Abstract
Translocator protein TSPO is an 18 kDa protein implicated in numerous cell functions and is highly expressed in secretory and glandular tissues, especially in steroidogenic cells. TSPO expression is altered in pathological conditions such as certain cancers and neurological diseases. In search of the factors regulating Tspo expression, we recently showed that high levels of TSPO in steroidogenic cells may be due to high constitutive expression of protein kinase Cε (PKCε), while phorbol-12-myristate 13-acetate (PMA) activation of PKCε drives inducible TSPO expression in non-steroidogenic cells, likely through activator protein 1 (AP1). In this study, we aimed to identify the signal transduction pathway through which PKCe regulates Tspo gene expression. The MEK1/2 specific inhibitor U0126, but not NFκB inhibitors, reduced basal Tspo promoter activity in TSPO-rich steroidogenic cells (MA-10 Leydig), as well as basal and PMA-induced Tspo promoter levels in TSPO-poor non-steroidogenic cells (NIH-3T3 fibroblasts). AP1 and signal transducer and activation of transcription 3 (STAT3) have binding sites in the Tspo promoter and are downstream targets of PKCε and MAPK (Raf-1 -ERK1/2) pathways. PKCe overexpression induced STAT3 phosphorylation in NIH-3T3 cells, while PKCe knockdown reduced STAT3 and c-Jun phosphorylation in Leydig cells. MEK1/2, ERK2, c-Jun, and STAT3 knockdown reduced Tspo mRNA and protein levels in Leydig cells. Additionally, Raf-1 reduced Tspo mRNA levels in the same cells. MEK1/2, c-Jun, and STAT3 knockdown also reduced basal as well as PMA-induced Tspo mRNA levels in NIH-3T3 cells. Together, these results demonstrate that PKCe regulates Tspo gene expression through a MAPK (Raf-1-MEK1/2-ERK1/2) signal transduction pathway, acting at least in part through c-Jun and STAT3 transcription factors.
Translocator protein (18 kDa) TSPO is a nuclear encoded, primarily mitochondrial, high affinity cholesterol- and drug-binding protein that has been implicated in numerous cell functions, including cholesterol transport and steroidogenesis, cellular respiration, oxidative stress, proliferation, and apoptosis (1;2). Among these functions, the role of TSPO in cholesterol transfer across the outer to the inner mitochondrial membrane, the rate-determining step in steroidogenesis, is the most well-studied (1;2). TSPO is a ubiquitous protein with diverse levels of expression across the body. Particularly high TSPO levels are found in secretory and glandular tissues, and especially in steroid hormone producing cells, whereas intermediate levels are observed in renal and myocardial tissues. Lower levels are detected in brain and liver (1-5). TSPO levels are also elevated in cancerous tissues of the breast, ovary, colon, prostate, and brain compared to normal human tissues, suggesting a role for TSPO in carcinogenesis (6-10). A positive correlation between TSPO levels and the metastatic potential of human breast and brain tumors further supports this hypothesis (2;6-8). TSPO expression is also upregulated in the brain at sites of injury and inflammation, as well as following a number of neuropathological conditions, including stroke, herpes and HIV encephalitis, and neurodegenerative disorders such as Alzheimer's disease, multiple sclerosis, amyotrophic lateral sclerosis, Parkinson's disease, and Huntington's disease (2;6-8;11).
A number of physiological and pharmacological modulators have been shown to alter TSPO levels. These include IL-1, TNF-α, serotonin, norepinephrine, dopamine, peroxisome proliferators, and ginkgolide B (12-14). Despite the numerous studies describing TSPO protein levels in various species and tissues, little is known about the mechanisms underlying its transcriptional regulation. Typical of housekeeping genes, sequence analysis of the mouse Tspo promoter revealed that it lacks TATA and CCAAT elements, but contains a series of proximal GC boxes and several putative binding sites for transcription factors such as v-Ets erythroblastosis virus E26 oncogene homolog (Ets), AP1, specificity protein 1/specificity protein 3 (Sp1/Sp3), AP2, Ik2, GATA, SOX, and SRY (5;15;16). Deletion and mutational studies of the Tspo promoter revealed a differential transcriptional regulation of Tspo in TSPO-rich steroidogenic mouse Leydig cells compared to non-steroidogenic mouse NIH-3T3 fibroblasts. Two proximal Sp1/Sp3 sites in the 123-15 bp region and members of the Ets family of transcription factors, as well as an AP1 factor were found to be important for basal transcriptional activity (5;15;16).
We previously analyzed Tspo transcriptional responses in steroidogenic (MA-10 mouse tumor Leydig cells) and non-steroidogenic cells (NIH-3T3 mouse fibroblasts and SV-40 transformed COS-7 monkey kidney cells). We found that high levels of endogenous protein kinase C epsilon (PKCε) regulated Tspo expression in TSPO-rich steroidogenic cells, while phorbol-12-myristate 13-acetate (PMA) induced Tspo expression in TSPO-poor nonsteroidogenic cells, with PKCε mediating this effect. Moreover, the PKCε-activated pathway likely targeted the AP1 and Ets transcription factors, whose binding sites are localized to the 805-515-bp region upstream of the transcription start site (16).
Protein kinase C is a family of serine/threonine-specific protein kinases consisting of at least 11 known isoforms (17). PKC isoform expression and distribution is cell-type and condition-specific. A member of the typical group of PKC isoforms, PKCε, has been shown to be involved in a variety of signaling events involved in cell proliferation, differentiation, apoptosis, nervous functions, and secretory vesicle trafficking (18). PKCε is abundant in endocrine, neuronal, and immune cells (19;20). Moreover, the presence of high levels of PKCε in high-grade tumors, together with studies indicating its participation in tumor development, metastasis, and invasion, have suggested that PKCε is an oncogene (19). Interestingly, the profile of TSPO expression in endocrine tissues and various cancers, as well as the role of TSPO in cell proliferation, tumor invasion, and metastasis, parallels that of PKCε (16). PKCε has been shown to exert its effects through multiple downstream signaling pathways including, but not limited to, RACK2, NFκB, STAT3, mitogen-activated protein kinase MAPK (Raf-ERK1/2), or MARCKS (18;21;22).
Here we used specific pathway inhibitors in steroidogenic (MA-10 mouse tumor Leydig) and non-steroidogenic (NIH-3T3 mouse fibroblasts) cells to show that the Raf-1 - ERK1/2 signaling pathway is the downstream target through which PKCε regulates Tspo gene transcription. In the classic MAPK (Raf-1 -ERK1/2) pathway, Ras activates Raf-1 to phosphorylate MEK1/2, which in turn activates ERK1/2 by phosphorylation. Since AP1 and STAT3 have binding sites in the Tspo promoter and are downstream targets of the PKCε and MAPK (Raf-1 -ERK1/2) pathways, we also investigated their possible regulation by these kinases and their participation in regulating TSPO expression. The results obtained demonstrate that PKCe regulates Tspo gene expression through a MAPK (Raf-1-MEK1/2-ERK1/2) signal transduction pathway, acting at least in part through c-Jun and STAT3 transcription factors.
EXPERIMENTAL PROCEDURES
Cell Culture
MA-10 cells were a gift from Dr. Mario Ascoli (University of Iowa, Ames, IA). Cells were cultured in T75-cm2 cell culture flasks (Dow Corning Corp., Corning, NY) and grown in DMEM/Ham-F12 nutrient mixture (Sigma Chemical Co., St. Louis, MO) supplemented with 5% fetal bovine serum and 2.5% heat-inactivated horse serum (GIBCO, Burlington, ON). NIH-3T3 cells (Lombardi Cancer Center Tissue Culture Facility, Washington, DC) were maintained in DMEM supplemented with 10% normal calf serum. Before experimentation, MA-10 and NIH-3T3 cells were allowed to recover from plating for 24 h.
Transfection and Luciferase Assays
MA-10 and NIH-3T3 cells were seeded in 6-well plates at a density of 150,000 cells per well. The Tspo promoter construct pGL3-805 (1.5 μg) reporter plasmid was used for transfections as described previously (5). In brief, cells were transfected for 24 h using Fugene 6 (Roche, Indianapolis, IN) for MA-10 cells and Polyfect (Qiagen, Valencia, CA) for NIH-3T3 cells. All cultures received equimolar amounts of experimental constructs, where PUC19 or empty vector were added where needed to keep the final amount of DNA constant. All cultures were co-transfected with a Renilla luciferase reporter under the control of the thymidine kinase promoter (PRLTK, Promega Corp., Madison, WI) to normalize for transfection efficiency. Twenty-four hours after transfection, cells were treated with PMA (Sigma-Aldrich) or dimethylsulfoxide (DMSO, vehicle) (Sigma-Aldrich) or MEK Inhibitors, 1,4-diamino-2,3-dicyano-1,4-bis (2-aminophenylthio)butadiene (U0126) or 2-(2-Amino-3-methoxyphenyl)-4H-1-benzopyran-4-one (PD98059), at indicated doses (Calbiochem, Gibbstown, NJ) for 24 h or as otherwise indicated. Cells were preincubated with inhibitors for 1 h before adding PMA. Cellular extracts were collected by incubating cells in 1X passive lysis buffer (Promega Corp.), followed by processing with the Dual-Luciferase Reporter System (Promega Corp.). Activity was measured using the Victor2 automated plate reader (Perkin Elmer, Waltham, MA).
