found to be mutated frequently in UM. Several groups have shown that approximately 46% of UMs carry mutations in the GNAQ gene [27, 81, 104] (Table 2) turning GNAQ into an oncogene. This oncogenic conversion leads to constitutive activation of the MAP-kinase pathway which results in a situation in which the cell is provided continuous growth signals in the absence of extracellular stimuli [113] and thus cell proliferation. GNAQ status was found not to be correlated with disease free survival; so it could represent an early event in tumorigenesis [27, 104]. This mutation is also found in 83% of blue naevi of the skin [81].
Table 2.
Commonest known genetic changes in CM and UM.
| CM | |||||
|---|---|---|---|---|---|
| Gene | Mechanism | Location | Cases (%) | Reference | |
|
| |||||
| Proto oncogenes | NRAS | mutation | 1p13 | 15–25 | [71, 72] |
| AKT3 | amplification | 1q44 | 40–67 | [73] | |
| BRAF | mutation | 7q34 | 36–61 | [74–76] | |
| NBS1 | amplification | 8q21 | * | [77] | |
| MYC | amplification | 8q24 | 1–40 | [78–80] | |
| DDEF1 | amplification | 8q24 | — | ||
| GNAQ | mutation | 9p21 | 83∗1 | [81] | |
| CCND1 | amplification | 11q13 | 6–44 | [82–84] | |
| HDM2 | amplification | 12q15 | — | ||
| BCL-2 | amplification | 18q21 | >90% | [85, 86] | |
|
| |||||
| Tumor suppressor genes | LZTS1 | deletion | 8p21 | — | |
| CDKN2A-sporadic | deletion, mutation | 9p21 | * | [87] | |
| CDKN2A-familial | deletion, mutation | 9p21 | 30–80 | [88–91] | |
| PTEN | deletion, mutation | 10q23 | 10–40 | [92–94] | |
|
| |||||
| UM | |||||
|
| |||||
| Gene | Mechanism | Location | Cases (%) | Reference | |
|
| |||||
| Proto oncogenes | NRAS | mutation | 1p13 | * | [95–98] |
| AKT3 | amplification | 1q44 | — | ||
| BRAF | mutation | 7q34 | 48∗2 | [81, 99–101] | |
| NBS1 | amplification | 8q21 | 50 | [62, 102] | |
| MYC | amplification | 8q24 | 43 | [103] | |
| DDEF1 | amplification | 8q24 | 50 | [63, 69] | |
| GNAQ | mutation | 9p21 | 46 | [81, 104] | |
| CCND1 | amplification | 11q13 | 65 | [69, 105–107] | |
| HDM2 | amplification | 12q15 | 97 | [69, 105, 106] | |
| BCL-2 | amplification | 18q21 | 100 | [105, 108, 109] | |
|
| |||||
| Tumor suppressor genes | LZTS1 | deletion | 8p21 | — | [23] |
| CDKN2A-sporadic | deletion, mutation | 9p21 | * | [110] | |
| CDKN2A-familial | deletion, mutation | 9p21 | * | [110] | |
| PTEN | deletion, mutation | 10q23 | 15 | [111, 112] | |
— no data available.
*Rarely observed or sporadic reports in literature.
∗1Observed in 83% of blue naevi.
∗2Observed in 48% of iris melanomas.
Furthermore, the DDEF1-gene has been described in UM. It is located on 8q24 and found to be mutated in 50% of UMs leading to overexpression [69]. High expression of DDEF1 was shown to result in more motile low-grade UM cells by Ehlers et al. [63] and could therefore be important in metastatic development [63, 114, 115]. The NBS1-gene is found to be overexpressed in 50% of UM [62]. The encoded protein product is postulated to be part of a complex involved in DNA-repair [102]. It is theorized that overexpressed NBS1 could allow UM progression by promoting the repair of DNA damage which occurs more frequently in advanced tumors with increased genetic instability. High expression of the HDM2-gene on 12q15 is found in 97% of UM [69]. High HDM2 expression was shown to inhibit p53 and its function of eliminating abnormal cells [105, 106]. An elevated expression of BCL-2, located on 18q21, is observed in UM but also in normal melanocytes. This overexpression is reported to block apoptosis [105, 106, 108, 109] and is suggested to be responsible for the resistance to chemotherapy or irradiation of melanocytes [69, 116]. In 65% of UM cases, CCND1 is reported to be overexpressed. Overexpression of CCND1 leads to activation of cyclin dependent kinases (CDKs) which consequently phosphorylate and inactivate Rb [69, 106, 107]. The CCND1 overexpression is associated with large tumor size, epitheloid cytology, and poor prognosis [106].
7. Chromosomal Aberrations in CM
CMs display a more complex karyotype compared to UM. The most frequently observed chromosomal aberration in CM is monosomy 10. This aberration is found in approximately 60% of CM cases and appears to be significantly more frequent compared to UM, where monosomy 10 is found in 27% of cases [42] (Figure 2). Because monosomy 10 could include loss of tumor suppressor genes, much research has been aimed at identifying possible tumor suppressor genes involved. Phosphatase and tensin homolog (PTEN) is one of the identified tumor suppressor genes, located on 10q23, with strong evidence for a role in CM tumorigenesis [92] (Table 2). PTEN is thought to be inactivated by deletion or mutation and through loss of its negative regulatory effect on AKT, lead to activation of the AKT-pathway, and consequently prevent apoptosis [85, 111]. The actual inactivation of PTEN is observed in up to 30%–40% of CM cell lines [92, 93], but only in 10% of primary CMs. PTEN inactivation or downregulation is mainly found in tumors with an increase in aneuploidy, suggesting that it is a late event in tumor progression [27, 111]. In UM, inactivation of PTEN is reported in 15% of cases and has been linked to an increase in aneuploidy but also poor clinical outcome [111, 112].
The other frequently reported chromosomal aberrations involved in CM are −1p, +1q, −4, −5, −6q, +7, −9p, −11q, −12q, −14, −15, −16, −17p, +18, +20, −21, and −22 [42]. Some of them will be discussed here along with the most well-known genes, involved in tumorigenesis and/ or metastatic development.
