Abstract
RNA interference plays a significant role in manipulating cellular and viral mechanisms to maintain latency during HIV-1 infection. HIV-1 produces several microRNAs including one from the TAR element which alter the host's response to infection. Since cyclin/cdk complexes are important for viral transcription, these studies focus on the possible cdk inhibitors that inhibit viral transcription, without affecting normal cellular mechanisms. Roscovitine and Flavopiridol are well-studied cdk inhibitors that are effective at suppressing their target cdks at a low IC50. These cdk inhibitors and possibly future generations of drugs are affected by microRNA mechanisms. From our studies, we developed a third generation derivative called CR8#13. In cells that lack Dicer there was a higher level of basal viral LTR-reporter transcription. When drugs, specifically Flavopiridol and CR8#13 were added, the transcriptional inhibition of the LTR was less potent in cells that lacked Dicer. Also, after transfection with HIV-1 clone (pNL4.3), CR8 and CR8#13 derivatives were shown to be more effective viral transcription inhibitors in cell lines that contained Dicer (T-cells) as compared to Dicer deficient lines (monocytes). We next asked whether the addition of CR8 or CR8#13 could possibly increase levels of TAR microRNA in HIV-1 LTR containing cells. We demonstrate that the 3'TAR microRNA is produced in higher amounts after drug treatment, resulting in microRNA recruitment to the LTR. MicroRNA recruitment results in chromatin alteration, changes in Pol II phosphorylation and viral transcription inhibition. In conclusion, our results indicate that viral microRNA, specifically the TAR microRNA produced from the HIV-1 LTR is responsible for maintaining latent infections by manipulating host cell mechanisms to limit transcription from the viral LTR promoter. With the microRNA machinery present, cdk inhibitors are able to significantly increase the amount of TAR microRNA, leading to downregulation of viral LTR transcription.
Keywords: microRNA, HIV-1, TAR, cdk inhibitor, ATP analogs, Tat transactivation
INTRODUCTION
Human immunodeficiency virus-1 (HIV-1) is the etiological agent of acquired immunodeficiency syndrome (AIDS). Highly active antiretroviral therapy (HAART) is currently the most effective HIV-1 treatment and has been shown to significantly reduce AIDS-related mortality. Since latently infected cells still produce viral RNA and even small amounts of infectious virus, there is a chance of mutational escape from cells treated with drugs.
Current HIV-1 therapies, including HAART, are mostly ineffective at eliminating the virus and also are the main cause of drug-resistant variants.
RNA interference (RNAi) is a regulatory mechanism conserved in higher eukaryotes, such as primates and mice. RNAi involves small RNA molecules that guide a protein effecter complex to a complementary or mostly complementary sequence of nucleic acid. The end result is the down regulation of protein expression through either transcriptional silencing, cleavage of target mRNA or inhibition of translation (Agrawal et al, 2003; Bartel, 2004). Exogenously introduced dsRNA is recognized by Dicer, and cleaved into characteristic 21 nucleotide segments with 2 nucleotide 3'overhangs (siRNAs and microRNAs) that direct the RNAi machinery for sequence-specific inhibition of mRNA expression. microRNAs are also produced from genomic DNA that is transcribed by RNA polymerase II. Endogenously expressed RNA can be involved in RNAi through a slightly different pathway involving Drosha-mediated cleavage of RNA stem-loops in the nucleus, followed by export to the cytoplasm by Exportin-5, and finally cleavage by Dicer to generate a small RNA duplex approximately 22 nucleotides in length with a two nucleotide 3' overhang on each strand (Hannon, 2002). One strand of the microRNA duplex is incorporated into Argonaute-containing effector complexes, which silence gene expression through two distinct mechanisms. In the first, the small RNA associates with the RNA-induced silencing complex (RISC) and guides the complex to a complementary sequence of mRNA where a member of the Argonaute family of proteins cleave the target mRNA, leading to silencing of a gene. Alternatively, the microRNA may guide the RISC complex to a somewhat complementary region in the 3'UTR of the mRNA. In addition to attaching the RISC complex, the RNA can associate with the RNA-induced initiation of transcriptional silencing (RITS) complex. Similar to the RISC mechanism, the microRNA guides this complex to a complementary region of chromosomal DNA and recruits factors that modify the chromatin structure and induce transcriptional silencing (Volpe et al, 2002; Matzke and Birchler, 2005).
Several viruses encoding microRNAs have already been identified, including human cytomegalovirus, human herpesevirus 8, Epstein Barr virus, and herpes simplex virus (Grey et al, 2005; Pfeffer et al, 2005; Umbach et al, 2008). The functions of a number of viral microRNA have been dissected and they appear capable of regulating both viral and cellular genes (Dykxhoorn, 2007). In terms of HIV-1, several previous studies have reported the production of microRNAs from the TAR, miR-H1, nef and env RNAs (Omoto et al, 2004; Provost et al, 2006; Klase et al, 2007; Kaul et al, 2009). All or few of the HIV-1 generated microRNA could potentially inhibit viral replication, block translation of viral proteins, or cause remodeling of the viral genome. Thus, RNAi-based strategies have considerable therapeutic potential against HIV-1 infection.
The majority of current therapies target viral proteins. There is a need for development of host gene-based therapies as these are most probably resistant to mutations. One attractive host candidate for antiviral therapeutics is the cell cycle machinery. The host cell cycle is dependent on the activity of cyclin-dependent kinases (cdks) and their catalytic cyclin subunits. The cdk/cyclin complexes aid in the advancement of eukaryotic cell through the G1/S and G2/M cell cycle checkpoints. For the G1/S checkpoint, the cdk2/cyclin E complex phosphorylates the retinoblastoma (Rb) protein (Athanassiou et al, 2004). HIV-1 has the ability to manipulate the cdk/cyclin mechanisms within a cell to support its own life cycle. For example, HIV-1 targets the cdk2/cyclin E complex to allow cells to pass through the G1/S checkpoint, enabling transcription of integral proliferative genes to increase HIV-1 genome replication (Nekhai et al, 2002). cdk/cyclin complexes are also linked to the viral proteins through interaction with the vital HIV-1 Tat (transactivator of transcription) protein. Tat is the main transcriptional activator of the HIV-1 LTR and also induces some cellular genes to help maintain virus production and/or cell survival (Bohan et al, 1992; Zhou et al, 2000). Tat binds the viral TAR element, and the Tat-TAR complex recruits viral and cellular components to initiate and elongate the viral promoter. For example, Tat recruits the pTEFb elongation complex to the promoter. The activated components of this complex, cdk9 and cyclin T1, then hyper-phosphorylate the large subunit of the RNA polymerase II C-terminal domain and other factors to activate transcription elongation (Kim et al, 2002). Therefore, cdk/cyclin inhibitors are potential HIV-1 therapeutics.
The two highly studied cdk inhibitors in relation to HIV are Roscovitine and Flavopiridol, which inhibit cdk1, 2, 5, 7, 9 and cdk1, 2, 4, and 9, respectively (Haesslein and Jullian, 2002; Vandromme et al, 2006; Oumata et al, 2008). Roscovitine is most effective against cdk2 and cdk9 at an average IC50 of <300nM and Flavopiridol inhibits cdk9 at an IC50 of 3nM. A lower IC50 enables these drugs to be more effective at suppressing the viral gene expression, rather than normal cellular promoters that may use either cdk2 or cdk9 for their transcription. More potent and specific analogs have been developed based on these two initial compounds. Cyc202 (R-roscovitine) targets the cdk2/cyclin E complex by binding to ATP pockets and allows apoptosis to occur in HIV-1 infected T-cells, monocytes, and peripheral blood mononuclear cells (Agbottah et al, 2005). Recently, we have investigated whether derivatives of Cyc202 could potentially inhibit viral transcription at a lower IC50. Treatment with Cyc202 was able to inhibit uploading of the cdk2/cyclin E and cdk9/cyclin T1 complexes onto HIV-1 DNA. A slight alteration at the purine ring of Cyc202 resulted in a second generation drug, CR8. Here, CR8 and its third generation derivatives have been tested for the potency and specificity of inhibiting viral transcription. Results related to these second and third generation drugs along with the potential need for functional microRNA machinery will be discussed here.
