Abstract
Meaningful RNAi-based data for target gene identification are strongly dependent on the use of a biologically relevant cell type and efficient delivery of highly functional siRNA reagents into the selected cell type. Here we report the use of the Amaxa® Nucleofector® 96-well Shuttle® System for siRNA screening in primary cells. Lonza's Clonetics® HUVEC-Human Umbilical Vein Endothelial Cells were transfected with Thermo Scientific Dharmacon siGENOME® siRNA Libraries targeting protein kinases and cell cycle related genes and screened for genes important for cell viability. Of the 37 primary hits, down-regulation of 33 led to reduced proliferation or increased cell death, while down-regulation of two allowed for better cell viability. The validated four genes out of the 16 strongest primary hits (COPB2, PYCS, CDK4 and MYC) influenced cell proliferation to varying degrees, reflecting differing importance for survival of HUVEC cells. Our results demonstrate that the Nucleofector® 96-well Shuttle® System allows the delivery of siRNA libraries in cell types previously considered to be difficult to transfect. Thus, identification and validation of gene targets can now be conducted in primary cells, as the selection of cell types is not limited to those accessible by lipid-mediated transfection.
Keywords: Nucleofection, RNAi, siRNA, primary cell, screening, transfection, HUVEC
INTRODUCTION
RNAi-based library screening has become a powerful in vitro tool to identify drug targets that play a role in disease development and progression (Martin and Caplen, 2007). Successful screening experiments using siRNA require efficient delivery of highly functional and specific siRNA molecules into appropriate cells. While lipid-mediated transfection is a common approach for siRNA delivery, many cell types, including suspension cell lines and primary cells, are not compatible with this technology (Merkerova et al, 2007). This limitation prevents analysis of many biologically relevant cell types and restricts siRNA library screenings mainly to transformed, adherent cells that often exhibit phenotypic and genetic anomalies after extended periods of culturing lines (MacKeigan et al, 2005; Bartz et al, 2006; Whitehurst et al, 2007). Ideally, the diversity of biological questions requires the use of appropriate cell types, typically primary cells. In addition to this issue, several of the lipid delivery reagents can cause cytotoxicity and are capable of inducing a potent interferon response and/or altering gene expression profiles (Marques and Williams, 2005; Fedorov et al, 2005; Wang, 2006). These unintended phenotypes can significantly affect experimental outcomes and drastically interfere with identifying relevant genes and understanding a gene's function. Human Umbilical Vein Endothelial Cells (HUVEC), a difficult-to-transfect cell type, were screened with an siRNA library delivered using the Amaxa® Nucleofector® 96-well Shuttle® System. The screen targeted protein kinases and genes associated with the cell cycle to identify target genes important for cell viability.
MATERIALS AND METHODS
The siRNA reagents used were Dharmacon Human siGENOME® SMARTpool® siRNA Libraries for Protein Kinases (targeting 779 genes) and Cell Cycle Regulation (targeting 111 genes) (Thermo Fisher Scientific). Clonetics® HUVEC Cells (Lonza) were cultured in Clonetics® EGM® Endothelial Growth Medium (Lonza) at 37oC, 5% (v/v) CO2 and transfected according to the recommendations in the respective Optimized Protocol for 96-well Nucleofection® (Amaxa). Briefly, 2 × 104 HUVEC cells were transfected with 20 pmol siRNA (if not noted differently). For optimal assay conditions, post-transfection HUVEC cells were plated in 96-well culture plates at a density of 2 × 103 cells per well (100 μl). Outer wells of culture plates were filled with media only in order to avoid edge effects in the phenotypic assays. HUVEC cells were analyzed 72 hrs post-transfection for cell viability. The QuantiGene® Branched DNA Assay (Panomics) was utilized to quantify transcript levels and correlate target knockdown with biological phenotype. Cyclophilin B served as reference mRNA and values were normalized to samples transfected with control siRNA. For the primary screen (n=3 independent experiments), Clonetics® HUVEC cells were transfected with the respective libraries or control siRNAs and analyzed for phenotypic effects (cell viability). Data from each screen were analyzed by statistical means: the Z' factors (Zhang et al, 1999) of controls were determined to evaluate the quality of the experiment and robust Z-score calculation (Chung et al, 2008) was used for hit identification. For target validation, selected hits were first re-evaluated with a higher number of samples using the siRNA utilized in the primary screen. Samples were randomly arranged across the plate to ensure independence of the phenotype from well positions. Subsequently, hits were further validated by demonstrating multiple knockdown reagents in different formats induced the same phenotypes (e.g., single or specificity-enhanced Thermo Scientific Dharmacon ON-TARGETplus siRNA Reagents).