7.1. Chromosome 1
Rearrangements of the distal part of the short arm of chromosome 1, leading to loss or gain of 1p, are reported in 28% and, respectively, 33% of CMs. Several regions along chromosome 1 are of specific interest because they harbor the NRAS- and AKT3-gene. NRAS is located in the 1p13-region and shown to be activated by mutation in 15%–25% of CMs [71, 72]. NRAS is believed to be also involved in the MAP-kinase pathway. Activation of NRAS leads to activation of the MAP-kinase pathway and as a result cellular proliferation. Additionally, NRAS binds and activates lipid kinase phosphoinositide-3 kinase (PI3K), thereby activating the AKT-pathway and preventing apoptosis [85]. A direct activating mutation of the AKT3-gene located on 1q44 is found in 40–67% of CMs [73]. Overexpression of AKT3 renders cells less sensitive to apoptotic stimuli and as mentioned before; PTEN inactivation can lead to the selective activation of AKT in CMs [92]. Different groups have shown NRAS mutations to be very rare in UM [95–98].
7.2. Chromosome 6
Alterations of chromosome 6 are reported in a total of 66% of CMs, of which +6p is observed in 24% and −6q in 42% [42]. Of these alterations, the 6q10–q27 region shows the highest frequency of rearrangements as a result of deletion, translocation, or due to the formation of an isochromosome of its short arm. The region on the short arm of chromosome 6 that frequently shows alterations spans from 6p21 to 6p25 and mainly results in gain of DNA material. Up till now, there have not been reports about possible over- or underexpressed genes on chromosome 6 involved in tumorigenesis. As mentioned before, both +6p and −6q are common in UM. The prognostic value of these alterations, however, proved to be lower than in CM [42, 46].
7.3. Chromosome 7
In 36% of CMs, gain of DNA-material on both arms of chromosome 7 is observed. Most frequently described are somatic mutations within the 7q34 region, where the BRAF-gene is located. Up to 60%–70% of CMs are characterized by activating mutations in BRAF [74]. The BRAF-gene encodes a kinase involved in the MAP-kinase pathway which, by mutation, is thought to lead to constitutive activation of the aforementioned pathway [117] and cell proliferation. A single substitution (p.V600E) appears to account for more than 90% of all BRAF mutations [118]. The same mutation is also found in 80% of benign naevi and is therefore believed to be an early event in melanomagenesis [75]. There is however evidence from another study that indicates a role in later stages of tumor growth and development [76]. Mutations of BRAF were shown to be absent in UMs [95, 99]. But in a small study, BRAF mutations were shown to occur in 48% of UM of the iris [100].
7.4. Chromosome 9
Chromosomal aberrations on chromosome 9 presenting as either deletions of the short arm, −9p10–24 (37% of CMs), or long arm, +9q22–34 (15% of CMs), have been reported. One of the best characterized genes in CM is CDKN2A, located on 9p21. Inactivating mutations, or loss, results in inactivation of the two encoding tumor suppressor genes p16 and p14. Both genes were already related to high susceptibility for CM and were found in a total of 30%–80% of familial CM [88–90]. These mutations are however rarely observed in sporadic CM [87] or UM [110].
8. Epigenetics
Over the last years, there have been growing interest for the role of epigenetics in CM and UM pathogenesis and metastasis. The most well-known epigenetic features are methylation and microRNAs (miRNAs). Both act through different mechanisms by which they are thought to alter normal gene transcription. Methylation is frequently reported to induce silencing of certain genes by direct methylation of DNA strands or hypermethylation of specific promoters. Because human cancers are theorized to cause global demethylation and promoter hypermethylation, it is thought that this could lead to activation of imprinted genes and the inactivation of genes [119]. In CM, several genes commonly hypermethylated have been identified such as RASSF1A, APC, PYCARD, RARB, MGMT, DAPK, 3-OST-2, HOXB13, SYK, TIMP3A, CDKN2A, FHIT, SOCS1, SOCS2, and PTEN. In UM, the studies regarding gene/promoter methylation status are still limited but CDKN2A is found to be methylated in 33% of cases [120, 121]. Similarly, RASSF1 appears to be methylated in 13%–70% [122] and hTERT in up to 52% [123]. It is not certain whether these methylated sites contribute to metastasis.
MiRNAs have recently come to the attention because of their inhibitory effect on translation of mRNAs into proteins. Although there are limited studies available on the role of miRNAs, several miRNAs have been marked as possibly involved in UM tumorigenesis and/or metastasis such as let-7b, miR18a, miR-199a, miR495, miR549, and more [124, 125]. Worley et al. [124] and Radhakrishnan et al. [125] reported differentially expressed sets of miRNAs that could accurately distinguish two different classes with a low- and high- risk potential for metastatic disease. These miRNAs were shown to bind to genes often found to be deleted in UM such as 8p22, but also 13q and 17p. In CM, many different miRNAs have been identified such as miR-137, miR-182, miR-221, miR-222, and different subtypes of the let-7 family [126–129]. These are thought to act as important factors in CM tumorigenesis and metastasis; further research is however required to analyse their exact role in CM.
9. The Relation between CM and UM
Although there are many differences between CM and UM, they do share some features. First of all, both tumors derive from neural crest melanocytes which migrated to the epidermic tissue or the eye. This common origin is still observed on morphologic and gross histopathologic research of tumor material from CM and UM. The chromosomal regions frequently observed to be amplificated or deleted in both melanotic tumors do resemble each other although the exact frequencies in which they occur differ. For instance, monosomy 3 is observed in around 50% of UMs and in 25% of CMs. The same holds for gene expression status: many of the genes found to be frequently overexpressed or underexpressed in CM are also observed in UM. Furthermore, both tumors are highly metastatic which is illustrated by the early initiation of metastases. UM, however, is not known to spread by lymphogenous route as CM is. This is an important difference and possibly due to the anatomical restrictions of the eye and the lymphatic system. Another difference concerns the role of UV-radiation, which appears to be an important risk factor for the development of CM but is not known as a risk factor for development of choroid-localized UM. There is however evidence regarding an interaction between UV-radiation and development of UM in the easily to sunlight exposed iris [7].