MATERIALS AND METHODS
Cell culture
TZM-bl cell lines were grown in Dulbecco's modified Eagle's medium supplemented with fetal bovine serum (FBS) (10%, v/v), 2mM L-glutamine, and antibiotics (penicillin 100U/ml, streptomycin 100mg/ml) (cDMEM).
HCT116 WT and HCT116 Dicer -/- cell lines were grown in McCoy's medium supplemented with FBS (10%, v/v), 2mM L-glutamine, and antibiotics (penicillin 100U/ml, streptomycin 100mg/ml). CEM, ACH2, Jurkat and U937 cells were grown in RPMI 1640 supplemented with FBS, L-glutamine, and antibiotics (penicillin 100U/ml, streptomycin 100mg/ml). All cell lines were maintained at 37°C in 5% (v/v) CO2. ACH2 cells are infected with HIV-1; TZM-bl cells contain a stably-integrated HIV-1 LTR-Luciferase reporter; CEM, Jurkat, and U937 cells are uninfected. Transfections were carried out using Attractene (Qiagen) lipid reagent. Cells were cultured to confluence and pelleted at 4°C for 15min at 3,000rpm. The cell pellets were washed twice with 25ml of phosphate-buffered saline (PBS) with Ca2+ and Mg2+ (Quality Biological) and centrifuged once more. Cell pellets were resuspended in lysis buffer (50mM Tris-HCl, pH 7.5, 120mM NaCl, 5mM EDTA, 0.5%, v/v, NP-40, 50mM NaF, 0.2mM Na3VO4, 1mM DTT, one complete protease cocktail tablet/50ml (Roche) and incubated on ice for 20min, with gentle vortexing every 5min. Cell lysates were transferred to eppendorf tubes and were centrifuged at 10,000rpm for 10min. Supernatants were transferred to a fresh tube where protein concentrations were determined using Bio-Rad protein assay (Bio-Rad, Hercules, CA).
Drug screening and cell counting
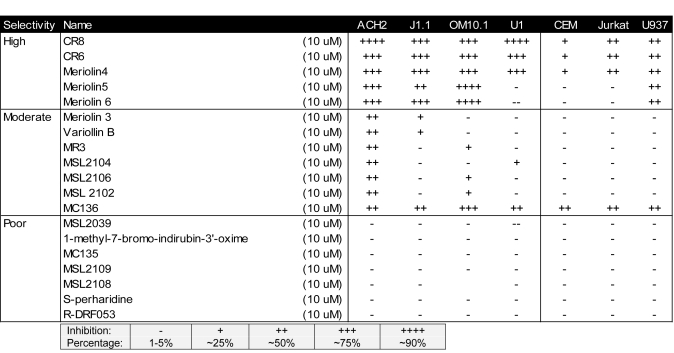
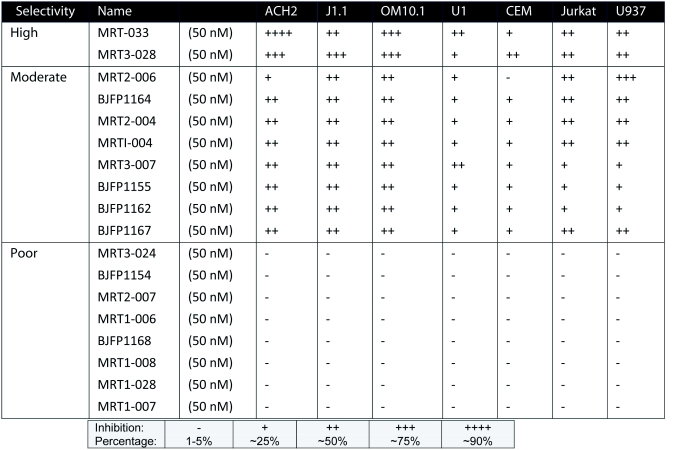
HIV-1 infected and uninfected cells were treated with nineteen inhibitors (Table 1) at 10μM concentration. Among the inhibitors were meriolins (Bettayeb et al, 2007) and variolins (Simone et al, 2005), which have exhibited distinct cdk inhibitory activities. Another set of drugs used were derived from 2,6,9-trisubstituted cdk inhibitory purines using a classical medicinal chemistry approach (Oumata et al, 2008). Some of these compounds are 6-aminomethylenebiaryl analogs of CYC202. Among these inhibitors, CR8, an analog bearing a 2-pyridyl on position 4 of its phenyl ring, was created (Galons et al, 2010). Table 2 includes HIV-1 infected and uninfected cells, which were treated with eighteen Roscovitine/CR8 derivatives at 50nM concentration. Forty-eight hours after treatment, cytotoxicity was primarily determined by trypan blue exclusion. Cells were counted to determine cell death after 48hrs.
Table 1.
Screening of various CDK inhibitors in HIV-1 cell killing assay.
Table 2.
Screening of Roscovitine/CR8 derivatives in HIV-1 cell killing assay.
RT-PCR and primers
For mRNA analysis of cdk9 related genes following drug treatments, total RNA was isolated from cells using Trizol (Invitrogen) according to the manufacturer's protocol. A total of 1μg of RNA from the RNA fraction was treated with 0.25mg/ml DNase I for 60min, followed by heat inactivation at 65°C for 15min. A total of 1μg of total RNA was used to generate cDNA with the iScript cDNA Synthesis kit (Bio-Rad) using oligo-dT reverse primers.
Electroporation and reverse transcriptase assay
For electroporations, Jurkat and U937 cells were resuspended at 3 million cells in 250μl of RPMI. Five microgram of pNL4-3 was then added to the cell suspension. Cells were pulsed a single time at 210V, 800μF, and low resistance. Electroporated cells were immediately transferred to RPMI-1640 with L-glutamine and Penicillin/Streptomycin with 10% (v/v) FBS and plated in 6-well plates. Twenty four hours-post electroporation, cells were treated with drugs for additional 48hrs and harvested for RT analysis. RT assays were performed as described in ((Guendel et al. 2009; Easley et al. 2010).
Poly-A RT-PCR
For poly-A RT-PCR detection of microRNAs, 500ng of RNA from the microRNA-enriched fraction was used to generate cDNA using the Quantimir kit (SBI) according to the manufacturer's protocol. Briefly, small RNA species are poly-adenylated and then reverse transcription reactions are performed with a company-provided RT primer. For PCR, a universal reverse primer is provided by the manufacturer. Specific microRNA forward primers are identical in sequence to the microRNA of interest. PCR products corresponding to the amplified microRNAs were separated in a 3.5% (w/v) agarose gel and quantified using the Kodak 1D software.
Chloramphenicol acetyltransferase (CAT) assay
Various cells lines were transfected with the HIV-1 LTR-CAT plasmid and pc-Tat to initiate the LTR transcription, and subsequently subjected to various drug treatments. After 48hrs, cells were lysed and CAT assays were perfomred as previously described (Guendel et al, 2009; Van Duyne et al, 2008; Easley et al, 2010).
Luciferase assay
TZM-bl cells were transfected with pc-Tat (0.5μg) using Attractene reagent (Qiagen) according to the manufacturers' instructions. Twenty-four hours later, cells were treated with DMSO or the indicated compound. Forty-eight hours-post drug treatment, luciferase activity of the firefly luciferase was measured with the BrightGlo Luciferase Assay (Promega).
RNase protection assay (RPA)
Total RNA was extracted from TNF-treated CEM and ACH2 cells follow by drug treatment with Cyc202 (500nM), CR8 (100μM), CR8#13 (50nM), and Flavopiridol (50nM) for 48hrs using Trizol reagent (Invitrogen). RNase protection assays were performed as previously described (Klase et al, 2007).
Chromatin immunoprecipitation assay (ChIP)
TZM-bl cells were treated with TSA for 7 days and processed for ChIP. For ChIP, approximately 5×106 cells were used per IP. ChIP assays were performed as previously described. PCR was performed using 0.1μM of HIV-1 LTR primers (Klase et al, 2007; Guendel et al, 2009; Easley et al, 2010).