Quantitative Real Time (QRT)-PCR
NIH-3T3 cells were seeded in 6-well plates, and after 24 h were treated with DMSO or PMA for an additional 24 h. In silencing experiments, MA-10 and NIH-3T3 cells were seeded and treated with siRNA as described below and then cells were collected for QRT-PCR analysis. RNA was extracted using the Rneasy kit (Qiagen) according to the manufacturer's instructions. RNA (1 μg) was reverse transcribed using the TaqMan reverse transcription reagents kit (Roche) according to manufacturer's instructions to a 20 μl final volume. The resulting cDNA was diluted to 60 μl final volume with RNase free water and used as a template for real time PCR using Tspo specific primers (forward 5′-CACCGCATACATAGTAGTTGAGCA-3′ and reverse 5′-CCCGCTTGCTGTACCCTTACC-3′), and SYBR GREEN dye. Amplification of 18s ribosomal RNA served as a control.
MEK and NFκB inhibitors
MA-10 and NIH-3T3 cells were seeded at 150,000 cells per well in 6-well plates. Twenty-four hours later, cells were transfected with pGL3-805 for 24 h before being preincubated with the MEK inhibitors U0126 (10 μM, or as indicated), PD98059 (10 μM), or with the NFkB SN50 Cell-Permeable Inhibitor Peptide (10 and 20 μM) (Calbiochem #481480) or the NFkB SN50M Cell-Permeable Inactive Control Peptide (Calbiochem #481486) (10 and 20 μM), for 1 h prior to PMA treatment (50 nM). Twenty-four hours later, cells were lysed and promoter activity was measured using the dual luciferase assay as described above. Data were normalized to Renilla and presented as fold activity over the untreated control.
Immunoblotting
NIH-3T3 cells were seeded at 150,000 cells per well in 6-well plates for 24 h. Cells were preincubated with MEK inhibitors U0126 (10 μM) or PD98059 (10 μM) for 1 h before treating with PMA for the indicated times. Cells were lysed in 1X cold RIPA buffer (Cell Signaling, Danvers, MA) supplemented with Halt protease inhibitor cocktail 1:100 (Pierce, Rockford, IL). Protein concentration was measured using a standard Bradford assay (Bio-Rad, Hercules, CA), and equal amounts of protein were electrophoresed on 4-20% tris-glycine polyacrylamide gels (Invitrogen), transferred onto PVDF membranes, and probed with antibodies to PKCε (1:800) and c-Jun (1:800) (Santa Cruz, Biotechnology, Santa Cruz, CA), TSPO (1:1000) (28), p-c-Jun (Ser73) (1:1000), ERK1/2 (1:2000) and pERK1/2 (1:2000), STAT3 (1:1000), pSTAT3 (Tyr705) (1:500), pSTAT3 (Ser727) (1:500) (Cell Signaling Technology, Danvers, MA), GAPDH (1:2000) (Trevigen, Gaithersburg, MD), HPRT (1:1000), Tubulin (1:2000), pMEK1 (1:1000) and tMEK (1:1000) (Abcam, Cambridge, MA), and p-Raf-1 (1:500) (Biosource, Camarillo, CA). Immunoreactive proteins were visualized by enhanced chemiluminescence (Amersham, Piscataway, NJ) using a FUJI image reader LAS4000 for capturing images or film processing. Densitometry of each band was measured using Multi-gauge V3.0 software.
PKCε overexpression
The pCMV6-XL4 expression plasmid containing Homo sapiens protein kinase C epsilon (PRKCE) was obtained from Origene (Rockville, MD). The plasmid was amplified and purified, and the PKCε insert was sequenced. pCMV6-XL4 containing PKCε or empty vector were then transfected into NIH-3T3 cells as described above. Twenty-four hours later, cells were treated with DMSO or PMA (50 nM) for an additional 24 h or as indicated. Cells were then harvested for protein studies as described above.
PKCε, c-Jun, Stat3, and MEK1/2 siRNA treatments
MA-10 and NIH-3T3 cells were seeded at 150,000 cells per well in 6-well plates. Cells were transfected 24 h later with 4 or 8 μg of the PKCε siRNA pool, 1 μg Raf-1, 2 μg of the Stat3 and MEK1/2 siRNA pool (Dharmacon, Lafayette, CO), or 2 μg c-Jun siRNA (Dharmacon) using X-treme GENE siRNA Transfection Reagent (Roche), as recommended by the manufacturer. For Raf-1 siRNA pool, MA-10 cells were cultured for 72 h before being harvested for QRT-PCR. MA-10 and NIH-3T3 cells were transfected with Jun siRNA pool, 24 h after seeding the cells and 48 h later. Cells were harvested at 72 h for QRT-PCR or 96 h for immunoblot analysis. Stat3 and MEK1/2 siRNA pools were transfected into MA-10 cells using the protocol described above for c-Jun, while NIH-3T3 cells were transfected only once and 72 h later mRNA was collected for QRT-PCR analysis. NIH-3T3 cells were treated with or without PMA (50 nM) at 48 h for an additional 24 h before being collected for QRT-PCR analysis. Target gene knockdown was verified by QRT-PCR and/or immunoblot analysis of cell extracts when indicated.
ERK2 shRNA plasmid treatment
MA-10 cells were seeded at 150,000 cells per well in 6-well plates for 24 h before being transfected for an additional 72 h with 800 ng of shRNA ERK2 plasmid or pKD-negative control-v1 (Millipore, Lake Placid, NY) using Fugene 6 (Roche), followed by collection for protein analysis. Target gene knockdown was verified by immunoblot analysis of cell extracts.
c-Jun overexpression
NIH-3T3 cells were seeded in 12 well plates at 10,000 cells/well for 24 h before transfection with 10 ng of either pCMV-6/c-Jun containing Homo sapiens c-Jun cDNA (bearing >93% homology with mouse c-Jun; Origene, Rockville, MD), or empty pCMV-6 vector as a control along with pGL3-805 or pGL3-805m (containing a double mutation at AP1 and Ets sites) or pGL3- Basic as a control. Cells were treated after 24 h with 50 nM PMA for an additional 24 h before being lysed and assayed for luciferase activity as described above.
Preparation of Nuclear Extracts and Electrophoretic Mobility Shift Assay (EMSA)
NIH-3T3 cells were transfected with pCMV-6 or pCMV-6 containing PKCε as described above for 24 h before treatment with or without 50 nM PMA for an additional 24 h. Nuclear extracts were prepared using NE-PER nuclear and cytosolic extraction reagents (Pierce Chemical Co., Rockford, IL). For EMSA, double-stranded, 5′-biotinylated oligonucleotides containing wild type and mutant AP1-binding sequences (Table I) were incubated with 5 μg of NIH/3T3 nuclear extracts. Unlabeled double-stranded AP1 and Ets oligonucleotides were used for competition experiments. EMSA was carried out using the LightShift Chemiluminescent EMSA kit (Pierce Chemical Co.) in the presence of EDTA and Mg2+. The manufacturer's protocol was slightly modified to optimize protein-DNA interactions in the presence of antibodies. DNA-protein complexes were separated on 6% nondenaturing polyacrylamide gels, transferred to nylon membranes, and processed for visualization. All antibodies used were purchased from Santa Cruz Biotechnology (c-Jun: sc-44-x, STAT3: sc-482x and rabbit IgG: sc-2027). DNA-protein binding was optimized by including 10 μg BSA and 4 mM DTT in the all reactions.
Table 1.