Maybe the two types of melanotic tumors are more similar than previously thought because of its common origin and the differences are merely a result of the exact location of the melanoma and its direct environment. Each location has its own array of carcinogens to which the tissue is exposed to. For instance, the retina is less intensely exposed to UV-radiation than the skin. The epithelial environment the cutaneous melanocytes reside in leads to the cells having more epithelial qualities of which downregulation of the molecule E-cadherin during local invasion is an example. Uveal melanocytes do not require this “mesenchymal to epithelial transition” because they are not in an epithelial environment. This could for part explain the differences in the spectrum of mutations between the two types of melanocytic tumors.
10. Conclusion
Despite all developments in diagnostics and therapeutics of primary UMs in the last 20–30 years, there have been no significant decrease in metastasis-related deaths [6, 130]. The prognosis for patients with metastasized disease still is 2–7 months, regardless of systemic therapy. This is probably due to the early initiation of metastasis in both CM and UM, which underlines the need for early prognostication. This could, at least for part, be achieved by continuing the search for prognostic factors in CM and UM through genetic research on tumor material. Genetic research has showed us that CM and UM have aberrations in common but that these differ in frequency between the two tumors. Even so, both express many of the same genes but not all. In CM, alterations of chromosomes 1, 6, 7, 9, 10, 14, 16, and 21 are frequently observed and already several candidate genes and proteins involved in the tumorigenesis of CM have been identified. UMs were shown to frequently display chromosomal aberrations on chromosomes 1, 3, and 8. Of these, monosomy 3, gain of 8q, and the combination of loss of 1p36 and monosomy 3 appeared to be significant prognostic factors for decreased survival; There have not been identified genes yet that are prognostically active in UM, and at this point developments in UM lag behind compared to CM. New insights in UM, however, came about by gene expression profiling of UMs which were shown to cluster naturally in two classes with different prognosis [52–54]. Generally, array technology has proven to outperform clinical and histopathological parameters in determining a patients' prognosis. This led to the frequent usage of gene expression testing in the current clinical setting in an attempt to identify high-risk patients. We do have to remind that we do not yet know whether monosomy 3 and classifier genes are truly involved in tumor progression and metastatic potential or that those are merely markers of the underlying cause. Additionally, we have to evaluate whether these results may aid clinicians in assessing eligibility of patients for future (adjuvant) systemic therapies. Most of the genetic research is conducted on relatively large UMs because small UMs are treated conservatively and this has biased UM research. Recent groups already reported about the suitability of fine needle aspiration biopsy in harvesting of tumor material from patients treated with eye-saving modalities [131–134]. Also for this diagnostic option we have to evaluate whether this will be beneficial for patient care and can lead to predictions about prognosis for the individual patient. Some genetic markers have already proven its value in predicting prognosis next to clinical and histopathological markers and could lead to selection for patient-tailored therapies in the near future. Also, the challenge will be to prove or disprove the cost-effectiveness of array technology and find additional genetic markers predictive of worse prognosis in CM and UM patients. Concluding, much information has been gained by genetic research of melanonomas and further research could augment our knowledge. Because there are similarities between the two tumors, research on one of two tumors could provide clues for research on the other. Epigenetics, the whole new field in genetic research, does look like a promising ally in our quest to understanding of pathogenesis and metastasis in CM and UM and might provide us with valuable prognostic information in the near future.
Acknowledgment
The authors would like to thank the Rotterdam Ocular Melanoma Research group (ROMS).
References
- 1.Garbe C, Blum A. Epidemiology of cutaneous melanoma in Germany and worldwide. Skin Pharmacology and Applied Skin Physiology. 2001;14(5):280–290. doi: 10.1159/000056358. [DOI] [PubMed] [Google Scholar]
- 2.Giblin A-V, Thomas JM. Incidence, mortality and survival in cutaneous melanoma. Journal of Plastic, Reconstructive and Aesthetic Surgery. 2007;60(1):32–40. doi: 10.1016/j.bjps.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Nambiar S, Mirmohammadsadegh A, Hengge UR. Cutaneous melanoma: fishing with chips. Current Molecular Medicine. 2008;8(3):235–243. doi: 10.2174/156652408784221333. [DOI] [PubMed] [Google Scholar]
- 4.Singh AD, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmology Clinics of North America. 2005;18(1):75–84. doi: 10.1016/j.ohc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Diener-West M, Hawkins BS, Markowitz JA, Schachat AP. A review of mortality from choroidal melanoma: II. A meta-analysis of 5-year mortality rates following enucleation, 1966 through 1988. Archives of Ophthalmology. 1992;110(2):245–250. doi: 10.1001/archopht.1992.01080140101036. [DOI] [PubMed] [Google Scholar]