RESULTS
Effect of inhibitors on HIV-1 infected and uninfected cells
We reasoned that the specific targeting of cell cycle and cdk regulators with small molecule therapeutics might provide better insights into how to inhibit HIV-1 infected cells. To that end, HIV-1 infected and uninfected cells were studied by culturing ACH2, J1.1, OM10.1, U1 and uninfected CEM, Jurkat T-cells, and U937 in media with inhibitor concentration of 10μM to compare different classes of inhibitors. Cells were treated for 48hr and cell viability was determined using trypan blue exclusion method. Results of such a screen from the 19 inhibitors are shown in Table 1 where percent of dead cells are indicated after various drug treatments. A number of drugs caused death in HIV-1 infected cells much more efficiently than uninfected cells. The inhibitors were classified into three categories: high, moderate or poor, according to their effect on cellular viability in both HIV-1 infected and uninfected cells. The top candidate that killed HIV-1 infected cells, while having little cell death in uninfected cells was CR8, a second generation drug derived from Cyc202.
In an attempt to test a third generation of drugs, we made 18 new derivatives of the CR8 with slight modifications for testing against HIV-1. Similar to the previously performed assay for cdk inhibitors, infected ACH2, J1.1, OM10.1, U1 and uninfected CEM, Jurkat T-cells, and U937 were cultured in media with low inhibitor concentration (50nM) to evaluate relative efficacies of the CR8 derivatives. Cells were treated for 48hrs and cell viability was determined using trypan blue exclusion method. Results of such a screen from the 18 CR8 derivatives are shown in Table 2 where percent of dead cells are indicated after various drug treatments. Interestingly, the two derivatives that caused maximum death in infected cells were minimally toxic to uninfected control cells.
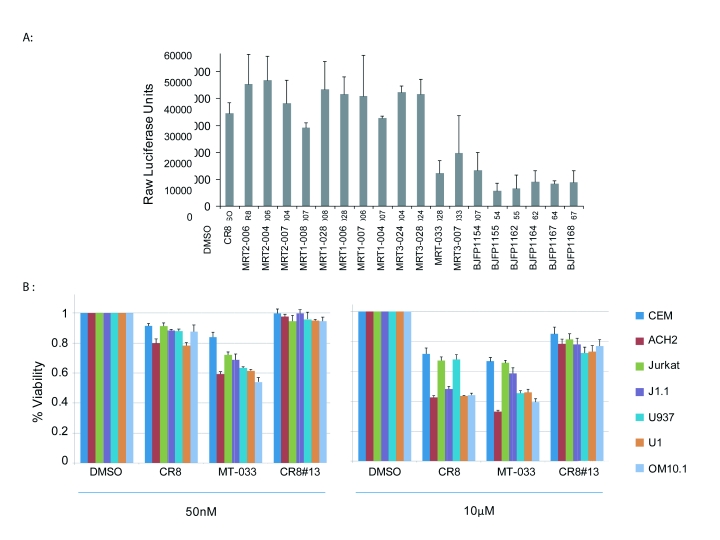
This prompted us to determine if derivatives exhibiting minimal cell killing of infected cells were able to inhibit viral transcription. TZM-bl cells that have an integrated HIV-1 LTR-luciferase reporter were transfected with pc-Tat (encodes HIV-1 Tat protein) and then treated with 18 CR8 derivatives. The addition of Tat enables activated transcription of HIV-1 promoter driving luciferase in these cells. Treated cells were assayed 48hrs after Tat transfection and drug treatment. Among the tested inhibitors indicated in Table 2, luciferase assays revealed 9 analogs as efficiently decreasing viral transcription of the fully chromatinized HIV-1 promoter (Figure 1A). The compound BJFP1154 (CR8#13), exhibited the greatest transcriptional inhibition of the HIV-1 LTR, although it did not affect cell viability as shown in Table 2.
Figure 1.
Screening of CR8 derivatives on Tat-dependent transcription HIV-1 LTR. A. TZM-bl cells were transfected with 1ug of Tat and treated the next day with DMSO, or the indicated CR8 derivative compounds at 50nM. 48 hrs-post drug treatment, luciferase activity of the firefly luciferase was measured with the BrightGlo Luciferase Assay and luminescence was read from a 96 well plate on an EG&G Berthold luminometer. Assays were performed in triplicate, average and standard deviations are shown. B. MTT assay of CR8, MRT-033, and BJFP1154 (CR8#13) were tested on all infected ACH2, J1.1, OM10.1, U1 and uninfected CEM, Jurkat T-cells, and U937 cells at 50nM and 10μM drug concentrations.
To further validate the cell death results for a few of the compounds, more quantitative MTT assay were performed (Figures 1B). CR8, MRT-033, and BJFP1154 (CR8#13) were tested on all infected ACH2, J1.1, OM10.1, U1 and uninfected CEM, Jurkat T-cells, and U937 cells. They were cultured in media with both low (50nM) and high (10μM) inhibitor. CR8 exhibited significant cell killing of infected cells over uninfected cells at both high and low concentrations. MRT-033 exhibited killing of infected cells, ACH2, U1 and OM10.1 cells. However, both compounds, CR8 and MRT-033 still exhibited some cell killing of uninfected cells. CR8#13 showed minimal cell killing even at the high concentration.
Data in Figure 1 show that there are a number of inhibitors that effectively decrease HIV-1 transcription and have no effect on cell viability or toxicity. For instance drugs, such as MRT3-028 exhibited significant cell killing in infected cells and were also proficient in HIV-1 transcription inhibition (Figure 1A, Lane 12). Similarly, BJFP1155 and BJFP1164 are capable of moderate cell killing of infected cells, while also having the ability to block HIV-1 transcription (Figure 1A, Lanes 16 and 18). BJFP1154 (CR8#13) was unique in that it displayed very little cytotoxicity, while causing the most significant decrease of HIV-1 transcription. This makes CR8#13 a novel new drug candidate to control HIV-1 transcription and infection.
Efficacy dependence of CR8#13 on microRNA machinery for transcription inhibition
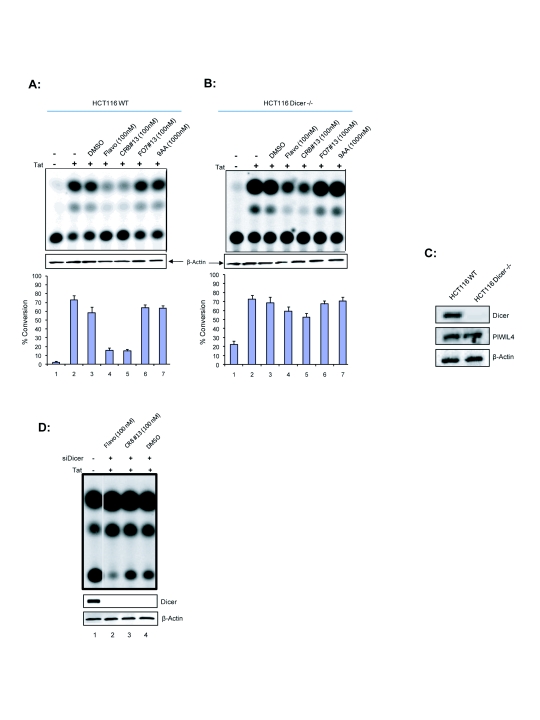
We previously observed that the parent, Cyc202, was an effective inhibitor against HIV-1 in T-cells but not in monocytes (Agbottah et al, 2005). Specifically, we observed that the integral microRNA machinery, Dicer and Drosha, were significantly decreased in monocytes leading us to hypothesize on the dependence of Cyc202 on the microRNA machinery for efficacy (Klase et al, 2007; Coley et al, Submitted). Since CR8#13 is derived from Cyc202, we looked at how the microRNA machinery, via the RITS or RISC complex would be involved in specifically inhibiting viral transcription. We proceeded to study the efficacy of the derived drugs in the presence or absence of Dicer, an integral part of cellular microRNA machinery. We implemented the use of the HCT116 colon carcinoma cell line that either contained a WT Dicer (HCT116 WT) or lacked the Dicer protein (HCT116 Dicer-/-). These cells were first transfected with the HIV-1 LTR-CAT reporter construct. HIV-1 LTR transcription was activated with pc-Tat and treated at 6hrs with DMSO (negative control), Flavopiridol (positive control), CR8#13, F07#13, or 9AA (other inhibitors that are not ATP analogs). Cells were harvested 48hrs-post treatment and processed for CAT assays (Figure 2A and B). There was greater activation of the HIV-1 LTR in cells lacking detectable Dicer, resulting in significantly more viral transcription than in cells containing Dicer. This is consistent with the previous data indicating that Dicer plays an inhibitory role in HIV-1 transcription (Triboulet et al. 2007).