RESULTS AND DISCUSSION
Viability assay optimization
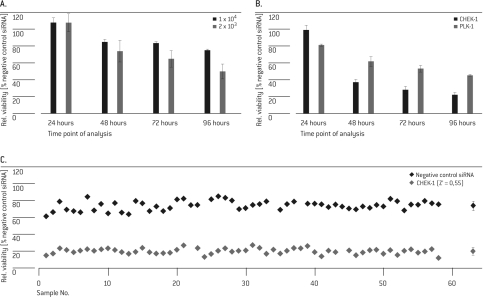
For the kinase and cell cycle screen in HUVEC Cells, siRNA reagents targeting polo-like kinase 1 (PLK-1) and Cell Cycle Check-point Kinase 1 (CHK-1 or CHEK-1) were selected as positive controls to set up the viability assay. PLK-1 is a key regulator of mitotic progression in mammalian cells and the knock-down of PLK-1 is known to induce apoptosis in cancer cells (Spänkuch-Schmitt et al, 2002; Reagan-Shaw and Ahmad, 2005). CHEK-1 is involved in the DNA damage response and is also required for cell proliferation and survival. CHEK-1 knockdown by siRNA has been reported to induce mitotic arrest (Tang et al, 2006). As such, down-regulation of PLK-1 and/or CHEK-1 is expected to decrease cell viability. Using PLK-1, post-transfection plating densities were adjusted to allow for significant discrimination of positive and negative control samples on the phenotypic level. This was achieved by plating HUVEC cells at a low cell density of 2 × 103 per well for 3-4 days after transfection (Figure 1A). As shown in Figure 1B, the phenotypic effect for PLK-1 silencing was weaker and built up slower than silencing of CHEK-1, thus representing potential differences expected for “strong” and “weak” library targets. It has been reported earlier that PLK-1 depletion by siRNA transfection exerts a strong effect on cancer cell lines, but not primary cells (Spänkuch-Schmitt et al, 2002; Reagan-Shaw and Ahmad, 2005). However, as the down-regulation of PLK-1 mRNA was not demonstrated in the previous reports, it could not be excluded that the underlying cause was more of an issue of inefficient transfection of the siRNA rather than the significance of PLK-1 roles in cell survival in primary cells. Nevertheless, despite a PLK-1 mRNA knockdown of more than 90% (data not shown), cell survival of HUVEC cells was diminished only to approximately 50% after 96 hrs, suggesting that persistent PLK-1 may indeed be a factor in survivability of cancer cells. An analysis time point of 72 hrs suited well for both targets. In pilot screens for further determination of assay robustness, controls were plated into the central 60 wells of a 96-well culture plate and analyzed for cell viability. Wells in the outer rows were filled with medium to avoid potential edge effects. Z' factors of both positive controls (CHEK-1: 0.55; PLK-1: 0.22; Figure 1C; data for PLK-1 not shown) reflected a suitable window for discrimination of potential hits with different phenotypic strength in the subsequent screen from background.
Figure 1.
Determination of optimal assay conditions. In three independent experiments, HUVEC cells were transfected with 20 pmol SMARTpool® siRNA targeting PLK-1 (A, B) or CHEK-1 (B, C) and siGENOME® non-targeting control. Cell viability was analyzed at different time points post Nucleofection® (A/B: 24, 48, 72 and 96 hrs; C: 72 hrs). Values were normalized to the negative control samples (A, B) or to untreated cells (C). The rightmost dots in C represents the mean and SD of the 60 individual values.