- 6.Harbour JW. Molecular prognostic testing and individualized patient care in uveal melanoma. American Journal of Ophthalmology. 2009;148(6):823–829. doi: 10.1016/j.ajo.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt-Pokrzywniak A, Jöckel K-H, Bornfeld N, Sauerwein W, Stang A. Positive interaction between light iris color and ultraviolet radiation in relation to the risk of uveal melanoma: a case-control study. Ophthalmology. 2009;116(2):340–348. doi: 10.1016/j.ophtha.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 8.Shields JA, Shields SC. Intraocular Tumors: An Atlas and Textbook. 2nd edition. Philadelphia, Pa, USA: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 9.Meyle KD, Guldberg P. Genetic risk factors for melanoma. Human Genetics. 2009;126(4):499–510. doi: 10.1007/s00439-009-0715-9. [DOI] [PubMed] [Google Scholar]
- 10.Singh AD, De Potter P, Fijal BA, Shields CL, Shields JA, Elston RC. Lifetime prevalence of uveal melanoma in white patients with oculo(dermal) melanocytosis. Ophthalmology. 1998;105(1):195–198. doi: 10.1016/s0161-6420(98)92205-9. [DOI] [PubMed] [Google Scholar]
- 11.Moan J, Porojnicu AC, Dahlback A. Ultraviolet radiation and malignant melanoma. Advances in Experimental Medicine and Biology. 2008;624:104–116. doi: 10.1007/978-0-387-77574-6_9. [DOI] [PubMed] [Google Scholar]
- 12.Saida T. Lessons learned from studies of the development of early melanoma. International Journal of Clinical Oncology. 2005;10(6):371–374. doi: 10.1007/s10147-005-0539-0. [DOI] [PubMed] [Google Scholar]
- 13.Shields CL. The hunt for the secrets of uveal melanoma. Clinical and Experimental Ophthalmology. 2008;36(3):277–280. doi: 10.1111/j.1442-9071.2008.01717.x. [DOI] [PubMed] [Google Scholar]
- 14.Miller AJ, Mihm MC., Jr. Melanoma. The New England Journal of Medicine. 2006;355(1):51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 15.Bedikian AY, Legha SS, Mavligit G, et al. Treatment of uveal melanoma metastatic to the liver: a review of the M. D. Anderson Cancer Center experience and prognostic factors. Cancer. 1995;76(9):1665–1670. doi: 10.1002/1097-0142(19951101)76:9<1665::aid-cncr2820760925>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 16.Kujala E, Mäkitie T, Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma. Investigative Ophthalmology and Visual Science. 2003;44(11):4651–4659. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- 17.Bedikian AY, Millward M, Pehamberger H, et al. Bcl-2 antisense (oblimersen sodium) plus dacarbazine in patients with advanced melanoma: the Oblimersen Melanoma Study Group. Journal of Clinical Oncology. 2006;24(29):4738–4745. doi: 10.1200/JCO.2006.06.0483. [DOI] [PubMed] [Google Scholar]
- 18.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. The New England Journal of Medicine. 2004;351(10):998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 19.Vahrmeijer AL, van de Velde CJH, Hartgrink HH, Tollenaar RAEM. Treatment of melanoma metastases confined to the liver and future perspectives. Digestive Surgery. 2009;25(6):467–472. doi: 10.1159/000184738. [DOI] [PubMed] [Google Scholar]
- 20.Bedikian AY. Metastatic uveal melanoma therapy: current options. International Ophthalmology Clinics. 2006;46(1):151–166. doi: 10.1097/01.iio.0000195852.08453.de. [DOI] [PubMed] [Google Scholar]
- 21.Leiter U, Meier F, Schittek B, Garbe C. The natural course of cutaneous melanoma. Journal of Surgical Oncology. 2004;86(4):172–178. doi: 10.1002/jso.20079. [DOI] [PubMed] [Google Scholar]
- 22.Gragoudas ES, Egan KM, Seddon JM, et al. Survival of patients with metastases from uveal melanoma. Ophthalmology. 1991;98(3):383–390. doi: 10.1016/s0161-6420(91)32285-1. [DOI] [PubMed] [Google Scholar]
- 23.Onken MD, Worley LA, Harbour JW. A metastasis modifier locus on human chromosome 8p in uveal melanoma identified by integrative genomic analysis. Clinical Cancer Research. 2008;14(12):3737–3745. doi: 10.1158/1078-0432.CCR-07-5144. [DOI] [PubMed] [Google Scholar]
- 24.Gragoudas ES. Proton beam irradiation of uveal melanomas: the first 30 years. The Weisenfeld lecture. Investigative Ophthalmology and Visual Science. 2006;47(11):4666–4673. doi: 10.1167/iovs.06-0659. [DOI] [PubMed] [Google Scholar]
- 25.Willson JKV, Albert DM, Diener-West M, et al. Assessment of metastatic disease status at death in 435 patients with large choroidal melanoma in the collaborative ocular melanoma study (coms) coms report no. 15. Archives of Ophthalmology. 2001;119(5):670–676. doi: 10.1001/archopht.119.5.670. [DOI] [PubMed] [Google Scholar]
- 26.Diener-West M, Reynolds SM, Agugliaro DJ, et al. Screening for metastasis from choroidal melanoma: the Collaborative Ocular Melanoma Study Group Report 23. Journal of Clinical Oncology. 2004;22(12):2438–2444. doi: 10.1200/JCO.2004.08.194. [DOI] [PubMed] [Google Scholar]
- 27.Landreville S, Agapova OA, Hartbour JW. Emerging insights into the molecular pathogenesis of uveal melanoma. Future Oncology. 2008;4(5):629–636. doi: 10.2217/14796694.4.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eskelin S, Pyrhönen S, Summanen P, Hahka-Kemppinen M, Kivelä T. Tumor doubling times in metastatic malignant melanoma of the uvea: tumor progression before and after treatment. Ophthalmology. 2000;107(8):1443–1449. doi: 10.1016/s0161-6420(00)00182-2. [DOI] [PubMed] [Google Scholar]
- 29.Shields CL, Shields JA, Kiratli H, De Potter P, Cater JR, McLean IW. Risk factors for growth and metastasis of small choroidal melanocytic lesions. Ophthalmology. 1995;102(9):1351–1361. [PubMed] [Google Scholar]
- 30.Shields CL, Cater J, Shields JA, Singh AD, Santos MCM, Carvalho C. Combination of clinical factors predictive of growth of small choroidal melanocytic tumors. Archives of Ophthalmology. 2000;118(3):360–364. doi: 10.1001/archopht.118.3.360. [DOI] [PubMed] [Google Scholar]