Figure 2.
Effect of Drugs on Tat-mediated transactivation in HCT116 WT and HCT116 Dicer-/- cells. pHIV-1 LTR–CAT (1μg) construct was transfected in 2×106 cells in the absence or presence of Tat (1μg). Six hours later, the transfected cells were treated with DMSO, Flavopirodol (100nM), CR8#13 (100nM), F07#13 (100nM), and 9AA (1000nM). Treated cells were incubated in complete DMEM for 48hrs at 37°C. Cells were harvested and cell extracts were used for CAT analysis. One tenth the amount of HCT116 Dicer-/- extract compared to HCT116 WT was used for CAT analysis. Values represent the percentage of conversion of the [14C] chloramphenicol substrate in the CAT assay. Panel A shows results from HCT116 WT cells and panel B are from HCT116 Dicer-/- cells. C. Confirmation western blot for Dicer and PIWIL4 in the HCT116 WT and Dicer -/- cells. One hundred microgram of total extracts were ran on a 4-20% (w/v) SDS/PAGE and western blotted for presence of Dicer, PIWIL4 and actin. D. CAT assay of Dicer WT HCT116 cells that had been treated with siRNA against Dicer, transfection with Tat followed by drug treatment.
Flavopiridol and CR8#13 inhibited viral transcription significantly better in cells that contained Dicer (Figure 2A, Lanes 4 and 5). F07#13 and 9AA, control drugs previously shown to inhibit viral transcription, had moderate effects (Figure 2A, Lanes 6-7). It has previously been shown that 9AA efficiently inhibited HIV-1 transcription (at higher concentrations) through the restoration of p53 and p21WAF1 functions (Guendel et al, 2009). F07#13 and 9AA likely do not utilize the TAR microRNA pathway for their inhibitory activity in these cells. Figure 2C substantiates that the presence of Dicer is significantly decreased in the HCT116 Dicer-/- cells. Importantly these cells contain miRNA which may be the product of PIWI expression. In order to provide further support that presence of Dicer has an effect on drug efficacy; a similar CAT assay was performed as in Figure 2A. siRNA against Dicer was transfected along with pther plasmids into HCT116 WT cells and CAT enzyme was detect in 2 days. When Dicer levels were decreased, the viral transcription inhibition caused by the drugs was also decreased (Figure 2D). The dependence of CR8#13 and Flavopiridol on Dicer for increased transcription inhibition indicated that microRNA machinery may be important in increasing efficacy and specificity toward the HIV-1 promoter.
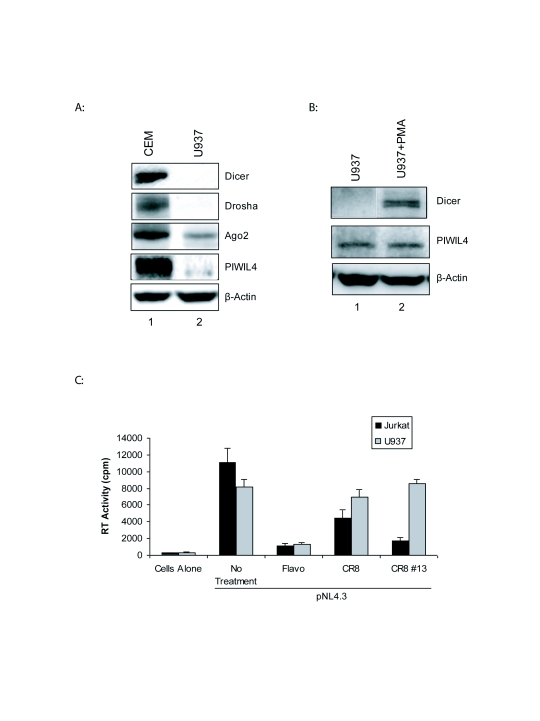
We next examined whether these drugs were effective in cell lines infected with an HIV-1 construct. To this effect Jurkat T-cells and promonocytic U937 cells were transfected with pNL4.3 and then treated with Flavopiridol, CR8 and CR8#13. We previously showed that Dicer levels were low to undetectable in monocytes (Klase et al, 2007). Here, these promonocytic cells show undetectably low levels of Dicer, as compared with T-cells (Figure 3A). The presence of Dicer protein was detectable only when cells had differentiated into macrophages with PMA treatment. Once again, PIWIL4 levels are constantly present in these cells and could contribute to microRNA formation either at monocyte or macrophage stages. The supernatants from these cells were collected and processed for exogenous reverse transcriptase (RT) levels. From the results shown in Figure 3B, Flavopiridol was able to decrease RT levels in both cell lines. However both CR8 and CR8#13 were effectively able to decrease RT levels in the Jurkat T-cell lines as compared to U937 monocytic cells. Along these lines, previous studies have shown that promonocytic cells expressed significantly less or nonexistent amounts of Dicer, as compared to T-cells (Klase et al, 2007). These results further suggest the dependence of CR8#13 effects on the microRNA machinery.
Figure 3.
RT assay to determine virus production in drug treated cells. A. Western blot for Dicer, Drosha, Ago2 and PIWIL4 in control T- cells (CEM) and monocytes (U937) are shown (50μg of total protein). Dicer protein expression becomes apparent only after PMA treatment resulting in differentiation of cells into macrophages. PIWIL4 are present in both cell types. B. Jurkat T-cell and promonocytic U937 cells were electroporated with 5μg pNL4.3 followed the next day by drug treatment of Flavopiridol (200nM), CR8 (100nM) or CR8#13 (50nM). Cell supernatants were collected at 48 hours post drug treatment. Viral supernatants (10μl) were incubated in a 96-well plate with reverse transcriptase (RT) reaction mixture overnight at 37°C, and 10μl of the reaction mix was spotted on a DEAE Filtermat paper, washed with 5% (w/v) Na2HPO4 followed by water wash, and then dried completely. RT activity was measured in a Betaplate counter.
CR8#13 does not affect cdk9 responsive cellular genes
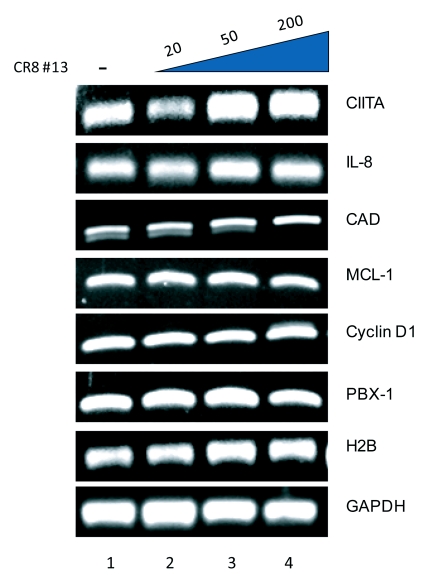
Flavopiridol at low concentrations is a known inhibitor of cdk9 (Chao et al, 2000); hence, we wished to determine if CR8#13 would have any effect on cdk9 responsive genes in vivo. Our ultimate goal has been to find an inhibitor that effects viral transcription at low IC50 with minimal inhibition on cellular genes necessary for normal cell development. We utilized a set of cellular and viral genes to test the effect of CR8#13 on their transcription using RT/PCR. Known cdk9 responsive cellular genes included: CIITA, IL-8, CAD, MCL-1, Cyclin D1, and PBX-1 (Carlson et al, 1999; Kanazawa et al, 2000; Barboric et al, 2001; Lam et al, 2001; Eberhardy and Farnham, 2002; Chao et al, 2003). Histone H2B gene served as a negative control for lack of cdk9 requirement for its gene expression (Medlin et al, 2005). Results in Figure 4 show that treatment of cells with CR8#13 at various concentrations (20, 50, 200nM) did not inhibit transcription of genes that were cdk9 responsive. This further reinforces the idea that the transcriptional inhibition caused by CR8#13 may be specific to HIV-1 promoter and not cellular genes that utilize cdk9 pathway.