Oigonucleotide Sequences Used in Electrophoretic Mobility Shift Assays
| Oligonucleotide Name | Sequence |
|---|---|
| S-Ap1 wt | CTCTAATTGACTCACAGGAAGAGGTT |
| AS-AP1 wt | AACCTCTTCCTGTGAGTCAATTAGAG |
| S-AP1 WT biotin5 | CTCTAATTGACTCACAGGAAGAGGTT |
| AS-AP1 WT biotin5 | AACCTCTTCCTGTGAGTCAATTAGAG |
| S-Ap1 MUT | CTCTAATTCTCTCACAGGAAGAGGTT |
| AS-Ap1 MUT | AACCTCTTCCTGTGAGAGAATTAGAG |
| S-Ets MUT | CTCTAATTGACTCACAGTCAGAGGTT |
| AS-Ets MUT | AACCTCTGACTGTGAGTCAATTAGAG |
Chromatin immunoprecipitation (ChIP)
ChIP was performed using the Magna ChIP G kit (Millipore) according to the manufacturer's protocol. Briefly, MA-10 and NIH-3T3 cells were grown in 150 mm dishes to 80% confluence and were cross-linked with medium containing 1% formaldehyde for 10 min at room temperature. Plates were washed 3 times with ice cold 1X PBS before being scraped and pelleted by centrifugation (800 × g) for 5 minutes at 4°C. Cells were lysed using lysis buffer (provided by manufacturer- Millipore) containing protease inhibitors and nuclei were collected by centrifugation (800 × g) for 5 minutes at 4°C. Nuclei were suspended in nuclear lysis buffer (provided by manufacturer- Millipore) containing protease inhibitors and kept on ice. Nuclear DNA was sheared to an average size of 200-1000 bp with eight 10-sec pulses at 25% power using a Vibracell VC 130 sonicator (Sonics and Materials Inc., Newtown, CT) fitted with a 3-mm stepped microtip. After shearing, chromatin was immunoprecipitated with 20 μl STAT3 (#9139 Cell Signaling Technology) or 2 μl c-Jun (# SC-44 X, Santa Cruz) antibodies overnight at 4°C with rotation, and complexes were captured using magnetic protein-G beads. Magnetic beads were washed with a low and high salt immune complex wash buffer, LiCl immune complex wash buffer and lastly TE buffer following the manufacturer's instructions. Cross-links were reversed by incubating beads in a rotating oven for 4 h at 62°C. DNA was purified and diluted 1:10 before being subjected to QRT-PCR using primers specific for the mouse Tspo distal promoter (forward 5′- TGGTTAAGAACAGGAAGCCcG-3′ and reverse 5′- TGGTGTCTGCAAGGTAAAGGTGA-3′). Immunoprecipitates obtained with normal 2 μl rabbit IgG (#2729s, Cell Signaling Technology) served as negative controls. NIH-3T3 cells were grown in 150 mm dishes to 70% confluence before being treated with DMSO or PMA for an additional 24 h, after which cells were crosslinked and subjected to the same procedure as described above.
Sequence and statistical analysis
Sequence analysis was performed using Vector NTI software (Informax, Invitrogen). Statistical analysis was performed using Prism version 4.0 (GraphPad Software, San Diego, CA). Group means were compared using the Student's t test or two-way ANOVA. Data are presented as mean ± SEM, where p < 0.05 was considered significant.
RESULTS
Promoter regions 805-515 and 123-15 harbor elements essential for basal Tspo promoter activity in MA-10 and NIH-3T3 cells
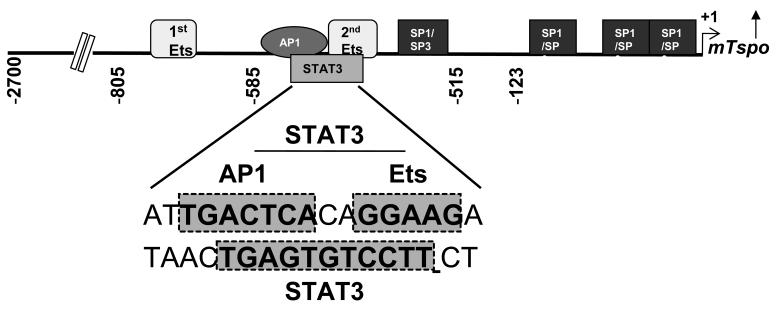
Detailed database motif analysis of the proximal promoter led to identification of multiple putative transcription factor binding sites (Figure 1). Of the multiple sites, three Sp1/Sp3, two Ets and one AP1 sites are dispersed within the first 805 bp of the promoter. Using mutational analysis and siRNA technology, we previously demonstrated that Sp1/Sp3 and Ets sites display differential importance for basal transcription in steroidogenic MA-10 Leydig and non-steroidogenic NIH/3T3 fibroblast cells (5;15). Interestingly, analysis of the complimentary strand revealed a STAT3 binding site that spans the AP1 and Ets sites in the 585-515 region. In this study we aimed to identify any potential role for the STAT3 and AP1 sites in regulating the basal and inducible activity of the Tspo gene promoter.
Figure 1. Promoter regions 805-515 and 123-1 harbor elements essential for basal Tspo promoter activity in MA-10 and NIH-3T3 cells.
Schematic representation of the mouse Tspo promoter showing transcription factor binding sites important for activity. The sequence harboring AP1, Ets and an overlapping STAT3 site is magnified. Shaded boxes indicate the nucleotide sequences of transcription factor binding sites.
PMA-inducible and basal Tspo promoter activity depends on the MAPK/ERK1/2 signaling pathway
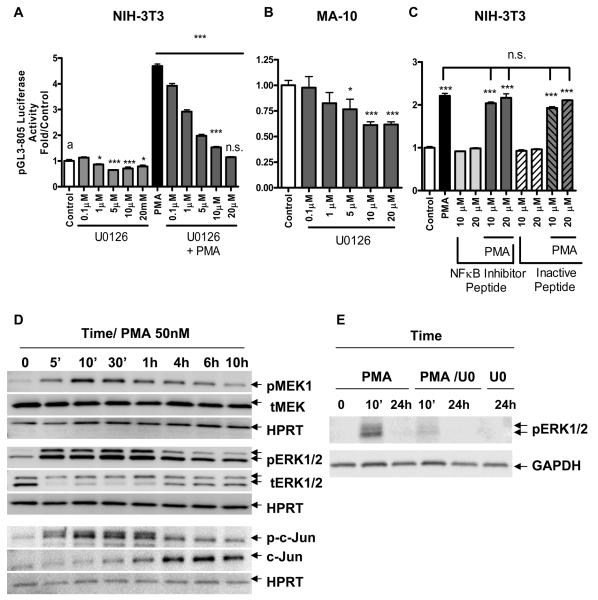
We previously reported that PMA-induced TSPO expression in non-steroidogenic NIH-3T3 fibroblasts and the high constitutive expression levels in steroidogenic MA-10 Leydig cells are under the control of PKCε. Since PMA induced Tspo transcription by activating PKCε in NIH-3T3 cells, which is known to act through multiple signaling pathways, we attempted to determine the involvement of the MAPK (Raf-ERK1/2) pathway in regulating Tspo promoter activity. As such, we transfected MA-10 and NIH-3T3 cells with the pGL3-805 promoter construct for 24 h before treatment with the MEK1/2 specific inhibitor U0126 for 1 h prior to adding PMA for an additional 24 h. As before, PMA induced Tspo promoter activity in NIH-3T3 (Figure 2A) but not in MA-10 cells (data not shown) (16). To further characterize the effect of U0126 on Tspo promoter activity, NIH-3T3 and MA-10 cells transfected with pGL3-805 were treated with a broad range of doses of the inhibitor or control DMSO. U0126 inhibited the effect of PMA in a dose-dependent manner in NIH-3T3 cells, where 20 μM of U0126 abolished the stimulatory effect of PMA (Figure 2A). In addition, U0126 reduced basal Tspo promoter activity in both NIH-3T3 and MA-10 cells in a dose-dependent manner (Figure 2A, B).
Figure 2. PMA-induced activation of the Tspo promoter is mediated by a MAPK pathway.
(A) Dose-dependent effect of U0126 on pGL3-805 luciferase activity, alone or with PMA (50 nM) or DMSO (control) in NIH-3T3 cells. (B) Dose-dependent effect of U0126 on pGL3-805 luciferase activity in MA-10 cells. (C) Dose-dependent effect of NFκB inhibitor peptide or its inactive peptide incubated in the presence or absence of PMA (50 nM) on NIH-3T3 cells. Cells were transfected with pGL3-805 for 24 h previous to treating with PMA. Cells were preincubated with the indicated concentrations of inhibitors or vehicle for 1 h prior to adding PMA up to 24 h after which they were lysed and luciferase activity was measured (D) Immunoblot of phosphorylated (p) and total (t) levels of MEK1, ERK1/2, and c-Jun in NIH-3T3 cells treated with or without PMA (50 nM) for the indicated time course. (E) Effect of U0126 (UO) on PMA-induced ERK1/2 phosphorylation. Cells were seeded for 24 h previous to treatment with PMA for the indicated time points. When treated with U0126 inhibitor, cells were preincubated for 1 h before addition of PMA. Blots shown are representative of at least two independent experiments. Results in (A), (B), and (C) are derived from three independent experiments (n = 9) and presented as fold over untreated control. *p < 0.05 and ***p < 0.001 vs. control; ns, non-significant.