- 31.Diener-West M, Reynolds SM, Agugliaro DJ, et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: collaborative Ocular Melanoma Study Group Report No. 26. Archives of Ophthalmology. 2005;123(12):1639–1643. doi: 10.1001/archopht.123.12.1639. [DOI] [PubMed] [Google Scholar]
- 32.Damato B. Legacy of the collaborative ocular melanoma study. Archives of Ophthalmology. 2007;125(7):966–968. doi: 10.1001/archopht.125.7.966. [DOI] [PubMed] [Google Scholar]
- 33.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Research. 2004;64(20):7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balch CM, Buzaid AC, Soong S-J, et al. New TNM melanoma staging system: linking biology and natural history to clinical outcomes. Seminars in Surgical Oncology. 2003;21(1):43–52. doi: 10.1002/ssu.10020. [DOI] [PubMed] [Google Scholar]
- 35.van Dekken H, Pizzolo JG, Reuter VE, Melamed MR. Cytogenetic analysis of human solid tumors by in situ hybridization with a set of 12 chromosome-specific DNA probes. Cytogenetics and Cell Genetics. 1990;54(3-4):103–107. doi: 10.1159/000132971. [DOI] [PubMed] [Google Scholar]
- 36.Trask BJ. Fluorescence in situ hybridization: applications in cytogenetics and gene mapping. Trends in Genetics. 1991;7(5):149–154. doi: 10.1016/0168-9525(91)90378-4. [DOI] [PubMed] [Google Scholar]
- 37.Damato B, Dopierala J, Klaasen A, van Dijk M, Sibbring J, Coupland SE. Multiplex ligation-dependent probe amplification of uveal melanoma: correlation with metastatic death. Investigative Ophthalmology & Visual Science. 2009;50(7):3048–3055. doi: 10.1167/iovs.08-3165. [DOI] [PubMed] [Google Scholar]
- 38.Horsman DE, Sroka H, Rootman J, White VA. Monosomy 3 and isochromosome 8q in a uveal melanoma. Cancer Genetics and Cytogenetics. 1990;45(2):249–253. doi: 10.1016/0165-4608(90)90090-w. [DOI] [PubMed] [Google Scholar]
- 39.Prescher G, Bornfeld N, Becher R. Nonrandom chromosomal abnormalities in primary uveal melanoma. Journal of the National Cancer Institute. 1990;82(22):1765–1769. doi: 10.1093/jnci/82.22.1765. [DOI] [PubMed] [Google Scholar]
- 40.Sisley K, Rennie IG, Cottam DW, Potter AM, Potter CW, Rees RC. Cytogenetic findings in six posterior uveal melanomas: involvement of chromosomes 3, 6, and 8. Genes Chromosomes and Cancer. 1990;2(3):205–209. doi: 10.1002/gcc.2870020307. [DOI] [PubMed] [Google Scholar]
- 41.Scholes AGM, Damato BE, Nunn J, Hiscott P, Grierson I, Field JK. Monosomy 3 in uveal melanoma: correlation with clinical and histologic predictors of survival. Investigative Ophthalmology and Visual Science. 2003;44(3):1008–1011. doi: 10.1167/iovs.02-0159. [DOI] [PubMed] [Google Scholar]
- 42.Höglund M, Gisselsson D, Hansen GB, et al. Dissecting karyotypic patterns in malignant melanomas: temporal clustering of losses and gains in melanoma karyotypic evolution. International Journal of Cancer. 2004;108(1):57–65. doi: 10.1002/ijc.11558. [DOI] [PubMed] [Google Scholar]
- 43.White VA, Chambers JD, Courtright PD, Chang WY, Horsman DE. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer. 1998;83(2):354–359. [PubMed] [Google Scholar]
- 44.Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jöckel K-H, Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347(9010):1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 45.Sisley K, Rennie IG, Parsons MA, et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes and Cancer. 1997;19(1):22–28. doi: 10.1002/(sici)1098-2264(199705)19:1<22::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 46.Sisley K, Parsons MA, Garnham J, et al. Association of specific chromosome alterations with tumour phenotype in posterior uveal melanoma. British Journal of Cancer. 2000;82(2):330–338. doi: 10.1054/bjoc.1999.0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horsthemke B, Prescher G, Bornfeld N, Becher R. Loss of chromosome 3 alleles and multiplication of chromosome 8 alleles in uveal melanoma. Genes Chromosomes and Cancer. 1992;4(3):217–221. doi: 10.1002/gcc.2870040305. [DOI] [PubMed] [Google Scholar]
- 48.Aalto Y, Eriksson L, Seregard S, Larsson O, Knuutila S. Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Investigative Ophthalmology and Visual Science. 2001;42(2):313–317. [PubMed] [Google Scholar]
- 49.Prescher G, Bornfeld N, Friedrichs W, Seeber S, Becher R. Cytogenetics of twelve cases of uveal melanoma and patterns of nonrandom anomalies and isochromosome formation. Cancer Genetics and Cytogenetics. 1995;80(1):40–46. doi: 10.1016/0165-4608(94)00165-8. [DOI] [PubMed] [Google Scholar]
- 50.Onken MD, Worley LA, Person E, Char DH, Bowcock AM, Harbour JW. Loss of heterozygosity of chromosome 3 detected with single nucleotide polymorphisms is superior to monosomy 3 for predicting metastasis in uveal melanoma. Clinical Cancer Research. 2007;13(10):2923–2927. doi: 10.1158/1078-0432.CCR-06-2383. [DOI] [PubMed] [Google Scholar]
- 51.Parrella P, Fazio VM, Gallo AP, Sidransky D, Merbs SL. Fine mapping of chromosome 3 in uveal melanoma: identification of a minimal region of deletion on chromosomal arm 3p25.1-p25.2. Cancer Research. 2003;63(23):8507–8510. [PubMed] [Google Scholar]
- 52.Tschentscher F, Hüsing J, Hölter T, et al. Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Research. 2003;63(10):2578–2584. [PubMed] [Google Scholar]
- 53.van Gils W, Lodder EM, Mensink HW, et al. Gene expression profiling in uveal melanoma: two regions on 3p related to prognosis. Investigative Ophthalmology and Visual Science. 2008;49(10):4254–4262. doi: 10.1167/iovs.08-2033. [DOI] [PubMed] [Google Scholar]