Figure 4.
Agarose gels showing a lack of effect on cellular genes controlled by cdk9 after treatment with CR8#13. 293T cells were treated with three different concentrations of CR8#13 (20 nM, 50 nM, and 200 nM). Cells were processed 48hrs-post treatment for RT-PCR. Effector cdk9 genes such as CIITA, IL-8, CAD, MCL-1, Cyclin D1, and PBX-1 were used in the RT-PCR.
Possible effect of TAR microRNA in increasing the effectiveness of HIV-1 transcription inhibitors
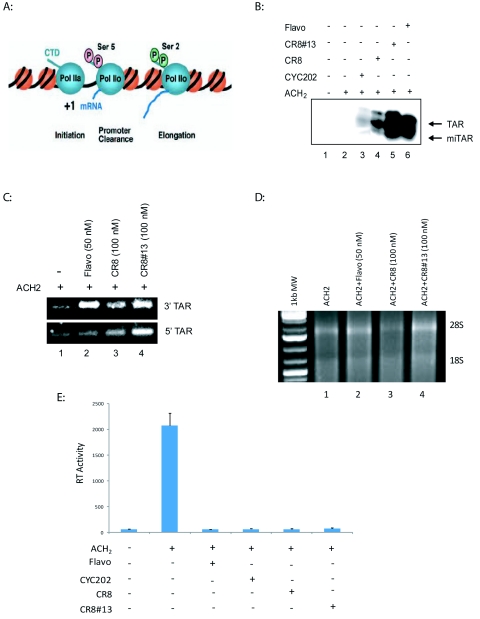
Latently infected cells produce a high level of short, abortive RNA transcripts only 50-100nt in length, that contain the HIV TAR stemloop (Adams et al, 1994). It has been shown that TAR serves as a substrate for Dicer resulting in short 21-22 RNA molecules capable of silencing HIV-1 transcription (Klase et al, 2007). Since these TAR-containing short transcripts are the dominant HIV-1 RNA produced in appreciable quantities during latency, it is possible that these RNA molecules could suppress viral gene expression. Our current studies point to cdk inhibitors functionally interacting with TAR microRNA, which affects RNA polymerase II processing, and may increase the efficacy of the drugs' inhibition on the HIV-1 promoter. As seen in Figure 5A, a model for transcription argues that RNA polymerase II is phosphorylated at Ser5 at the initiation complex and Ser2 on the elongating complex. RNA polymerase II associated with HIV-1 promoter is, however, phosphorylated on both Ser2 and 5 in the presence of Tat (Zhou et al, 2004). In order to ascertain whether or not cells treated with CDK inhibitor produce additional TAR derived miRNA, we utilized RNase protection assays to detect small RNA fragments corresponding to TAR sequence (Klase et al, 2007). Briefly, a probe complementary to the entire length of the 5' portion of the TAR stem loop was designed, which would detect the generation of ~21nt RNAs from any position within that sequence. The results were considered positive if the 32nt probe was cleaved to ~21nt, indicating protection by a microRNA. The latently infected T-cell clone, ACH2, was treated with TNF (for viral induction) followed by treatment with Cyc202, CR8, and CR8#13. Thirty micrograms of total RNA from each condition was used for RPA analysis to detect the presence of the 5' TAR miRNA. Higher levels of TAR and miTAR RNA were observed in CR8#13 treated cells as compared to Flavopiridol treatment (Figure 5B, Lanes 5 and 6). This lead to further experimentation to determine levels of each of the microRNA produced from TAR region (3'TAR stem and 5'TAR stem) in presence of these drugs.
Figure 5.
Increased production of TAR microRNA due to drug treatment. A. Model of the effect of RNA polymerase II phosphorylation on transcription. RNA polymerase II CTD is hypo-phosphorylated at the initiation complex; Ser5 is only phosphorylated at the promoter clearance stage; and Ser2 is mostly phosphorylated at the elongation phase. HIV-1 genome is unique in that it contains both Ser2 and Ser5 phosphorylation at the elongation stage (Zhou et al, 2004). Phosphorylation of Ser2 and Ser5 could be seen by multiple cyclin/cdk complexes. B. Ten micrograms of total RNA from TNF treated CEM (lane 1) and TNF treated ACH2 cells (lanes 2-6) were hybridized to a radiolabeled TAR 5' probe and then treated with RNase A. Arrows indicate the probe protected by TAR at 27 nucleotides and the probe protected by a TAR miRNA at approximately 22nt. Cyc202 concentration at 500nM, CR8 at 100nM, CR8#13 at 50nM, and Flavopiridol at 50nM were used for these experiments. C. ACH2 cells were treated with Flavopiridol (50nM), CR8 (100nM) and CR8#13 (100nM). RNA was extracted 48hrs-post drug treatment. 500ng of RNA from the microRNA-enriched fraction was used to generate cDNA using the Quantimir kit (SBI). RT reactions are performed followed by PCR in which a universal reverse primer is provided by the manufacturer. Specific microRNA forward primers are identical in sequence to the microRNA of interest. PCR products corresponding to the amplified microRNAs were resolved in a 3.5% (w/v) agarose gel. The PCR products are at around 67bp as compared with the Fermentas 1kb DNA Plus Ladder. Increased amounts of 3' and 5' TAR microRNA were observed post drug treatment. D. Total RNA (1ug) from each samples was separated in a 1% (w/v) agarose gel. The location of both 18S and 28S are shown. E. RT assay was performed to detect viral levels in ACH2 cells after TNF and drug treatments. ACH2 cells were treated with Flavopiridol (50nM), Cyc202 (500nM), CR8 (100nM) and CR8#13(100nM). Supernatents were collected 48hrs later and used for RT assay. TNF treatment significantly increased RT levels in ACH2 cells and drug treatment was able to decrease RT levels.
Total RNA from the ACH2 cells treated with Flavopiridol, CR8, and CR8#13 were extracted and processed for RT-PCR detection of microRNAs, specifically for the 3'TAR and 5'TAR stems regions. The QuantiMir RT Kit provides a simple and sensitive method to detect small RNA molecules. After total RNA is extracted, the microRNAs are polyA tagged and then an oligo-dT adaptor is annealed. At this point, RT is used to create first strand cDNAs. From here, standard end-point PCR can be used to detect specific microRNAs. The product would be the adaptor (46bp) plus the miRNA (~2bp). Results in Figure 5C indicate that there is an increase in 3'TAR with each drug treatment over the untreated control Flavopiridol and CR8#13 treatment exhibited a similar increase in 3'TAR microRNA levels. In contrast, there was an increase in the 5'TAR microRNA levels only with the cells treated with CR8#13. The results from Figure 5C could potentially explain the higher levels of miTAR observed in Figure 5B (Lane 5). As control we ran the RNA on an agarose gel to check for integrity of the total RNA (Figure 5D). Finally, we looked at the RT levels in the supernatant of these drug treated cells and found almost complete inhibition of virus replication in these cells (Figure 5E). Collectively, these results lead us to hypothesize that the 5'TAR microRNA could potentially be responsible for the effective inhibitory effects of CR8#13. Finally, this suggests that inhibitors of RNA polymerase II Ser2/Ser5 phosphorylation may slow down RNA polymerase II moving toward the 3' end of the HIV-1 genome and create more short TAR transcripts than normally present in these cells, thus providing a mechanistic explanation of transcription inhibition observed with inhibitors, such as CR8#13.