To confirm the data obtained with U0126, we next performed a series of experiments using the MEK1/2 inhibitor PD98059 (data not shown). In contrast to what we expected, PD98059 at 10 μM, a dose commonly used to block MEK 1/ 2 activation, not only failed to inhibit PMA-induced Tspo promoter activity in NIH-3T3 cells, but it enhanced basal transcription in both NIH-3T3 and MA-10 cells, and the effect of PMA in NIH-3T3 cells. We next investigated this finding in detail using QRT-PCR to examine the effect of PD98059 on Tspo mRNA levels. PD98059 at 10 μM induced Tspo mRNA levels significantly after a 48 h treatment. When used at 100 μM PD98059 reduced Tspo promoter activity. Since PD98059 is a flavone with a possible effect on the antioxidant capacity of cells, resulting from the modulation of antioxidant enzyme activity as well as inhibition of enzymes that generate reactive metabolites, we investigated a possible effect on glutathione GSH levels. PD98059 reduced GSH levels only at the higher dose (100 μM). To examine whether the low concentrations (10 μM) of PD98059 could affect the cellular oxidative machinery we used a mouse oxidative stress and antioxidant defense profiler PCR array that identified 30 target genes down regulated after the 24 h treatment (data not shown). These findings suggest that PD98059 at low doses (10 μM) acts as a weak inducer of oxidative stress, which is sufficient to drive Tspo expression. At high concentrations PD98059 is a strong inducer of oxidative stress leading to cytotoxicity and reduction of Tspo levels.
To determine whether the effect of PMA on the Tspo promoter was specific to the MAPK pathway, we next examined the role of the NFκB pathway in this process. NIH-3T3 cells transfected with pGL3-805 were treated with PMA for an additional 24 h in the presence or absence of the NFκB inhibitor peptide SN-50, or its mutated inactive negative control. Cells were preincubated with the inhibitors for 1 h before adding PMA. Figure 2C shows that the NFκB inhibitor did not affect either the stimulatory effect of PMA on the Tspo promoter, or basal Tspo promoter activity. NIH-3T3 cells were also treated with the small molecule NFkB Activation Inhibitor showing similar negative results (data not shown). Treating MA-10 cells with either of the NFκB inhibitors did not reduce basal Tspo promoter activity (data not shown).
To investigate if PMA indeed activated the MAPK (Raf-1 -ERK1/2) pathway, NIH-3T3 cells were seeded for 24 h and treated with PMA for different lengths of time (Figure 2D). PMA phosphorylated MEK1 and ERK1/2 in a time-dependent manner, with ERK2 sustaining higher activation, up to 10 h, normalized to the HRPT control (Figure 2D). We previously reported that PMA induced the up-regulation of c-Jun protein levels, as well as its phosphorylation (16). Data shown in Figure 2D confirm and extend these findings by showing that PMA induces c-Jun phosphorylation as early as 5 min, and significantly induces c-Jun protein levels starting at 1 h after treatment. In agreement with the promoter data presented above, treatment of the cells with U0126 inhibited ERK1/2 phosphorylation (Figure 2E).
PKCε affects Raf-1, ERK1/2 and STAT3 phosphorylation and prolongs the activation of the ERK1/2 pathway
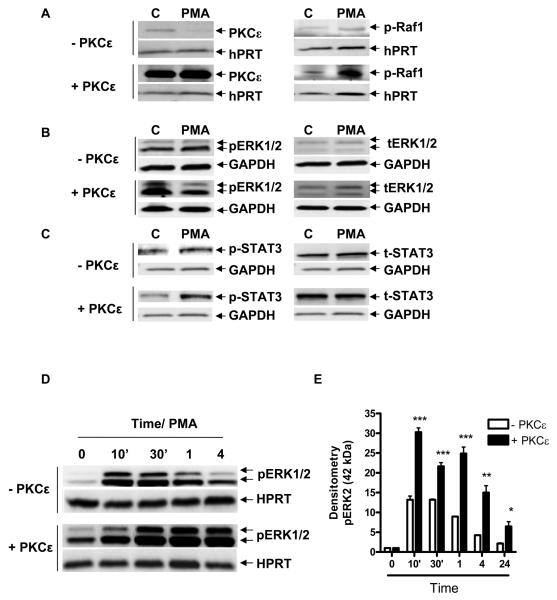
We previously demonstrated that PKCε induced TSPO and c-Jun mRNA and protein levels. To investigate whether PKCε exerts its effect through the MAPK pathway, we transfected NIH-3T3 cells with either a PKCε-expressing CMV vector or an empty vector for 24 h as previously described (16). Cells were then treated with PMA for an additional 24 h before harvesting for immunoblot analysis. Immunoblotting with an anti-p-Raf-1 antibody indicated increased levels of p-Raf-1, even after treatment with PKCε and PMA for 24 h, compared to controls (Figure 3A). In the same samples, ERK1/2 phosphorylation in PKCε-transfected cells was moderately higher than control where as in PMA treated samples ERK1/2 phosphorylation was slightly higher by 24 h (Figure 3B&E).
Figure 3. PKCε affects Raf-1, ERK1/2 and STAT3 phosphorylation and prolongs and maximizes activation of the ERK1/2 pathway.
NIH-3T3 cells were transfected with the PKCε expression vector or empty CMV vector for 24 h, then treated with or without PMA for 24 h and examined by immunoblot for the effect on p-Raf-1 (A), pERK1/2 (B), and p-STAT3 Tyr705 (C). (D) Immunoblot analysis of pERK1/2 levels in NIH-3T3 cells treated with PMA and overexpressing the PKCε expression vector or empty CMV vector for the indicated times. (E) Densitometry of the data shown in (D), obtained from three independent experiments. In (E), immunoreactive bands were normalized to HPRT, and are expressed as an increase relative to control, (CMV samples were normalized to CMV at time zero (white bars), and PKCε samples were normalized to PKCε time zero (black bars). *p < 0.05, **p < 0.01, and ***p < 0.001 vs. control. Data are representative of at least three independent experiments.
In addition, the PKCε-transfected cells treated with PMA showed higher STAT3 phosphorylation levels compared to control cells transfected with the CMV empty vector (Figure 3C). A PMA time-course treatment of NIH-3T3 cells transfected with CMV empty vector (-PKCε) or PKCε showed a maximized activation of ERK1/2 as well as a prolonged basal ERK1/2 phosphorylation in PKCε control compared to the CMV control at 24 h in representative blots (Figures 3B&D). A densitometry analysis of three blots indicated a significantly higher induction of ERK2 in PKCε-transfected samples compared to empty CMV vector alone (Figure 3E).
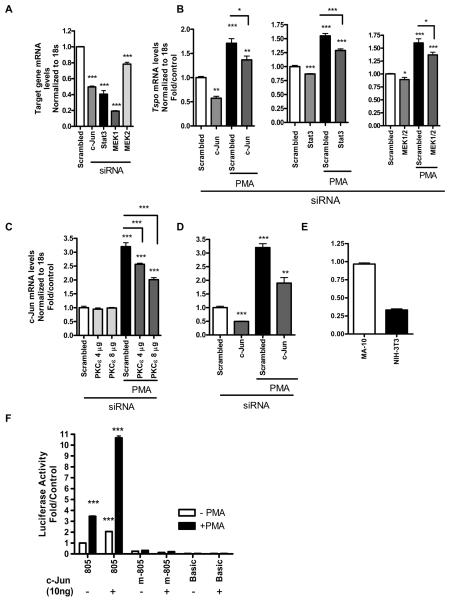
Raf-1, MEK1/2, Stat3, and c-Jun siRNA and ERK2 shRNA significantly reduce TSPO expression levels in MA-10 cells
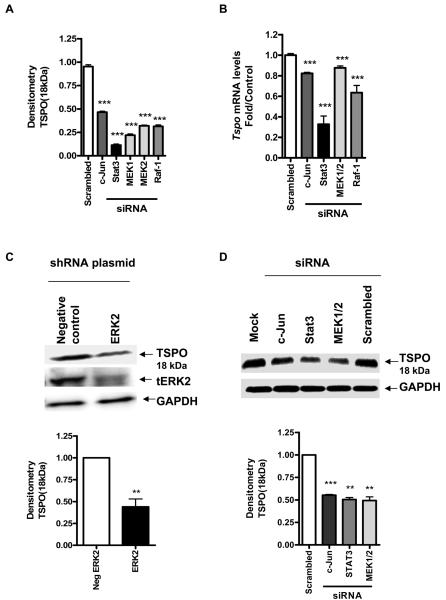
To confirm the putative role of the MAPK pathway in the regulation of TSPO expression, a pool of Raf-1 and MEK1/2 siRNA and ERK2 shRNA were used to lower Raf-1 and MEK1/2 mRNA levels and ERK2 protein levels, in TSPO-rich MA-10 cells. Figure 4A shows that the siRNA pool significantly reduced c-Jun, Stat3, both MEK1& MEK2 and Raf-1 mRNA levels. Knockdown of both Raf-1 and MEK1/2 significantly reduced Tspo mRNA levels (Figure 4B). ERK2 shRNA significantly reduced ERK2 protein levels compared to the negative control, and knockdown of ERK2 and both MEK1/2 significantly reduced TSPO protein levels (Figure 4C&D). We previously linked c-Jun to regulation of TSPO expression through binding to its corresponding AP1 site on the Tspo promoter (16). siRNA for c-Jun significantly reduced Tspo mRNA levels (Figure 4B) as well as TSPO protein levels (Figure 4D). Reducing Stat3 mRNA levels also resulted in reduction of Tspo mRNA (Figure 4B) and protein levels in MA-10 cells (Figure 4D).