- 54.Onken MD, Ehlers JP, Worley LA, Makita J, Yokota Y, Harbour JW. Functional gene expression analysis uncovers phenotypic switch in aggressive uveal melanomas. Cancer Research. 2006;66(9):4602–4609. doi: 10.1158/0008-5472.CAN-05-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worley LA, Onken MD, Person E, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clinical Cancer Research. 2007;13(5):1466–1471. doi: 10.1158/1078-0432.CCR-06-2401. [DOI] [PubMed] [Google Scholar]
- 56.Petrausch U, Martus P, Tönnies H, et al. Significance of gene expression analysis in uveal melanoma in comparison to standard risk factors for risk assessment of subsequent metastases. Eye. 2008;22(8):997–1007. doi: 10.1038/sj.eye.6702779. [DOI] [PubMed] [Google Scholar]
- 57.Horsman DE, White VA. Cytogenetic analysis of uveal melanoma: consistent occurrence of monosomy 3 and trisomy 8q. Cancer. 1993;71(3):811–819. doi: 10.1002/1097-0142(19930201)71:3<811::aid-cncr2820710325>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 58.Kilic E, van Gils W, Lodder E, et al. Clinical and cytogenetic analyses in uveal melanoma. Investigative Ophthalmology and Visual Science. 2006;47(9):3703–3707. doi: 10.1167/iovs.06-0101. [DOI] [PubMed] [Google Scholar]
- 59.Sisley K, Tattersall N, Dyson M, Smith K, Mudhar HS, Rennie IG. Multiplex fluorescence in situ hybridization identifies novel rearrangements of chromosomes 6, 15, and 18 in primary uveal melanoma. Experimental Eye Research. 2006;83(3):554–559. doi: 10.1016/j.exer.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 60.Hughes S, Damato BE, Giddings I, Hiscott PS, Humphreys J, Houlston RS. Microarray comparative genomic hybridisation analysis of intraocular uveal melanomas identifies distinctive imbalances associated with loss of chromosome 3. British Journal of Cancer. 2005;93(10):1191–1196. doi: 10.1038/sj.bjc.6602834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parrella P, Caballero OL, Sidransky D, Merbs SL. Detection of c-myc amplification in uveal melanoma by fluorescent in situ hybridization. Investigative Ophthalmology and Visual Science. 2001;42(8):1679–1684. [PubMed] [Google Scholar]
- 62.Ehlers JP, Harbour JW. NBS1 expression as a prognostic marker in uveal melanoma. Clinical Cancer Research. 2005;11(5):1849–1853. doi: 10.1158/1078-0432.CCR-04-2054. [DOI] [PubMed] [Google Scholar]
- 63.Ehlers JP, Worley L, Onken MD, Harbour JW. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clinical Cancer Research. 2005;11(10):3609–3613. doi: 10.1158/1078-0432.CCR-04-1941. [DOI] [PubMed] [Google Scholar]
- 64.Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Frontiers in Bioscience. 1998;3:D250–D268. doi: 10.2741/a239. [DOI] [PubMed] [Google Scholar]
- 65.Kilic E, Naus NC, van Gils W, et al. Concurrent loss of chromosome arm 1p and chromosome 3 predicts a decreased disease-free survival in uveal melanoma patients. Investigative Ophthalmology and Visual Science. 2005;46(7):2253–2257. doi: 10.1167/iovs.04-1460. [DOI] [PubMed] [Google Scholar]
- 66.van Gils W, Mensink HW, Kilic E, et al. Expression of APITD1 is not related to copy number changes of chromosomal region 1p36 or the prognosis of uveal melanoma. Investigative Ophthalmology and Visual Science. 2007;48(11):4919–4923. doi: 10.1167/iovs.07-0061. [DOI] [PubMed] [Google Scholar]
- 67.Häusler T, Stang A, Anastassiou G, et al. Loss of heterozygosity of 1p in uveal melanomas with monosomy 3. International Journal of Cancer. 2005;116(6):909–913. doi: 10.1002/ijc.21086. [DOI] [PubMed] [Google Scholar]
- 68.Parrella P, Sidransky D, Merbs SL. Allelotype of posterior uveal melanoma: implications for a bifurcated tumor progression pathway. Cancer Research. 1999;59(13):3032–3037. [PubMed] [Google Scholar]
- 69.Ehlers JP, Harbour JW. Molecular pathobiology of uveal melanoma. International Ophthalmology Clinics. 2006;46(1):167–180. doi: 10.1097/01.iio.0000195855.31324.db. [DOI] [PubMed] [Google Scholar]
- 70.Ghazvini S, Char DH, Kroll S, Waldman FM, Pinkel D. Comparative genomic hybridization analysis of archival formalin-fixed paraffin-embedded uveal melanomas. Cancer Genetics and Cytogenetics. 1996;90(2):95–101. doi: 10.1016/s0165-4608(96)00076-3. [DOI] [PubMed] [Google Scholar]
- 71.Papp T, Pemsel H, Zimmermann R, Bastrop R, Weiss DG, Schiffmann D. Mutational analysis of the N-ras, p53, p16(INK4a), CDK4, and MC1R genes in human congenital melanocytic naevi. Journal of Medical Genetics. 1999;36(8):610–614. [PMC free article] [PubMed] [Google Scholar]
- 72.Demunter A, Stas M, Degreef H, De Wolf-Peeters C, van den Oord JJ. Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. Journal of Investigative Dermatology. 2001;117(6):1483–1489. doi: 10.1046/j.0022-202x.2001.01601.x. [DOI] [PubMed] [Google Scholar]
- 73.Stahl JM, Sharma A, Cheung M, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Research. 2004;64(19):7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 74.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 75.Pollock PM, Meltzer PS. A genome-based strategy uncovers frequent BRAF mutations in melanoma. Cancer Cell. 2002;2(1):5–7. doi: 10.1016/s1535-6108(02)00089-2. [DOI] [PubMed] [Google Scholar]
- 76.Greene VR, Johnson MM, Grimm EA, Ellerhorst JA. Frequencies of NRAS and BRAF mutations increase from the radial to the vertical growth phase in cutaneous melanoma. Journal of Investigative Dermatology. 2009;129(6):1483–1488. doi: 10.1038/jid.2008.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meyer P, Stapelmann H, Frank B, et al. Molecular genetic analysis of NBS1 in German melanoma patients. Melanoma Research. 2007;17(2):109–116. doi: 10.1097/CMR.0b013e3280dec638. [DOI] [PubMed] [Google Scholar]