TAR microRNA induce formation of repressive chromatin markers on the HIV-1 LTR
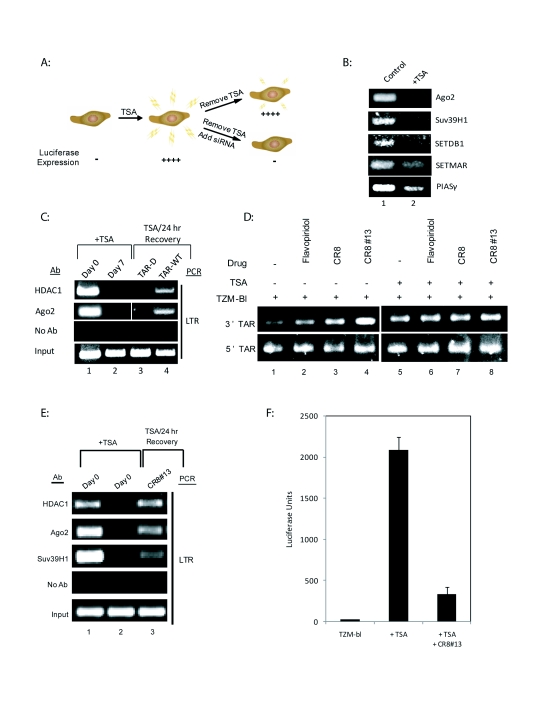
Recent studies have suggested that TAR derived microRNA may have effects on chromatin structure (Klase et al, 2009). To test the ability of TAR derived microRNA to direct chromatin remodeling at the viral LTR, chromatin immunoprecipitation (ChIP) assays were performed to examine the recruitment of factors to the HIV-1 LTR. One such factor, HDAC-1, is a histone deacetylase shown to be involved in silencing of HIV-1 promoter (Easow et al, 2007). TZM-bl cells carrying integrated HIV-1 LTR were utilized for these experiments (Figure 6A). In these cells LTR is already silenced. Previous work in the field has shown that chronic treatment of HeLa cells with the HDAC inhibitor TSA abolishes the heterochromatic state (Cobb et al, 2005). We therefore treated the cells for seven days with a sub-lethal dose of the HDAC inhibitor TSA and assayed for factor occupancy on the promoter. Chromatin changes were verified by performing ChIP assays before and after TSA treatment using antibodies specific for inhibitory factors including the components of the RNAi machinery (Figure 6B). The rationale here was to see if silenced state (absence of TSA) would have differing factor occupancy as compared to active state (presence of TSA). The results in panel B established that the RNAi protein Argonaute and the histone modifiers Suv39H1 and SETDB1 are present at the latent LTR and are removed once they are treated with TSA.
Figure 6.
HIV microRNAs specifically induce formation of repressive chromatin markers on the HIV-1 LTR. A. Model of TZM-bl cells suppression and activation. Trichostatin-A (TSA), a widely used HDAC inhibitor were used to activate the integrated HIV-1 LTR-Luc transcription in TZM-bl and abolish the repressive heterochromatic state. Seven days post treatment of TSA, the TZM-bl were transfected with the TAR microRNA. B. ChIP assays were performed with the TSA treated TZM-bls. Primers specific for the HIV-1 LTR were used to amplify DNA that was precipitated with each antibody. MicroRNA machinery (Ago2), histone methyltransferases (Suv39H1), chromatin remodeling markers (SETDB1, SETMAR), and transcription repressors (PIAS?) were downregulated after TSA treatment on the integrated HIV-LTR. C. ChIP assays were performed on several markers of chromatin repression (HDAC1) and microRNA machinery (Ago2) in TZMb1 cells. Primers specific for the HIV-1 LTR were used to amplify DNA that was precipitated with each antibody. Lane 1 shows basal levels of repressive markers on the HIV-1 LTR. Lane 2 shows that seven days of TSA treatment removes the markers of repressive chromatin. Lane 3 shows that the TAR-D mutant does not initiate a recruitment of repressive enzymes. Lane 4 demonstrates that addition of the WT-TAR molecule is sufficient to recruit Ago2 and HDAC1 back to the HIV-1 LTR region. D. TZM-bl cells were treated with Flavopiridol (50nM), CR8 (100nM) and CR8#13 (100nM) after 7-day TSA treatment. RNA was extracted 48hrs-post drug treatment. 500ng of RNA from the microRNA-enriched fraction was used to generate cDNA using the Quantimir kit (SBI) in order to poly-adenylate small RNA species. RT reactions were performed followed by PCR in which a universal reverse primer was provided by the manufacturer. Specific microRNA forward primers are identical in sequence to the microRNA of interest. PCR products corresponding to the amplified microRNAs were separated in a 3.5% (w/v) agarose gel. The PCR products are at around 67bp as compared with the Fermentas 1kb DNA Plus Ladder. Increased levels of 3'TAR microRNA were produced post CR8#13 treatment. E. ChIP assays were performed on several markers of chromatin repression (HDAC1, Suv39H1) and microRNA machinery (Ago2) in TZMb1 cells. Primers specific for the HIV-1 LTR were used to amplify DNA that was precipitated with each antibody. Lane 1 indicates basal levels of repressive markers on the HIV-1 LTR. Lane 2 indicates that seven days of TSA treatment removes the markers of repressive chromatin and Lane 3 shows results that the CR8#13 treatment is sufficient to recruit HDAC1, Ago2 and Suv39H1 back to the HIV-1 LTR region. F. Luciferase assays were performed on the cells used in Figure 6E. Luciferase activity increased with TSA treatment and then decreased post-CR8#13 treatment.
To evaluate the effect of TAR microRNA on recruitment of repressive chromatin remodeling factors to the HIV-1 LTR, we TSA treated the TZM-bl cells for 7 days followed by removal of TSA and transfection of these cells with either TAR-WT or TAR-D RNA control. On day 7 TSA was removed and replaced with complete media, and on Day 8 cells were used for ChIP for presence of either HDAC-1 and/or Argonaute (Ago2) (Figure 6C). Results indicated that prior to TSA treatment, HDAC-1 and Ago2 were associated with the LTR and this association was lost upon treatment with TSA (compare lanes 1 and 2). Transfection of the cells with TAR-WT RNA led to an increase in the reassociation of HDAC-1 and Ago2 to the LTR after 24hrs as compared to the control RNA (compare lane 3 to 4). HDAC-1 and Ago2 recruitment to an integrated LTR verifies that heterochromatin formation at the HIV-1 LTR is driven by RNAi mediated TAR microRNA.
We wished to determine if Flavopiridol, CR8, and CR8#13 treatments increased levels of TAR microRNA in the TSA-treated and control TZM-bl cells. Total RNA was extracted from both sets of cells treated with Flavopiridol, CR8, and CR8#13 and processed for RT-PCR detection of microRNAs, specifically for the 3'TAR and 5'TAR molecules. The results from Figure 6D validate the presence of both 3' and 5' TAR microRNA in these cells. The TZM-bl cells treated with TSA expressed higher amounts of both 3' and 5' TAR microRNA when treated with Flavopiridol and CR8. The addition of Flavopirodol, CR8, and especially CR8#13 significantly increased amount of the 3' TAR microRNA in the non-TSA treated TZM-bl cells. This was expected since previous results have shown that integral microRNA machinery (i.e., Ago2) can be found near the HIV-1 LTR before TSA treatment. Finally, we observed a similar increase in the 3' and 5'TAR microRNA from cells that had been treated first with TSA and then CR8#13.
In order to observe the drug effects on heterochrmatin formation, a ChIP assay was performed on TZM-bl cells treated with TSA similar to Figure 6B and C, and then treated with CR8#13. The results from Figure 6E show that treatment with CR8#13 results in the re-recruitment of heterochromatin markers, such as HDAC1, Ago2, and Suv39H1 to the HIV-1 promoter. In addition to the ChIP assay, a Luciferase assay was performed on the TSA and CR8#13 treated cells. Results in Figure 6F show that TSA treatment resulted in increased Luciferase activity of the intergrated HIV-1-Luc in TZM-bl cells. Treatment with CR8#13 was able to decrease Luciferase activity in these cells. Collectively, these results indicate that the HIV-1 TAR microRNA is responsible for the enhanced effectiveness of cdk inhibitors, especially CR8#13, on the HIV-1 promoter by recruiting both microRNA and chromatin remodeling complexes to the promoter proximal region.