Figure 4. Raf-1, MEK1/2, Stat3, and c-Jun siRNA and ERK2 shRNA significantly reduce TSPO expression levels in MA-10 cells.
(A) Effect of gene-specific siRNA treatment on c-Jun, Stat3, and MEK1/2, Raf-1 mRNA levels compared to control (scrambled siRNA). (B) Effect of down-regulation of c-Jun, Stat3, and MEK1/2, Raf-1 on Tspo mRNA levels compared to control cells incubated with scrambled siRNA. (C) Effect of down-regulation of ERK2 by shRNA on TSPO protein levels (top panel) and densitometric analysis of the immunoreactive bands (bottom panel). Cells were transfected with shRNA for ERK2 or pKD-negative control for 72 h before immunoblot analysis. (D) Effect of down-regulation of MEK1/2, Stat3, and c-Jun by siRNA on TSPO protein levels (top panel) and densitometric analysis of the immunoreactive bands (bottom panel). Cells were treated with the siRNA pool specific for c-Jun, Stat3, and MEK1/2, Raf-1 or scrambled oligonucleotides as described under materials and methods before analysis of Tspo levels using QRT-PCR and immunoblotting. Immunoreactive bands were normalized to GAPDH, and values are expressed as a decrease relative to controls incubated with scrambled siRNA. Immunoblots are representative of at least three independent experiments. Data in (A) were normalized to control (scrambled siRNA) and are derived from three independent experiments (n = 9) **p < 0.01, and ***p < 0.001 vs. control.
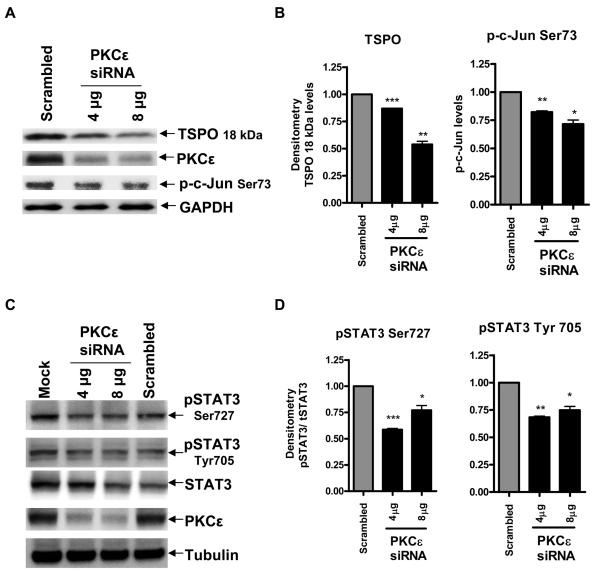
PKCε regulates TSPO expression as well as c-Jun and STAT3 activity in MA-10 cells
To determine whether PKCε regulates STAT3, we tested for the phosphorylation status of STAT3 in MA-10 cells after knockdown of PKCε. PKCε siRNA was used to lower PKCε protein levels in TSPO-rich MA-10 cells. Figure 5A shows that the pool of PKCε siRNA used inhibited PKCε protein expression reaching an 80% inhibition in the presence of 4 μg of siRNA. In agreement with our previous findings, PKCε knockdown reduced TSPO protein levels as well as p-c-Jun phosphorylation (Figure 5A&B) (16). Under the same conditions, the siRNA used also reduced the levels of STAT3 phosphorylated at Tyr705 (responsible for activation of the protein in the cytosol) as well as Ser727 (responsible for binding to DNA) (Figure 5 C&D). Normalizing the levels of phosphorylated STAT3 to Tubulin and dividing them by the total STAT3 levels indicates that PKCε significantly reduced the phosphorylation levels of a large portion of phosphorylated STAT3 at both the Tyr and Ser sites (Figure 5C&D), thus implicating STAT3 as a downstream target of PKCε in MA-10 cells and adding a new element to the machinery regulating TSPO expression.
Figure 5. PKCε mediates TSPO protein levels and regulates c-Jun and STAT3 activity in MA-10 cells.
Effect of PKCε knockdown on TSPO expression, phosphorylation levels of c-Jun at Ser73 and STAT3 at Ser727 and Tyr705. Cells were seeded and treated with the siRNA pool specific for PKCε, or with scrambled siRNA as mentioned in materials and methods. Cells were analyzed for PKCε, c-Jun pSer73 (A), STATpSer727, STATpTyr705, and STAT3 by immunoblotting 96 h after siRNA transfection (C). Densitometric analysis of the blots was performed and immunoreactive bands were normalized to GAPDH (B) or Tubulin (D) controls, and data are expressed as decrease relative to control. Immunoblots are representative of at least two independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 vs. control.
MEK1/2, STAT3, and c-Jun mediate the effect of PMA on Tspo mRNA levels in NIH-3T3 cells
Figure 6A presents mRNA levels of endogenous c-Jun, Stat3, and MEK1/2 following treatment with the respective siRNAs. Treating NIH-3T3 cells with a siRNA pool for c-Jun, Stat3, and MEK1/2 for 72 h before QRT-PCR analysis revealed that all were able to slightly reduce basal Tspo mRNA levels to a significant level (Figure 6B). Treatment with PMA for an additional 24 h following treatment for 48 h with the siRNAs listed above induced Tspo mRNA levels in the controls (scrambled) as we previously reported (16). We previously demonstrated that PKCε overexpression induced c-Jun protein levels. Treating NIH-3T3 with a PKCε siRNA pool reduced the PMA-induced c-Jun mRNA levels in a dose-dependent manner (Figure 6C). PMA induced c-Jun mRNA levels in the control (scrambled) by 3.5-fold, which was reduced by c-Jun siRNA (Figure 6D). Figure 6E shows a comparative representation of c-Jun mRNA levels in MA-10 vs. NIH-3T3 cells, were its levels are approximately 1/3 of those found in MA-10 cells. In order to confirm the participation of c-Jun in the regulation of TSPO expression, NIH-3T3 cells were transfected with pCMV-6/c-Jun cDNA or empty pCMV-6 along with Tspo promoter constructs (wt pGL3-805 or pGL3-805-m) or Basic pGL-3 vector as a control (Figure 6F). After 24 h cells were treated with 50 nM PMA for an additional 24 h before luciferase activity was measured. The results obtained showed that PMA induced pGL3-805 activity by 3.5-fold, as previously reported, an effect that was absent when treating cells containing pGL3-805m, which harbors a mutation in the AP1 and Ets binding sites (16). Interestingly, c-Jun alone was able to double basal Tspo activity in wt pGL3-805 and further potentiate the PMA effect to 11-fold, an effect which was abolished by the introduced mutations (Figure 6F), further confirming the role of c-Jun and its ability to act on Tspo promoter through its potential AP1 binding site (Figure 1).
Figure 6. MEK1/2, STAT3, and c-Jun mediate the effect of PMA on Tspo mRNA levels in NIH-3T3 cells.
(A) MEK1/2, Stat3, and c-Jun mRNA knockdown levels following treatment with gene-specific siRNAs compared to scrambled siRNA as described in materials and methods. (B) Effect of down-regulation of MEK1/2, Stat3, and c-Jun on Tspo mRNA levels in NIH-3T3 cells on Tspo mRNA levels. Cells were treated with the siRNA pool specific for MEK and Stat3 for 48 h or c-Jun for 48 h, where new siRNA was added at 24 h after the first transfection, followed by treatment with PMA for an additional 24 h. For control, cells were treated with scrambled siRNA for 48 h before treatment with PMA (50 nM) for an additional 24 h. Cells were then harvested and RNA was extracted and used for QRT-PCR analysis. (C) Effect of PKCε siRNA on newly synthesized c-Jun mRNA levels induced by PMA. (D) PMA induces c-Jun mRNA levels in NIH-3T3 cells, an effect reduced by c-Jun siRNA. (E) A comparative representation of c-Jun mRNA levels in MA-10 cells vs. NIH-3T3 cells. (F) Effect of overexpressing c-Jun on Tspo pGL3-805 or pGL3-m805 (harboring mutated AP1 and Ets sites) promoter constructs in NIH-3T3 cells. NIH-3T3 cells were seeded for 24 h prior to transfecting with either 10ng pCMV6-c-Jun or empty pCMV6 vector along with Tspo promoter constructs or basic as a control for an additional 24 h. Cells were then treated with 50 nM PMA for 24 h and luciferase activity was measured. Data were normalized to control (incubation with scrambled siRNA) and are derived from three independent experiments (n = 9). *p < 0.05, **p < 0.01 and ***p < 0.001 vs. control.