Primary screen and hit validation
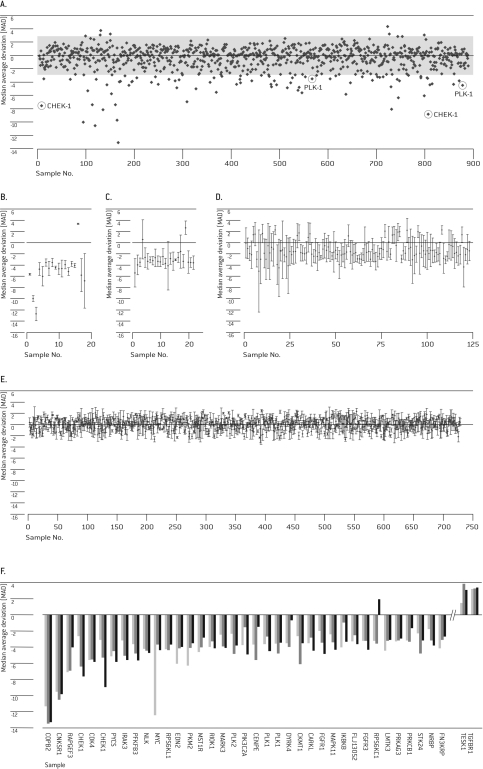
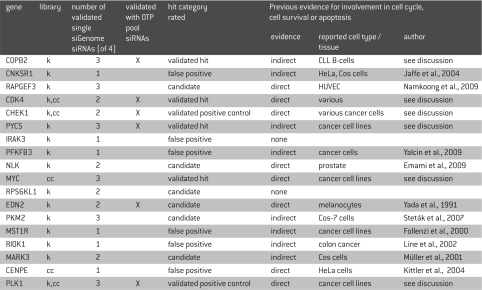
HUVEC Cells were transfected with siRNA pools targeting individual genes in the Human siGENOME® siRNA Libraries for protein kinases and cell cycle regulation. Multiple independent screening experiments (n=3) were performed to confirm the reproducibility of individual primary hits. Robust Z-score for cell viability was calculated for each of the 890 targets in the three independent experiments. As an example, the robust Z-scores of one screening experiment are shown in Figure 2A. A substantial proportion of targets displayed a median absolute deviation (MAD) below -3 or above 3 (MAD >3) including our positive controls PLK-1 and CHEK-1, which are members of both libraries. Thirty-five targets plus CHEK-1 and PLK-1 had a mean MAD greater than 3 in the three screening experiments and thus were considered as potential hits (Figure 2F). Eighteen of these targets showed a robust phenotype with MAD of greater than 3 in all three screening experiments (Figure 2B), while 24 targets were significant in two of the three experiments (Figure 2C) and 123 targets showed MAD greater than 3 in just one experiment (Figure 2D). The remaining 725 targets showed no response beyond the threshold in any experiment. The categories of targets with one or two experiments with MAD greater than 3 showed the highest standard deviations due to the occurrence of outliers. These categories also contained targets with a small standard deviation, suggesting that their importance for survival of HUVEC was not sufficiently strong for the chosen assay conditions (Figure 2B and C). Every sixth target fell into these categories of unclear importance, thus five of six targets were reproducibly classified as either important or not relevant for survival of HUVEC (Figure 2C and D). Generally, a considerable degree of variation was seen in the data, including the most significant hit category (Figure 2B). The most striking examples were MYC, which nevertheless could be validated, and CHEK1, which was a member of the library and also served as a positive control (Figure 2B and F). The degree of data variation might be attributed to not fully standardized cell culture conditions during the preparation of the experiments and thus argues for our strategy to select the primary hits from repeated screening experiments. As a consequence, we chose all 35 primary hits despite their standard deviation for further validation experiments. While a higher number of primary hits for validation lowered the probability for erroneously classified false negatives, it also lowered the validation rate. Of the 35 identified primary hits, 33 had a pro-proliferative/anti-apoptotic function, as their down-regulation led to increased cell death, while 2 had an anti-proliferate effect as their knockdown allowed for better cell viability. The 16 strongest of the 33 pro-proliferative hits plus the two positive controls were selected for further evaluation (Table 1).
Figure 2.