- 78.Kraehn GM, Utikal J, Udart M, et al. Extra c-myc oncogene copies in high risk cutaneous malignant melanoma and melanoma metastases. British Journal of Cancer. 2001;84(1):72–79. doi: 10.1054/bjoc.2000.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Treszl A, Ádány R, Rákosy Z, et al. Extra copies of c-myc are more pronounced in nodular melanomas than in superficial spreading melanomas as revealed by fluorescence in situ hybridisation. Cytometry Part B. 2004;60(1):37–46. doi: 10.1002/cyto.b.20016. [DOI] [PubMed] [Google Scholar]
- 80.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Research. 2007;67(6):2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 81.van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457(7229):599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. The New England Journal of Medicine. 2005;353(20):2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 83.Jönsson G, Dahl C, Staaf J, et al. Genomic profiling of malignant melanoma using tiling-resolution arrayCGH. Oncogene. 2007;26(32):4738–4748. doi: 10.1038/sj.onc.1210252. [DOI] [PubMed] [Google Scholar]
- 84.Sauter ER, Yeo U-C, Von Stemm A, et al. Cyclin D1 is a candidate oncogene in cutaneous melanoma. Cancer Research. 2002;62(11):3200–3206. [PubMed] [Google Scholar]
- 85.Ibrahim N, Haluska FG. Molecular pathogenesis of cutaneous melanocytic neoplasms. Annual Review of Pathology. 2009;4:551–579. doi: 10.1146/annurev.pathol.3.121806.151541. [DOI] [PubMed] [Google Scholar]
- 86.Jansen B, Schlagbauer-Wadl H, Brown BD, et al. BCL-2 antisense therapy chemosensitizes human melanoma in SCID mice. Nature Medicine. 1998;4(2):232–234. doi: 10.1038/nm0298-232. [DOI] [PubMed] [Google Scholar]
- 87.Pons M, Mancheño-Corvo P, Martín-Duque P, Quintanilla M. Molecular biology of malignant melanoma. Advances in Experimental Medicine and Biology. 2008;624:252–264. doi: 10.1007/978-0-387-77574-6_20. [DOI] [PubMed] [Google Scholar]
- 88.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 89.Kamb A, Gruis NA, Weaver-Feldhaus J, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264(5157):p. 436. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 90.Sharpless NE, Chin L. The INK4a/ARF locus and melanoma. Oncogene. 2003;22(20):3092–3098. doi: 10.1038/sj.onc.1206461. [DOI] [PubMed] [Google Scholar]
- 91.Goldstein AM, Chan M, Harland M, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. Journal of Medical Genetics. 2007;44(2):99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22(20):3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 93.Tsao H, Zhang X, Benoit E, Haluska FG. Identification of PTEN/MMAC1 alterations in uncultured melanomas and melanoma cell lines. Oncogene. 1998;16(26):3397–3402. doi: 10.1038/sj.onc.1201881. [DOI] [PubMed] [Google Scholar]
- 94.Tsao H, Zhang X, Fowlkes K, Haluska FG. Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. Cancer Research. 2000;60(7):1800–1804. [PubMed] [Google Scholar]
- 95.Mooy CM, van der Helm MJ, van der Kwast TH, De Jong PTVM, Ruiter DJ, Zwarthoff EC. No N-ras mutations in human uveal melanoma: the role of ultraviolet light revisited. British Journal of Cancer. 1991;64(2):411–413. doi: 10.1038/bjc.1991.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kiliç E, Brüggenwirth HT, Verbiest MMPJ, et al. The RAS-BRAF kinase pathway is not involved in uveal melanoma. Melanoma Research. 2004;14(3):203–205. doi: 10.1097/01.cmr.0000130006.46885.a0. [DOI] [PubMed] [Google Scholar]
- 97.Saldanha G, Purnell D, Fletcher A, Potter L, Gillies A, Pringle JH. High BRAF mutation frequency does not characterize all melanocytic tumor types. International Journal of Cancer. 2004;111(5):705–710. doi: 10.1002/ijc.20325. [DOI] [PubMed] [Google Scholar]
- 98.Janssen CS, Sibbett R, Henriquez FL, McKay IC, Kemp EG, Roberts F. The T1799A point mutation is present in posterior uveal melanoma. British Journal of Cancer. 2008;99(10):1673–1677. doi: 10.1038/sj.bjc.6604731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cohen Y, Goldenberg-Cohen N, Parrella P, et al. Lack of BRAF mutation in primary uveal melanoma. Investigative Ophthalmology and Visual Science. 2003;44(7):2876–2878. doi: 10.1167/iovs.02-1329. [DOI] [PubMed] [Google Scholar]
- 100.Henriquez F, Janssen C, Kemp EG, Roberts F. The T1799A BRAF mutation is present in iris melanoma. Investigative Ophthalmology and Visual Science. 2007;48(11):4897–4900. doi: 10.1167/iovs.07-0440. [DOI] [PubMed] [Google Scholar]
- 101.Spendlove HE, Damato BE, Humphreys J, Barker KT, Hiscott PS, Houlston RS. BRAF mutations are detectable in conjunctival but not uveal melanomas. Melanoma Research. 2004;14(6):449–452. doi: 10.1097/00008390-200412000-00003. [DOI] [PubMed] [Google Scholar]
- 102.Falck J, Petrini JHJ, Williams BR, Lukas J, Bartek J. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nature Genetics. 2002;30(3):290–294. doi: 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- 103.Parrella P, Caballero OL, Sidransky D, Merbs SL. Detection of c-myc amplification in uveal melanoma by fluorescent in situ hybridization. Investigative Ophthalmology and Visual Science. 2001;42(8):1679–1684. [PubMed] [Google Scholar]
- 104.Bauer J, Kilic E, Vaarwater J, Bastian BC, Garbe C, De Klein A. Oncogenic GNAQ mutations are not correlated with disease-free survival in uveal melanoma. British Journal of Cancer. 2009;101(5):813–815. doi: 10.1038/sj.bjc.6605226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brantley MA, Jr., Harbour JW. Deregulation of the Rb and p53 pathways in uveal melanoma. American Journal of Pathology. 2000;157(6):1795–1801. doi: 10.1016/s0002-9440(10)64817-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Coupland SE, Anastassiou G, Stang A, et al. The prognostic value of cyclin D1, p53, and MDM2 protein expression in uveal melanoma. Journal of Pathology. 2000;191(2):120–126. doi: 10.1002/(SICI)1096-9896(200006)191:2<120::AID-PATH591>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 107.Coupland SE, Bechrakis N, Schüler A, et al. Expression patterns of cyclin D1 and related proteins regulating G1-S phase transition in uveal melanoma and retinoblastoma. British Journal of Ophthalmology. 1998;82(8):961–970. doi: 10.1136/bjo.82.8.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ghana JS, Wilson GD, Cree IA, et al. c-myc, p53, and Bcl-2 expression and clinical outcome in uveal melanoma. British Journal of Ophthalmology. 1999;83(1):110–114. doi: 10.1136/bjo.83.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jay V, Yi Q, Hunter WS, Zielenska M. Expression of bcl-2 in uveal malignant melanoma. Archives of Pathology and Laboratory Medicine. 1996;120(5):497–498. [PubMed] [Google Scholar]