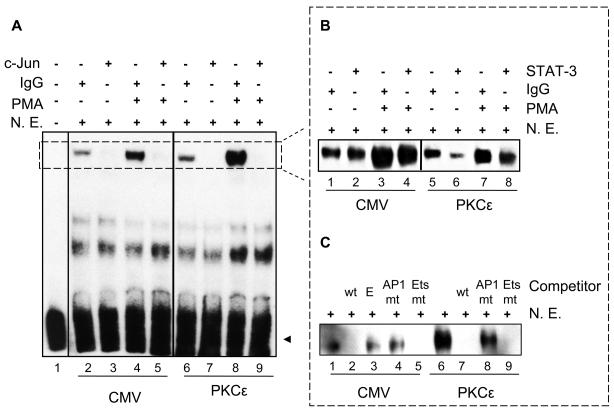
PMA induces binding of c-Jun and STAT3 to Tspo promoter sequences
EMSA was performed to investigate the effect of PMA on protein interactions at the AP1, Ets and the overlapping STAT-3 sites suspected to mediate the PMA effect on Tspo promoter regulation (Figure 7). It is important to note that the AP1 and Ets sites are separated by 3 bp resulting into identical wild-type and labeled probes, referred to as the AP1 oligonucleotide. Incubation of NIH-3T3 nuclear extracts transfected with CMV or PKCε with labeled AP1 oligonucleotide (Table I) generated the previously reported two protein-DNA complexes (16); only the more abundant specific complex 1 is visible under the exposure presented herein (Figure 7A, B and C). In Figure 7A, using nuclear extracts from CMV transfected NIH-3T3 cells treated with or without PMA, the phorbol ester induced complex 1 formation compared to control (lanes 4 vs 2); this increased DNA-protein binding was successfully competed using a c-Jun antibody (lanes 3 and 5). In cells transfected with PKCε, the same complex was formed (lanes 6 & 8), and treatment with PMA further potentiated complex 1 formation compared to PKCε alone; this DNA-protein binding interaction was also competed by c-Jun (lane 7&9). In order to investigate whether STAT3 is able to bind to the Tspo promoter, similar experiments were conducted and the specific complex formed is shown in Figure 7B. Incubation with a STAT3 antibody reduced the PMA-induced DNA-protein complex 1 formation (Figure 7B, lanes 4 & 8) as well as the DNA-protein binding induced by PKCε. The specificity of these interactions in nuclear extracts from transfected and non-transfected cells was confirmed by the addition of 200-fold excess of an unlabeled AP1 oligonucleotide (wt), which competed the formation of the complexes (Figure 7C, lanes 2 & 7), or an unrelated oligonucleotide (Epstein-Barr virus nuclear antigen recognition site (E) (Figure 7, lane 3). The addition of a 200-fold excess unlabeled oligonucleotide carrying a mutation on the putative AP1 site decreased the formation of both DNA-protein complexes in CMV and PKCε transfected cells (Figure 7C, lanes 4 & 8). On the other hand, a mutated Ets binding sequence (Figure 7C, lanes 5 & 9) successfully competed with nuclear protein binding to DNA in both complexes. The Ets mutated site was more effective than the AP1 mutated site in decreasing the formation of these complexes.
Figure 7. PMA induces c-Jun and STAT3 binding to their putative binding sites on the Tspo promoter.
EMSA of biotinylated AP1 oligonucleotide incubated without (A, lane 1) or with NIH-3T3 nuclear extract (N.E.) (all other lanes). EMSAs were performed on nuclear extracts from NIH-3T3 cells transfected with CMV and treated without (A, lanes 2, 3, 6 and 7) and with PMA (lanes 4, 5, 8 and 9). Supershift/immunodepletion binding reactions were performed in the presence of IgG (A, lanes 2, 4, 6 and 8) or c-Jun antibody (A, lanes 3, 5, 7 and 9). Small dotted box in (A) indicates specific DNA-protein complexes. Dotted box on the right indicates the same specific DNA-protein complexes formed under different conditions. In (B) EMSAs were performed on nuclear extracts from NIH-3T3 cells transfected with CMV (B, lanes 1-4) or PKCε (B, lanes 5-8) and treated without (B, lanes 1, 2, 5 and 6) and with PMA (lanes 3, 4, 7 and 8). Supershift/immunodepletion binding reactions were performed in the presence of IgG (B, lanes 1, 3, 5 and 7) or STAT3 antibody (B, lanes 2, 4, 6 and 8). Competition experiments were performed using nuclear extracts from NIH-3T3 cells transfected with CMV (C, lanes 1-5) or PKCε (C, lanes 6-9). Nuclear extracts were incubated with unlabeled AP1 oligonucleotide (wt) (C, lanes 2 & 7), AP1 mutant oligonucleotide (AP1 mut) (C, lanes 4 & 8), Ets mutant oligonucleotide (Ets mut) (C, lanes 5 & 9), or unrelated oligonucleotide (E) (C, lane 3). The free probe is indicated with an arrowhead. DNA-protein binding was optimized by including 10 μg BSA and 4 mM DTT in the all reactions. Results shown are representative of three independent experiments.
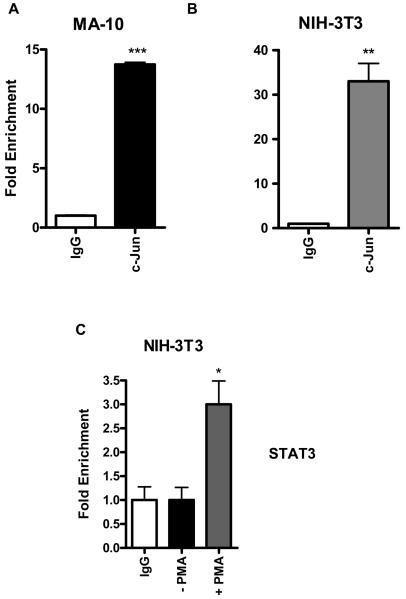
c-Jun is bound to the endogenous Tspo promoter, and PMA recruits STAT3 binding in NIH/3T3 cells
We next used a ChIP assay to determine whether STAT3 and c-Jun transcription factors are bound to the endogenous Tspo promoter in intact cells. Data shown in Figure 8A and B demonstrate that c-Jun binds to the endogenous promoter in both MA-10 and NIH-3T3 cells, whereas the IgG negative control does not. Based on this data and because a STAT3 binding site was identified in this sequence, which partially overlaps with the c-Jun and Ets sites that regulate the Tspo promoter in response to PMA (Figure 1), we examined whether STAT3 binds to the Tspo promoter by ChIP analysis. Figure 8C shows that PMA induced STAT3 binding to the Tspo promoter in NIH-3T3 cells, indicating that both STAT3 and c-Jun are important players in regulating Tspo gene expression.
Figure 8. c-Jun is bound to the endogenous Tspo promoter in intact MA-10, and PMA recruits STAT3 binding in NIH/3T3 cells.
DNA from MA-10 and NIH/3T3 cells was precipitated with antibodies specific for c-Jun (A & B) by ChIP analysis under basal conditions. NIH-3T3 cells were treated with or without 50 nM PMA for 24 h and DNA was precipitated with STAT3 specific antibodies (C) by ChIP analysis as described in materials and methods. DNA was amplified by QRT-PCR using primers specific for the distal area of the Tspo promoter spanning the binding sites for AP1, Ets and STAT3 shown in Figure 1. Normal rabbit IgG served as a negative control. Data are presented as fold enrichment of the ChIP antibody signal versus the negative control IgG using the ddCT method, and are derived from three independent experiments (n = 9). *p < 0.05, **p < 0.01 and ***p < 0.001.
DISCUSSION
The data presented in this manuscript implicate the MAPK (Raf-1/ERK2) signal transduction pathway as the downstream target of PKCε, responsible for mediating the effect of the kinase on Tspo gene promoter activity and expression. We also demonstrate that the PKCε-MAPK effect on TSPO expression is at least in part mediated by the c-Jun and STAT3 transcription factors. In previous studies, functional characterization of the Tspo promoter revealed the presence of two Sp1/Sp3 sites and two Ets sites in the proximal promoter; variable expression between the cell lines examined, as well as differences in the binding of these transcription factors to the Tspo promoter, were shown to result in differential regulation of the basal Tspo promoter activity between steroidogenic and non-steroidogenic cells (5;15). An AP1 site, adjacent to the second Ets site, was also identified in the Tspo promoter, and its role in regulating basal Tspo promoter activity was shown (16). We then investigated the molecular mechanisms governing Tspo regulation by exploring Tspo transcriptional responses to a tumor-promoting agent (PMA) in three cell lines with varying TSPO levels, and showed that only cell lines expressing low endogenous levels of TSPO were responsive to PMA, and that AP1 and the second Ets sites mediated this effect (16). The stimulatory effect of PMA in the low-expressing TSPO non-steroidogenic NIH-3T3 mouse fibroblasts was subsequently shown to be mediated by PKCε (16). PKCε was also found to be expressed at high levels in the steroidogenic MA-10 mouse tumor Leydig cells levels, where it maintained the high constitutive expression of TSPO in these cells (16). Thus, PKCε levels, activity, and targets seem to be the key drivers of Tspo promoter activity and protein expression.