Primary Screen. HUVEC cells were transfected with 20 pmol of the combined Human siARRAY® SMARTpool® siRNA Libraries for Kinases (targeting 779 genes) and Cell Cycle Regulators (targeting 111 genes). Cell viability was analyzed 72 hrs post-Nucleofection®. (A) Representation of robust Z-scores of cell viability measures from 1 screening experiment. (B-E) Robust Z-scores of all primary hits with MAD of >3 in all three (B), two of three (C), or one of three (D) independent experiments and with MAD of <3 in all three experiments (E). (F) Robust Z-scores of the top 37 primary hits (with MAD of >3) from three independent experiments.
Table 1.
Protein kinases and cell cycle regulators in HUVEC cells. Top hits were selected from the primary screen. Hits are shown in descending order of MADs. “k” indicates members of the siRNA library against kinases, “cc” indicates members of the siRNA library against cell cycle related genes.
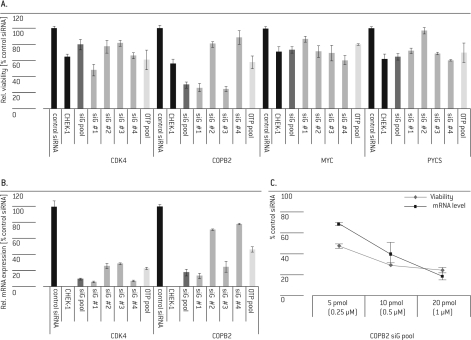
Five of the 18 selected targets (COPB2, CDK4, PYCS, MYC and PLK-1) were validated by demonstrating that the phenotype could be reproduced with multiple individual siGENOME® siRNAs from the original SMARTpool® and further with different siRNAs, i.e., an ON-TARGETplus® SMARTpoolV (Figure 3A, Table 1). The phenotypes could be correlated to the knockdown on mRNA level (Figure 3B) and to the amount of transfected siRNA (Figure 3C; only COPB2 shown). Both results, redundant phenotypic effects with an alternative siRNA pool and linking these phenotypes to proven specific mRNA knockdown, suggest that these were not off-target effects.
Figure 3.
Hit validation. HUVEC cells were transfected with 20 pmol (if not indicated differently) siGENOME® (siG) SMARTpool® or single siRNA #1 - 4 (from the de-convoluted pool) targeting CDK4, COPB2, MYC or PYCS. CHEK-1 and siGENOME® Non-Targeting siRNA #1 (control siRNA) and siRNA targeting CHEK-1 (CHEK-1) served as controls. 72 hrs post- Nucleofection® cell viability was analyzed and normalized to control the siRNA (A, C) and mRNA levels were determined for CDK4 (B) and COPB2 (B, C) and normalized to cyclophilin B mRNA and the control siRNA.
The reduction in cell survival with the ON-TARGETplus® SMARTpool® was mostly comparable to the corresponding siGENOME® SMARTpool®, but weaker than the strongest single siGENOME® siRNAs (Figure 3A and B). In the case of CDK4, the phenotypic effect of the ON-TARGETplus® SMARTpool® was comparable to siGENOME® siRNA duplex 4 despite a lower knock-down of the target mRNA (Figure 3A and B). For siRNAs of the same type, e.g., siGENOME® siRNAs, the phenotypic effects correlated well with the observed knock-down efficiencies (Figure 3A and B). Six of the 18 selected targets were confirmed with two of four single siGENOME® siRNAs, three of which could not be confirmed with the ON-TARGETplus® SMARTpool®, still suggesting them as potential hits that require further efforts for validation, such as expanding the set of tested single siRNAs. As discussed above for ON-TARGETplus® siRNAs, the insufficient phenotypic effect of the siRNAs, which could not be validated, may be explained by lower knock-down efficiencies or the reduction of off-target effects that contributed to the overall phenotype.
Five of the eighteen selected targets were considered “false positives”, because neither the ON-TARGETplus® SMARTpool®, nor more than one of four single siRNAs reproduced the phenotype seen with the original siGENOME® SMARTpool®. Thus, it is likely that these were the result of off-target effects of individual siRNA sequences. As for the above mentioned primary hits that showed a higher rate of validated single siRNAs, testing further single siRNAs with proven mRNA knockdown may allow a more definitive hit stratification.