- 110.Singh AD, Wang MX, Donoso LA, Shields CL, De Potter P, Shields JA. Genetic aspects of uveal melanoma: a brief review. Seminars in Oncology. 1996;23(6):768–772. [PubMed] [Google Scholar]
- 111.Ehlers JP, Worley L, Onken MD, Harbour JW. Integrative genomic analysis of aneuploidy in uveal melanoma. Clinical Cancer Research. 2008;14(1):115–122. doi: 10.1158/1078-0432.CCR-07-1825. [DOI] [PubMed] [Google Scholar]
- 112.Abdel-Rahman MH, Yang Y, Zhou X-P, Craig EL, Davidorf FH, Eng C. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. Journal of Clinical Oncology. 2006;24(2):288–295. doi: 10.1200/JCO.2005.02.2418. [DOI] [PubMed] [Google Scholar]
- 113.Wan PTC, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 114.Randazzo PA, Andrade J, Miura K, et al. The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(8):4011–4016. doi: 10.1073/pnas.070552297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu Y, Loijens JC, Martin KH, Karginov AV, Parsons JT. The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Molecular Biology of the Cell. 2002;13(6):2147–2156. doi: 10.1091/mbc.E02-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109(6):707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 117.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nature Reviews Molecular Cell Biology. 2004;5(11):875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 118.Platz A, Egyhazi S, Ringborg U, Hansson J. Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Molecular Oncology. 2008;1(4):395–405. doi: 10.1016/j.molonc.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatric Research. 2007;61(5, part 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 120.Merbs SL, Sidransky D. Analysis of p16 (CDKN2/MTS-1/INK4A) alterations in primary sporadic uveal melanoma. Investigative Ophthalmology and Visual Science. 1999;40(3):779–783. [PubMed] [Google Scholar]
- 121.van der Veiden PA, Metzelaar-Blok JAW, Bergman W, et al. Promoter hypermethylation: a common cause of reduced p16INK4a expression in uveal melanoma. Cancer Research. 2001;61(13):5303–5306. [PubMed] [Google Scholar]
- 122.Maat W, van der Velden PA, Out-Luiting C, et al. Epigenetic inactivation of RASSF1a in uveal melanoma. Investigative Ophthalmology and Visual Science. 2007;48(2):486–490. doi: 10.1167/iovs.06-0781. [DOI] [PubMed] [Google Scholar]
- 123.Moulin AP, Clément G, Bosman FT, Zografos L, Benhattar J. Methylation of CpG island promoters in uveal melanoma. British Journal of Ophthalmology. 2008;92(2):281–285. doi: 10.1136/bjo.2007.127035. [DOI] [PubMed] [Google Scholar]
- 124.Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Research. 2008;18(3):184–190. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 125.Radhakrishnan A, Badhrinarayanan N, Biswas J, Krishnakumar S. Analysis of chromosomal aberration (1, 3, and 8) and association of microRNAs in uveal melanoma. Molecular Vision. 2009;15:2146–2154. [PMC free article] [PubMed] [Google Scholar]
- 126.Bemis LT, Chen R, Amato CM, et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Research. 2008;68(5):1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 127.Segura MF, Hanniford D, Menendez S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(6):1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Felicetti F, Errico MC, Segnalini P, Mattia G, Carè A. MicroRNA-221 and -222 pathway controls melanoma progression. Expert Review of Anticancer Therapy. 2008;8(11):1759–1765. doi: 10.1586/14737140.8.11.1759. [DOI] [PubMed] [Google Scholar]
- 129.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 130.Singh AD, Topham A. Survival rates with uveal melanoma in the United States: 1973–1997. Ophthalmology. 2003;110(5):962–965. doi: 10.1016/S0161-6420(03)00077-0. [DOI] [PubMed] [Google Scholar]
- 131.Shields CL, Ganguly A, Materin MA, et al. Chromosome 3 analysis of uveal melanoma using fine-needle aspiration biopsy at the time of plaque radiotherapy in 140 consecutive cases: the Deborah Iverson, MD, Lectureship. Archives of Ophthalmology. 2007;125(8):1017–1024. doi: 10.1001/archopht.125.8.1017. [DOI] [PubMed] [Google Scholar]
- 132.Midena E, Bonaldi L, Parrozzani R, Tebaldi E, Boccassini B, Vujosevic S. In vivo detection of monosomy 3 in eyes with medium-sized uveal melanoma using transscleral fine needle aspiration biopsy. European Journal of Ophthalmology. 2006;16(3):422–425. doi: 10.1177/112067210601600310. [DOI] [PubMed] [Google Scholar]
- 133.Young TA, Rao NP, Glasgow BJ, Moral JN, Straatsma BR. Fluorescent in situ hybridization of monosomy 3 via 30-gauge fine-needles aspiration biopsy of choroidal melanoma in vivo. Ophthalmology. 2007;114(1):142–146. doi: 10.1016/j.ophtha.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 134.Shields CL, Materin MA, Teixeira L, Mashayekhi A, Ganguly A, Shields JA. Small choroidal melanoma with chromosome 3 monosomy on fine-needle aspiration biopsy. Ophthalmology. 2007;114(10):1919–1924. doi: 10.1016/j.ophtha.2007.04.054. [DOI] [PubMed] [Google Scholar]