PKCε signal transduction is complicated, and it is known to act through several cellular pathways such as NFκB, MAPK, p38, or by binding to RACK2. To identify the signaling pathway through which PMA and thus PKCε act on Tspo gene expression, we used pathway-specific inhibitors for the MAPK component MEK1/2, as well as specific inhibitors for the NFκB pathway. The MEK1/2 phosphorylation inhibitor U0126, which exerts its effect by blocking both new and existing MEK phoshophorylation and thus inhibiting MEK's ability to phosphorylate ERK1/2, completely abolished the activation of the Tspo promoter by PMA and reduced basal promoter activity in a dose-dependent manner. In contrast, both peptide and small molecule inhibitors of NFκB did not affect PMA-induced Tspo transcription, indicating the effect of PMA on Tspo promoter is NFκB-independent and is specific to the MEK1/2 signaling pathway.
We obtained contradictory findings using the MEK1/2-specific inhibitor PD898059, which unlike U0126 blocks only newly phosphorylated MEK, where a low dose (10 μM) induced Tspo promoter activity and mRNA levels and a high dose (100 μM) reduced Tspo mRNA levels. In search of the mechanisms underlying this biphasic effect of PD898058 we observed that high concentrations of the compound reduced a glutathione levels. However, although at 10 μM PD98059 did not affect glutathione levels, it altered the expression of 30 out of 84 genes involved in cellular oxidative stress defense. Taken together these results suggest that PD98059 at low doses acts as a weak inducer of oxidative stress altering the homeostasis of the cell antioxidant defense system and generating oxidative stress sufficient to drive Tspo expression (data not shown). This finding could be explained by the fact the PD898059 is a flavonoid previously shown to reduce antioxidant molecule levels in various cell types (23). Nevertheless, these data identified reactive oxygen species as a regulator of Tspo expression, a finding that will be investigated in detail in future studies.
Raf-1 was significantly phosphorylated at 24 h in NIH-3T3 cells transfected with PKCε and treated with PMA. On the other hand, PKCε increased ERK1/2 phosphorylation levels at 24 h. In these cells, PMA treatment of cells transfected with PKCε also increased STAT3 Tyr705 phosphorylation, suggesting that STAT3 may be a member of the transcriptional machinery regulating Tspo expression. Although ERK1/2 phosphorylation levels were induced with PMA in both empty vector- and PKCε-transfected cells, phosphorylation of ERK1/2 was significantly higher in PKCε-transfected cells; the presence of increased levels of PKCε were found to prolong ERK2 activation in agreement with previous findings in PNAC-1 cells (24). It is important to note that aberrant expression and/or activity of PKCε, MAPK/ERK1/2, AP1, STAT3, and TSPO have been linked to many diseases and cancers (21;25;26). Moreover, given the fact that STAT3 knockdown in vitro reduces TSPO levels, one could speculate that the STAT3-driven reduction in TSPO levels in vivo could be a contributing factor to the embryonic lethality seen in both STAT3 (27) and TSPO knockout mice (28).
Knockdown of Raf-1 and MEK1/2 with siRNAs significantly reduced TSPO mRNA and protein expression levels in MA-10 cells. MEK1/2 knockdown significantly reduced both basal and PMA-induced Tspo mRNA levels in NIH-3T3 cells. In addition, ERK2 shRNA significantly reduced TSPO levels in MA-10 cells, further confirming the role of the MAPK pathway in the regulation of TSPO expression.
Knockdown of c-Jun levels significantly reduced constitutive TSPO mRNA and protein levels in MA-10 cells, and both basal and PMA-induced Tspo mRNA levels in NIH-3T3 cells. This finding is in agreement with the previously proposed hypothesis that the effect of PKCε on TSPO expression is mediated at least in part through c-Jun (16). To confirm previous findings showing that PMA regulates c-Jun expression, in NIH-3T3 cells, we assessed c-Jun mRNA levels in cells treated with PMA. Interestingly, PMA induced new c-Jun synthesis in control cells treated with scrambled siRNA by 3.5-fold, an effect that was reduced in a dose-dependent manner by c-Jun and PKCε siRNA. Newly synthesized c-Jun may be compensating for the reduced c-Jun levels and thus allowing the induction of Tspo expression. A comparative study of c-Jun mRNA levels in MA-10 vs. NIH-3T3 cells showed that c-Jun mRNA levels in NIH-3T3 cells were approximately a third of those present in MA-10 cells, an observation that correlates well with the 3.5-fold induction of c-Jun mRNA levels in NIH-3T3 cells following PMA treatment. We previously reported that basal level of Tspo promoter activity in MA-10 cells is double to that found in NIH-3T3 cells. Following PMA treatment of NIH-3T3 cells Tspo promoter activity reach the basal levels found in MA-10 cells, an effect that could be mediated through c-Jun induction. Overexpression of c-Jun in NIH-3T3 cells along with Tspo promoter constructs was able to double the basal Tspo promoter activity and further potentiate the effect of PMA only when the AP1 binding site was intact, further confirming that this is the site through which c-Jun acts on Tspo promoter.
STAT3 knockdown by siRNA reduced Tspo mRNA and protein levels in MA-10 cells, as well as basal and PMA-induced Tspo mRNA levels in NIH-3T3 cells. We previously showed that PKCε levels correlate with TSPO levels, and much higher levels of both PKCε and TSPO are present in MA-10 cells compared to NIH-3T3 cells. Since PKCε has been linked to STAT3 regulation, and we demonstrated higher activation of STAT3 when overexpressing PKCε in NIH-3T3 cells, we investigated the effect of PKCε knockdown on STAT3 phosphorylation in MA-10 cells. Knockdown of PKCε in MA-10 cells reduced both TSPO expression and p-c-Jun levels at Ser73, a phosphorylation site important for c-Jun binding to DNA, (16) as well as STAT3 phosphorylation at Tyr705 and Ser727, in agreement with reports that PKCε is a regulator of STAT3 (21;22). To examine whether c-Jun and STAT3 bind to their DNA sequences in cells overexpressing PKCε, we performed EMSAs using NIH-3T3 nuclear extracts from cells transfected with PKCε and CMV, and examined the ability of c-Jun and STAT3 to shift/deplete the DNA-protein complexes formed. In agreement with previous data, c-Jun was able to efficiently deplete the complexes, while STAT3 was able to decrease complex formation in CMV/PMA treated cells as well as complex formation in PKCε transfected cells and PKCε/PMA treated cells. These data suggest that PKCε recruits STAT3 to the complex, an effect accomplished by treating with CMV transfected cells with PMA.
The specificity of the DNA-protein complexes formed was confirmed using a 200x abundant wt probe as well as ones containing mutations in the AP1 and Ets sites. The Ets mutated probe was more successful at competing the DNA-protein complex compared to AP1 mutated probe. This finding suggests two possible explanations: i. an intact Ets site is not important for the binding of proteins while an AP1 site is. ii. Due to the close proximity of the Ets and AP1 sites, an oligo containing a mutated Ets site, harbors an intact AP1 site allowing the binding of factors at the AP1 site and thus competing with the complex formation, whereas an oligonucleotide with a mutated AP1 site does not allow binding and is not as successful in depleting the complex.
To examine whether the effect of c-Jun and STAT3 on Tspo gene expression is direct, through promoter binding, or indirect, we performed a ChIP assay using primers spanning a 770 bp region of the Tspo promoter, where STAT3 and c-Jun factors are expected to bind. We previously identified both AP1 and Ets binding sites in the region of 585-515 bp. Interestingly, the complimentary strand to the two sites harbors a STAT3 binding site. Synergistic binding between c-Jun and STAT3 has been previously reported in the literature (29;30). This suggests that STAT3 and c-Jun could bind to the Tspo promoter at the same time, thus keeping the strands open and more accessible to transcription factors to induce gene expression. c-Jun has also been reported to recruit STAT3 to bind to the promoter, where c-Jun has been found to be present on the promoter before the arrival of phosphorylated STAT3 (31).We used a ChIP assay to test for binding by STAT3 and c-Jun to the Tspo promoter, and found that c-Jun indeed binds to the Tspo promoter in both MA-10 cells and NIH-3T3 cells. This finding confirms and extends previous data, where gel shift assays of PMA-treated NIH-3T3 cell extracts indicated increased binding of c-Jun to the Tspo promoter (16). Additional ChIP assays demonstrated that PMA induced binding of STAT3 to the Tspo promoter in NIH-3T3 cells. Because the Ras-MAPK pathway is not fully activated in NIH-3T3 cells (32), and this study demonstrated that PMA activation of Tspo promoter is mediated through a MAPK pathway, which also regulates STAT3 activity as a downstream target, we hypothesize that the increased activation of the MAPK pathway induced STAT3 activation and recruitment to the Tspo promoter. Indeed, a Ras/ERK1/2/STAT3 Ser727 pathway has been documented in literature (33). It is also possible that PMA induces changes of the activity of the Tspo promoter-bound proteins, allowing them to recruit additional factors and modifying the content of the factors binding to the AP-1 and Ets sites that span the STAT3 site.