Previous reports demonstrate the involvement of most of the identified primary hits in cell cycle regulation, cell survival or apoptosis (Table 1). No such evidence could be found for the kinases IRAK3, considered as false positive, and RPS6KL1, which has been validated with two of four single siGENOME® siRNAs. Hence, five of the classified “false positives” have been reported in earlier literature, but their importance for HUVEC cell survival and proliferation remain unclear. For a majority of the identified primary hits, earlier reports describe their importance for cancer cells, while little information is available with respect to primary cells.
Cyclins, cyclin-dependent kinases and their substrates play pivotal roles during cell cycle progression and control proliferation of normal cells. Cyclin-dependent kinase 4 (CDK4) phosphorylates retinoblastoma protein (Rb) and other Rb-related proteins (Ewen et al, 1993; Kato et al, 1993; Leng et al, 2002) ultimately promoting the expression of various genes essential for cell cycle phase G1-S transition (Nevins, 2001). Low molecular-weight inhibitors of CDK4 lead to a delayed G2/M progression with reduced cell growth and mitosis rates in a number of cell lines (Burgess et al, 2006). However, CDK4, as well as the functionally redundant CDK6 and the associated D-type cyclins (D1, D2 and D3) are not essential for cell proliferation in mammalian cell types (Kozar and Sicinski, 2005; Malumbres, 2004). This explains the moderate, albeit consistent, reduction in cell viability after CDK4 knockdown in our experiments. Nevertheless, inhibitors specific for CDK4 and CDK6 can show significant antiproliferative activity against Rb-positive tumor cells (Fry, 2004).
The proto-oncogene c-MYC has been intensively studied and its deregulation by various mitogens leads to the genesis of diverse human cancers (Oster, 2002). The gene product of c-MYC is a transcription factor, involved in the regulation of cell cycle related genes, such as CDK4 or Cyclin B1 (Menssen, 2002). While deregulation of c-MYC results in hyperproliferation, antisense oligo-mediated knockdown of c-MYC leads to growth inhibition, i.e., in human smooth muscle cells (Shi et al, 1993). The relevance of CDK4 and c-MYC for cell cycle progression has been evident from previous studies (Hermeking et al, 2000), but neither COPB2 nor PYCS have been directly related to cell cycle regulation or described as survival factors.
COPB2 is a subunit of the so called “coatomer” complex, which is essential for budding of cargo vesicles at the endoplasmatic reticulum and recognition of transport signals present on their surface for travelling to the Golgi apparatus (Reviewed in: Béraud-Dufour and Balch, 2002; Bonifacino and Glick, 2004). A putative importance of COPB2 for cell survival and as a drug target is underlined by the fact that novel cancer drugs have been designed based on Brefeldin A, an inhibitor of ADP ribosylation factor (ARF), which is indispensable for the assembly of the coatomer complex. (Carew et al, 2006; Donaldson et al, 1991, 1992a, 1992b; Helms and Rothman, 1992; Orci et al, 1993). The severe effect of COPB2 down-regulation on proliferation of HUVEC cells underlines its importance for cell survival and as a promising drug target for anti-tumor reagents.
PYCS is a mitochondrial enzyme pivotal for the synthesis of proline (Csukai et al, 1997). Reduction of proline content by over-expression of proline dehydrogenase leads to formation of reactive oxygen species and reduced cell viability in a number of cancer cell lines (Liu et al, 2006). Down-regulation of PYCS by siGENOME® SMARTpool® or single siRNAs consistently reduced HUVEC survival by some 40%, similar to CHEK1, pointing towards PYCS as being important but not essential for cell survival (Figure 3A).
CONCLUSIONS
The presented data demonstrate that the Nucleofector® 96-well Shuttle® System allows the delivery of siRNA libraries, e.g., Dharmacon siGENOME® Libraries, in cell types previously considered difficult to transfect. Preserved cell functionality and efficient mRNA knockdown allow the identification and validation of gene targets in primary cells, which reflect a higher biological relevance for certain pathways.
Most of the identified and validated targets have been reported earlier to be implicated in the cell cycle, cell survival or apoptosis in cancer cells, while little has been known about their importance in normal primary cells.