DISCUSSION
A viral microRNA generated from full-length, doubly spliced or singly spliced HIV-1 transcripts could potentially inhibit viral replication, block translation of viral proteins or cause remodeling of the viral genome. Since the integrated HIV-1 genome is associated with chromatin remodeling complexes and histone acetyl-transferases activity is involved with activation of the virus, it makes sense that a viral microRNA could have a significant role in the control of viral transcription.
The TAR element, about 50 nucleotides long, at the 5' end of the HIV-1 viral mRNA was one of the five structures within HIV that had a possibility of being processed by Dicer. The TAR element has been shown to be involved with activation of promoter proximal region and not so much elongation of transcription. It would be interesting if the TAR microRNA, rather than the entire hairpin, is actually the functioning sequence in regulating transcription. Further support of a role for TAR hairpins loaded into microRNA machinery is that TAR-RNA Binding Protein (TRBP) was identified as the human homologue of the Drosophila loquacious protein which is required for efficient loading of the microRNA into the RISC complex (Chendrimada et al, 2005). The fact that RNAi components, such as TRBP, can be found associated with the TAR element is strong evidence that TAR may be processed to yield microRNA. Previously, we successfully detected a TAR derived HIV-1 microRNA using RNase protection and subsequent pyrosequencing. We showed the presence of the 5' and 3' TAR microRNA in cell lines and de novo infection of primary cells and have cloned the TAR microRNAs from infected cells (Klase et al, 2009). The production of this TAR microRNA and binding to complementary genes could explain the downregulation of many cellular genes (Liang et al, 2005; Zhao et al, 2005). From various studies, latent cells have been shown to produce a high amount of short, abortive RNA transcripts only 50-100 nucleotides in length, that contain the HIV-1 TAR hairpin (Adams et al, 1994). Of the entire HIV-1 genome, these TAR-containing short transcripts are the only HIV RNA produced in large quantities during latency. Therefore, it is possible that the microRNAs generated from TAR may act to suppress viral gene expression and alter host-cell proteins levels in order to maintain the latent state.
While current anti-retroviral drugs can halt viral replication, latent HIV-1 infections persist in infected patients. Any halt or interruption to the therapy quickly results in a resurgence of viral titers due to the reservoir of latent infections. Consequently, research into the mechanisms underlying viral latency has taken on increasing significance and few labs have developed numerous models of latent infections. Understanding how latently infected cells function, enable virus replication, and avoid immune response is the key to finding an effective treatment for HIV-1 infection.
Since HIV-1 TAR microRNA plays significant roles in manipulating both cellular and viral mechanisms, it is no surprise that this viral microRNA could also be involved with drug efficacy. As previously reported, cdk/cyclin complexes play an important role in viral replication. Effective cdk inhibitors could play a significant role in suppressing viral replication and possibly better therapeutics for HIV-1 infection. As previously shown, Cyc202 (R-Roscovitine), was able to prevent cdk2/cyclin E binding to the HIV-1 LTR and inhibit HIV-1 in T-cells, monocytes, and peripheral blood mononuclear cells at a low IC50 (Agbottah et al, 2005). These results also showed that Cyc202 was able to inhibit HIV-1 more effectively in T-cells as compared to monocytes at equivalent drug concentrations. When using co-crystal structures, the purine ring of Cyc202 and of CR8, a derivative of Cyc202, is sandwiched between the side chain of Ile10 and of Leu134 of cdk2. Compared to the Cyc202 phenyl group, the phenyl ring of CR8 is positioned further away from Phe82 side chain. The CR8 isomers were roughly 25 times more potent than Cyc202. Through our initial drug screens, CR8 was able to effectively eliminate HIV-1 infected cells better than the uninfected cells.
We then attempted to find a derivative of CR8 that could potentially eliminate HIV-1 transcription completely without affecting essential cellular genes that incorporate the use of certain cdks. The most promising was CR8#13, which decreased viral transcription significantly more than the other roscovitine derivatives. Not only did CR8#13 downregulate viral transcription, it also did not affect cell viability or downstream cdk9 effector genes. This suggests that CR8#13 is capable of specifically targeting HIV-1 transcripts.
To understand the mechanism behind this cdk inhibitor, we attempted to examine possible reasons why this class of drugs is effective against HIV-1 transcription. The combined observations that the parental Cyc202 was more effective in T-cells rather than monocytes (Agbottah et al, 2005), and that monocytes expressed significantly less Dicer (Klase et al, 2007) led us to predict that microRNA was involved in drug efficacy. In fact, both Flavopiridol and the CR8#13 exhibited a dependence on the presence of the viral TAR microRNA. These two drugs exhibited significantly more downregulation of viral transcription compared to other drugs when Dicer is present. The difference between Flavopiridol and CR8#13 is also highlighted by the forms of microRNA that are produced following treatment. When looking specifically at the TAR microRNA produced from drug treated infected cells, the CR8#13 caused production of more viral microRNA than Flavopiridol. In addition, CR8#13 caused a significant increase in both the 3'TAR and 5'TAR, while Flavopiridol only increased the 3'TAR microRNA. The drugs that are more effective in the presence of Dicer utilize microRNAs to downregulate viral transcription; however, Flavopiridol and CR8#13 seems to utilize different microRNA mechanisms to inhibit HIV-1.
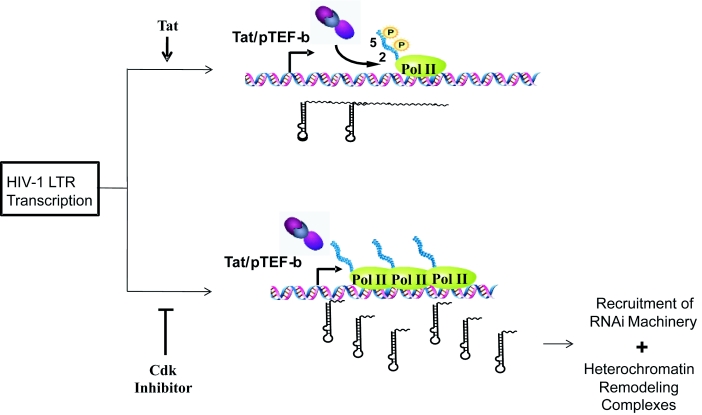
To understand mechanistically how the TAR microRNA is inhibiting HIV-1 transcription, we incorporated the use of TZM-bl cells. Using the TZM-bl cells, we are able to show that chromatin remodeling complexes and microRNA machinery are not bound to the HIV-1 LTR when the LTR becomes activated. However, when the TAR microRNA is reapplied, these complexes are re-linked to the LTR to terminate RNA polymerase II transcription and more TAR microRNA is created. Once again, CR8#13 was able to induce the production of both the 3' and 5'TAR microRNA, due to this premature termination of RNA polymerase II and recruitment of microRNA machinery to the HIV-1 LTR. Overall, a cdk inhibitor at low concentrations could potentially inhibit Pol II phosphorylation and elongation, increase TAR microRNA levels and therefore result in a higher specificity of inhibition toward the HIV-1 promoter as compared to other cellular or viral promoters. Along these lines Figure 7 shows such a model and further points to where an increase in TAR levels leads to the recruitment of RNAi machinery and heterochromatin remodeling complexes to the HIV-1 promoter.
Figure 7.
Model for cdk inhibitor-mediated viral microRNA production and transcriptional inhibition. During viral transcription, Tat/pTEF-b complexes increase phosphorylation of RNA polymerase II, leading to increased transcriptional elongation. In contrast, cdk inhibitors reduce phosphorylation of RNA polymerase II (at either Ser 2, 5 or both), consequently decreasing elongation. As a result, increased TAR transcripts are produced which aid in the recruitment of RNA interference machinery and heterochromatin remodeling complexes to the HIV-1 promoter, inhibiting transcription. This form of inhibition may ultimately lead to DNA methylation as a permanent epigenetic mark on HIV-1 LTR.