Our study suggests that overcoming the limitation of using transformed cell lines for functional screens and rather studying the relevant primary cells pledge the potential to identify novel drug target candidates.
Acknowledgments
This work was funded by Lonza Cologne AG and Thermo Fisher Scientific, Dharmacon Products.
COMPETING INTERESTS
MZ, LMA NUES, SS, SMO, SBSD, AT and HAMH work for Lonza Cologne AG R&D. ASA and DL work for and/or have financial interests in Thermo Fisher Scientific.
REFERENCES
- Bartz SR, Zhang Z, Burchard J, et al. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol. 2006;26:9377–9386. doi: 10.1128/MCB.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béraud-Dufour S, Balch W. A journey through the exocytic pathway. J Cell Sci. 2002;115:1779–1780. doi: 10.1242/jcs.115.9.1779. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Burgess A, Wigan M, Giles N, et al. Inhibition of S/G2 phase CDK4 reduces mitotic fidelity. J Biol Chem. 2006;281:9987–9995. doi: 10.1074/jbc.M512714200. [DOI] [PubMed] [Google Scholar]
- Calvin S, Emch J, Wang, et al. FuGENE®HD Transfection Reagent: Choice of a Transfection Reagent with Minimal Off-Target Effect as Analyzed by Microarray Transcriptional Profiling. Biochemica. 2006;4:22–25. [Google Scholar]
- Carew JS, Nawrocki ST, Krupnik YV, et al. Targeting endoplasmic reticulum protein transport: a novel strategy to kill malignant B cells and overcome fludarabine resistance in CLL. Blood. 2006;107:222–231. doi: 10.1182/blood-2005-05-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung N, Locco L, Huff KW, et al. An efficient and fully automated high-throughput transfection method for genome-scale siRNA screens. J Biomol Screen. 2008;13:149–158. doi: 10.1177/1087057107312032. [DOI] [PubMed] [Google Scholar]
- Csukai M, Chen CH, De Matteis MA, et al. The coatomer protein β'-COP, a selective binding protein (RACK) for protein kinase Cε. J Biol Chem. 1997;272:29200–29206. doi: 10.1074/jbc.272.46.29200. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Kahn RA, Lippincott-Schwartz J, et al. Binding of ARF and β-COP to Golgi membranes: possible regulation by a trimeric G protein. Science. 1991;254:1197–1199. doi: 10.1126/science.1957170. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Cassel D, Kahn RA, et al. ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein β-COP to Golgi membranes. Proc Nat Acad Sci USA. 1992a;89:6408–6412. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Finazzi D, Klausner RD. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992b;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- Emami KH, Brown LG, Pitts TE, et al. Nemo-like kinase induces apoptosis and inhibits androgen receptor signaling in prostate cancer cells. Prostate. 2009;69:1481–1492. doi: 10.1002/pros.20998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen ME, Sluss HK, Sherr CJ, et al. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- Fedorov Y, King A, Anderson E, et al. Different delivery methods-different expression profiles. Nat Methods. 2005;2:241. doi: 10.1038/nmeth0405-241. [DOI] [PubMed] [Google Scholar]
- Follenzi A, Bakovic S, Gual P, et al. Cross-talk between the proto-oncogenes Met and Ron. Oncogene. 2000;19:3041–3049. doi: 10.1038/sj.onc.1203620. [DOI] [PubMed] [Google Scholar]
- Fry DW, Harvey PJ, Keller PR, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, et al. Identification of CDK4 as a target of c-MYC. Proc Nat Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Aspenström P, Hall A. Human CNK1 Acts as a Scaffold Protein, Linking Rho and Ras Signal Transduction Pathways. Mol Cell Biol. 2004;24:1736–1746. doi: 10.1128/MCB.24.4.1736-1746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun P, Aral B, Saudubray JM. A new inherited metabolic disease: delta1-pyrroline 5-carboxylate synthetase deficiency. Bull Acad Natl Med. 1998;182:131–137. [PubMed] [Google Scholar]
- Kato J, Matsushime H, Hiebert SW, et al. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- Kittler R, Putz G, Pelletier L, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Kozar K, Sicinski P. Cell cycle progression without cyclin D-CDK4 and cyclin D-CDK6 complexes. Cell Cycle. 2005;4:388–391. doi: 10.4161/cc.4.3.1551. [DOI] [PubMed] [Google Scholar]
- Leng X, Noble M, Adams PD, et al. Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4. Mol Cell Biol. 2002;22:2242–2254. doi: 10.1128/MCB.22.7.2242-2254.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Line A, Slucka Z, Stengrevics A, et al. Characterisation of tumour-associated antigens in colon cancer. Cancer Immunol Immunother. 2002;51:574–582. doi: 10.1007/s00262-002-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Borchert GL, Surazynski A, et al. Proline oxidase activates both intrinsic and extrinsic pathways for apoptosis: the role of ROS/superoxides, NFAT and MEK/ERK signaling. Oncogene. 2006;25:5640–5647. doi: 10.1038/sj.onc.1209564. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Sotillo R, Santamaría D, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Marques JT, Williams RG. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- Martin SE, Caplen NJ. Applications of RNA interference in mammalian systems. Annu Rev Genomics Hum Genet. 2007;8:81–108. doi: 10.1146/annurev.genom.8.080706.092424. [DOI] [PubMed] [Google Scholar]
- Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Nat Acad Sci USA. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkerova M, Klamova H, Brdicka R, et al. Targeting of gene expression by siRNA in CML primary cells. Mol Biol Rep. 2007;34:27–33. doi: 10.1007/s11033-006-9006-x. [DOI] [PubMed] [Google Scholar]
- MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- Müller J, Ory S, Copeland T, et al. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol Cell. 2001;8:983–993. doi: 10.1016/s1097-2765(01)00383-5. [DOI] [PubMed] [Google Scholar]
- Namkoong S, Kim CK, Cho YL, et al. Forskolin increases angiogenesis through the coordinated cross-talk of PKA-dependent VEGF expression and Epac-mediated PI3K/Akt/eNOS signaling. Cell Signal. 2009;21:906–915. doi: 10.1016/j.cellsig.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Nevins JR. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- Orci L, Palmer DJ, Ravazzola M, et al. Budding from Golgi membranes requires the coatomer complex of non-clathrin coat proteins. Nature. 1993;362:648–652. doi: 10.1038/362648a0. [DOI] [PubMed] [Google Scholar]
- Oster SK, Ho CS, Soucie EL, et al. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- Reagan-Shaw S, Ahmad N. Silencing of polo-like kinase (Plk) 1 via siRNA causes induction of apoptosis and impairment of mitosis machinery in human prostate cancer cells: implications for the treatment of prostate cancer. FASEB J. 2005;19:611–613. doi: 10.1096/fj.04-2910fje. [DOI] [PubMed] [Google Scholar]
- Shi Y, Hutchinson HG, Hall DJ, et al. Downregulation of c-myc expression by antisense oligonucleotides inhibits proliferation of human smooth muscle cells. Circulation. 1993;88:1190–1195. doi: 10.1161/01.cir.88.3.1190. [DOI] [PubMed] [Google Scholar]
- Spänkuch-Schmitt B, Bereiter-Hahn J, Kaufmann M, et al. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst. 2002;94:1863–1877. doi: 10.1093/jnci/94.24.1863. [DOI] [PubMed] [Google Scholar]
- Tang J, Erikson RL, Liu X. Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1) Proc Nat Acad Sci USA. 2006;103:11964–11969. doi: 10.1073/pnas.0604987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst AW, Bodemann BO, Cardenas J, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Yada Y, Higuchi K, Imokawa G. Effects of Endothelins on Signal Transduction and Proliferation in Human Melanocytes. J Biol Chem. 1991;266:18352–18357. [PubMed] [Google Scholar]
- Yalcin A, Clem BF, Simmons A, et al. Nuclear targeting of 6-Phosphofructo-2-kinase (PFKFB3) increases proliferation via cyclin-dependent kinases. J Biol Chem. 2009;284:24223–24232. doi: 10.1074/jbc.M109.016816. [DOI] [PMC free article] [PubMed] [Google Scholar]