In conclusion, the HIV-1 field has progressed rapidly in developing an effective therapy, HAART, to prevent infection from spreading to other healthy cell. However, new treatments need to be developed in order to destroy latently infected cells and eliminate infection. It appears that the production of viral TAR microRNA, allows HIV-1 to both suppress apoptotic cellular mechanisms and also keep viral genome activation at a minimum until these latently infected cells are stimulated to produce more virus and potentially infect other healthy cells. These microRNA pathways also enable certain drugs to have greater efficacy and can play vital roles in drug functional pathways. Understanding latency and how microRNA mechanisms contribute is integral to finding a therapeutic to effectively treat HIV-1 infection.
CONCLUSIONS
We have demonstrated that CR8#13, a third generation cdk inhibitor, is an effective inhibitor of HIV-1 LTR at the viral promoter.
We showed that CR8#13 requires the cellular microRNA machinery to achieve significant transcriptional inhibition of HIV-1.
CR8#13 exerts its inhibitory effect only on HIV-1 LTR expression, as we demonstrated that normal cellular gene expression is unaffected following drug treatment.
We provide a mechanistic explanation for CR8#13 based HIV-1 inhibition by showing that CR8#13 increases 3' and 5' TAR microRNA production in HIV-1 infected cells.
We demonstrated that the increased TAR microRNA in infected cells causes formation of repressive chromatin structures on the HIV-1 promoter, thus inducing viral transcriptional inhibition.
Acknowledgments
We would like to thank the members of the Kashanchi lab for experiments and assistance with the manuscript. Most of the data on the current manuscript was generated using funds from NIH grants AI078859, AI074410 and AI043894. LC is a research assistant in the National Center for Biodefense and Infectious Diseases at The George Mason University.
LIST OF ABBREVIATIONS
- HAART
Highly Active Antiretroviral Therapy
- LTR
Long Terminal Repeat
- TAR
Transactivation Response Element
- pTEFb
Positive Transcription Elongation Factor b
- CDK
Cyclin-dependent Kinase
- CAT Assay
Chloramphenicol Acetyltransferase Assay
- ChIP
Chromatin Immunoprecipitation
- RT
Reverse transcriptase
COMPETING INTERESTS
None declared
REFERENCES
- Adams M, Sharmeen L, Kimpton J, et al. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc Natl Acad Sci USA. 1994;91:3862–3866. doi: 10.1073/pnas.91.9.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbottah E, de LaFuente C, Nekhai S, et al. Antiviral activity of CYC202 in HIV-1-infected cells. J Biol Chem. 2005;280:3029–3042. doi: 10.1074/jbc.M406435200. [DOI] [PubMed] [Google Scholar]
- Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanassiou Z, Dias RL, Moehle K, Dobson N, Varani G, Robinson JA. Structural mimicry of retroviral tat proteins by constrained beta-hairpin peptidomimetics: ligands with high affinity and selectivity for viral TAR RNA regulatory elements. J Am Chem Soc. 2004;126:6906–6913. doi: 10.1021/ja0497680. [DOI] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bettayeb K, Tirado OM, Marionneau-Lambot S, et al. Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases. Cancer Res. 2007;67:8325–8334. doi: 10.1158/0008-5472.CAN-07-1826. [DOI] [PubMed] [Google Scholar]
- Bohan CA, Kashanchi F, Ensoli B, Buonaguro L, Boris-Lawrie KA, Brady JN. Analysis of Tat transactivation of human immunodeficiency virus transcription in vitro. Gene Expr. 1992;2:391–407. [PMC free article] [PubMed] [Google Scholar]
- Carlson B, Lahusen T, Singh S, et al. Down-regulation of cyclin D1 by transcriptional repression in MCF-7 human breast carcinoma cells induced by flavopiridol. Cancer Res. 1999;59:4634–4641. [PubMed] [Google Scholar]
- Chao SH, Fujinaga K, Marion JE, et al. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- Chao SH, Walker JR, Chanda SK, Gray NS, Caldwell JS. Identification of homeodomain proteins, PBX1 and PREP1, involved in the transcription of murine leukemia virus. Mol Cell Biol. 2003;23:831–841. doi: 10.1128/MCB.23.3.831-841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Nesterova TB, Thompson E, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley W, Van Duyne R, Carpio L, et al. Absence of Dicer in monocytes and its regulation by HIV-1. J Biol Chem, submitted. 2010 doi: 10.1074/jbc.M110.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM. MicroRNAs in viral replication and pathogenesis. DNA Cell Biol. 2007;26:239–249. doi: 10.1089/dna.2006.0559. [DOI] [PubMed] [Google Scholar]
- Easley R, Carpio L, Guendel I, et al. HTLV-1 transcription and chromatin remodeling complexes. J Virol. 2010;84:4755–4768. doi: 10.1128/JVI.00851-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- Galons H, Oumata N, Meijer L. Cyclin-dependent kinase inhibitors: a survey of recent patent literature. Expert Opin Ther Pat. 2010;20:377–404. doi: 10.1517/13543770903524284. [DOI] [PubMed] [Google Scholar]
- Grey F, Antoniewicz A, Allen E, et al. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79:12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guendel I, Carpio L, Easley R, et al. 9-Aminoacridine inhibition of HIV-1 Tat dependent transcription. Virol J. 2009;6:114. doi: 10.1186/1743-422X-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesslein JL, Jullian N. Recent advances in cyclin-dependent kinase inhibition. Purine-based derivatives as anti-cancer agents. Roles and perspectives for the future. Curr Top Med Chem. 2002;2:1037–1050. doi: 10.2174/1568026023393291. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Kanazawa S, Okamoto T, Peterlin BM. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- Kaul D, Ahlawat A, Gupta SD. HIV-1 genome-encoded hiv1-mir-H1 impairs cellular responses to infection. Mol Cell Biochem. 2009;323:143–148. doi: 10.1007/s11010-008-9973-4. [DOI] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol. 2002;22:4622–4637. doi: 10.1128/MCB.22.13.4622-4637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, Kale P, Winograd R, et al. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, Winograd R, Davis J, et al. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology. 2009;6:18. doi: 10.1186/1742-4690-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001:2. doi: 10.1186/gb-2001-2-10-research0041. research0041.1-0041.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Maddukuri A, Teslovich TM, et al. Therapeutic targets for HIV-1 infection in the host proteome. Retrovirology. 2005;2:20. doi: 10.1186/1742-4690-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Medlin J, Scurry A, Taylor A, Zhang F, Peterlin BM, Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 2005;24:4154–4165. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekhai S, Zhou M, Fernandez A, et al. HIV-1 Tat-associated RNA polymerase C-terminal domain kinase, CDK2, phosphorylates CDK7 and stimulates Tat-mediated transcription. Biochem J. 2002;364:649–657. doi: 10.1042/BJ20011191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S, Ito M, Tsutsumi Y, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oumata N, Bettayeb K, Ferandin Y, et al. Roscovitine-derived, dual-specificity inhibitors of cyclin-dependent kinases and casein kinases 1. J Med Chem. 2008;51:5229–5242. doi: 10.1021/jm800109e. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Provost P, Barat C, Plante I, Tremblay MJ. HIV-l and the microRNA-guided silencing pathway: an intricate and multifaceted encounter. Virus Res. 2006;121(2):107–15. doi: 10.1016/j.virusres.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone M, Erba E, Damia G, et al. Variolin B and its derivate deoxy-variolin B: new marine natural compounds with cyclin-dependent kinase inhibitor activity. Eur J Cancer. 2005;41:2366–2377. doi: 10.1016/j.ejca.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne R, Easley R, Wu W, et al. Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology. 2008;5:40. doi: 10.1186/1742-4690-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandromme L, Piguel S, Lozach O, Meijer L, Legraverend M, Grierson DS. Suzuki-type Pd(0) coupling reactions in the synthesis of 2-arylpurines as Cdk inhibitors. Bioorg Med Chem Lett. 2006;16:3144–3146. doi: 10.1016/j.bmcl.2006.03.060. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Zhao RY, Bukrinsky M, Elder RT. HIV-1 viral protein R (Vpr) and host cellular responses. Indian J Med Res. 2005;121:270–286. [PubMed] [Google Scholar]
- Zhou M, Deng L, Lacoste V, et al. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J Virol. 2004;78:13522–13533. doi: 10.1128/JVI.78.24.13522-13533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Halanski MA, Radonovich MF, et al. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]