In summary, the data presented herein show that a PKCε-dependent signal transduction pathway activating the MAPK (Raf-1/MEK1/2/ ERK2) pathway acts, at least in part, through c-Jun and STAT3 transcription factors to drive Tspo transcription and protein expression (Figure 9). This pathway is constitutively activated in steroidogenic MA-10 cells and induced by PMA in non-steroidogenic NIH-3T3 cells. Because differential expression of TSPO across the body has been associated with numerous physiological functions, ranging from steroidogenesis to mitochondrial function and cell proliferation, and changes in TSPO expression have been linked to various diseases, such as cancer and neurological disorders, understanding how Tspo expression is regulated in health and disease may prove beneficial for diagnostic, therapeutic, and prognostic purposes. Identifying factors and cofactors participating in the regulation of TSPO expression, and linking Tspo gene regulation to a specific signal transduction pathway, not only provides new insights into understanding the TSPO regulation in steroidogenic versus nonsteroidogenic cells, where the focus of our research lies, but also gives clues regarding the factors and mechanism that could regulate TSPO expression in disease states.
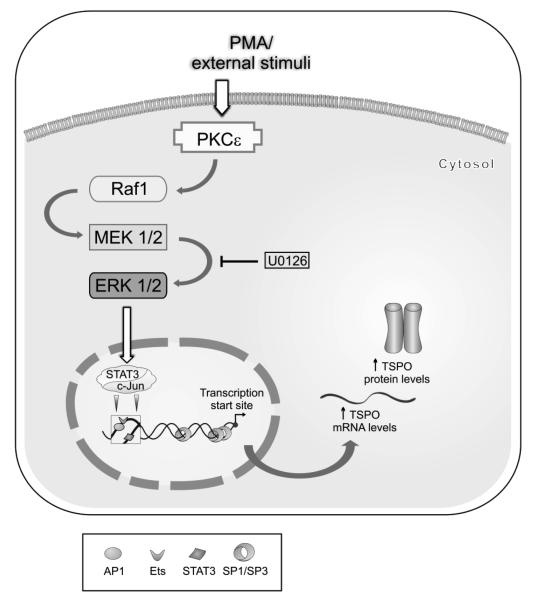
Figure 9. PKCε acts on the Tspo promoter through a MAPK (Raf-1 -ERK1/2) pathway and STAT3 and c-Jun transcription factors.
STAT3 has a binding site on the complimentary strand spanning the AP1 (a c-Jun binding site) and Ets binding sites.
ABBREVIATIONS
- AP1
activator protein 1
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- PKCε
protein kinase C epsilon
- PMA
phorbol-12-myristate 13-acetate
- Sp1
specificity protein 1/Sp1 transcription factor
- TSPO
translocator protein (18 kDa)
- STAT3
signal transducer and activator of transcription 3
Footnotes
This work was supported by Grant R01 ES07747 from the National Institutes of Health (to V.P.). V.P. was also supported by a Canada Research Chair in Biochemical Pharmacology. The Research Institute of MUHC is supported in part by a Center grant from Le Fonds de la recherche en santé du Québec.
REFERENCES
- 1.Casellas P, Galiegue S, Basile AS. Peripheral benzodiazepine receptors and mitochondrial function. Neurochem. Int. 2002;40:475–486. doi: 10.1016/s0197-0186(01)00118-8. [DOI] [PubMed] [Google Scholar]
- 2.Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacapere JJ, Lindemann P, Norenberg MD, Nutt D, Weizman A, Zhang MR, Gavish M. Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Braestrup C, Squires RF. Specific benzodiazepine receptors in rat brain characterized by high-affinity (3H)diazepam binding. Proc. Natl. Acad. Sci. U. S. A. 1977;74:3805–3809. doi: 10.1073/pnas.74.9.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gavish M, Bachman I, Shoukrun R, Katz Y, Veenman L, Weisinger G, Weizman A. Enigma of the peripheral benzodiazepine receptor. Pharmacol. Rev. 1999;51:629–650. [PubMed] [Google Scholar]
- 5.Giatzakis C, Papadopoulos V. Differential utilization of the promoter of peripheral-type benzodiazepine receptor by steroidogenic versus nonsteroidogenic cell lines and the role of Sp1 and Sp3 in the regulation of basal activity. Endocrinology. 2004;145:1113–1123. doi: 10.1210/en.2003-1330. [DOI] [PubMed] [Google Scholar]
- 6.Galiegue S, Casellas P, Kramar A, Tinel N, Simony-Lafontaine J. Immunohistochemical assessment of the peripheral benzodiazepine receptor in breast cancer and its relationship with survival. Clin. Cancer Res. 2004;10:2058–2064. doi: 10.1158/1078-0432.ccr-03-0988. [DOI] [PubMed] [Google Scholar]
- 7.Han Z, Slack RS, Li W, Papadopoulos V. Expression of peripheral benzodiazepine receptor (PBR) in human tumors: relationship to breast, colorectal, and prostate tumor progression. J. Recept. Signal. Transduct. Res. 2003;23:225–238. doi: 10.1081/rrs-120025210. [DOI] [PubMed] [Google Scholar]
- 8.Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res. 1999;59:831–842. [PubMed] [Google Scholar]
- 9.Katz Y, Ben-Baruch G, Kloog Y, Menczer J, Gavish M. Increased density of peripheral benzodiazepine-binding sites in ovarian carcinomas as compared with benign ovarian tumours and normal ovaries. Clin. Sci. (Lond) 1990;78:155–158. doi: 10.1042/cs0780155. [DOI] [PubMed] [Google Scholar]
- 10.Maaser K, Grabowski P, Sutter AP, Hopfner M, Foss HD, Stein H, Berger G, Gavish M, Zeitz M, Scherubl H. Overexpression of the peripheral benzodiazepine receptor is a relevant prognostic factor in stage III colorectal cancer. Clin. Cancer Res. 2002;8:3205–3209. [PubMed] [Google Scholar]
- 11.Papadopoulos V, Lecanu L. Translocator protein (18 kDa) TSPO: an emerging therapeutic target in neurotrauma. Exp. Neurol. 2009;219:53–57. doi: 10.1016/j.expneurol.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amri H, Drieu K, Papadopoulos V. Transcriptional suppression of the adrenal cortical peripheral-type benzodiazepine receptor gene and inhibition of steroid synthesis by ginkgolide B. Biochem. Pharmacol. 2003;65:717–729. doi: 10.1016/s0006-2952(02)01603-9. [DOI] [PubMed] [Google Scholar]
- 13.Gazouli M, Yao ZX, Boujrad N, Corton JC, Culty M, Papadopoulos V. Effect of peroxisome proliferators on Leydig cell peripheral-type benzodiazepine receptor gene expression, hormone-stimulated cholesterol transport, and steroidogenesis: role of the peroxisome proliferator-activator receptor alpha. Endocrinology. 2002;143:2571–2583. doi: 10.1210/endo.143.7.8895. [DOI] [PubMed] [Google Scholar]
- 14.Veenman L, Papadopoulos V, Gavish M. Channel-like functions of the 18-kDa translocator protein (TSPO): regulation of apoptosis and steroidogenesis as part of the host-defense response. Curr. Pharm. Des. 2007;13:2385–2405. doi: 10.2174/138161207781368710. [DOI] [PubMed] [Google Scholar]
- 15.Giatzakis C, Batarseh A, Dettin L, Papadopoulos V. The role of Ets transcription factors in the basal transcription of the translocator protein (18 kDa) Biochemistry. 2007;46:4763–4774. doi: 10.1021/bi062208o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batarseh A, Giatzakis C, Papadopoulos V. Phorbol-12-myristate 13-acetate acting through protein kinase Cepsilon induces translocator protein (18-kDa) TSPO gene expression. Biochemistry. 2008;47:12886–12899. doi: 10.1021/bi8012643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Rocha AB, Mans DR, Regner A, Schwartsmann G. Targeting protein kinase C: new therapeutic opportunities against high-grade malignant gliomas? Oncologist. 2002;7:17–33. doi: 10.1634/theoncologist.7-1-17. [DOI] [PubMed] [Google Scholar]
- 18.Garczarczyk D, Toton E, Biedermann V, Rosivatz E, Rechfeld F, Rybczynska M, Hofmann J. Signal transduction of constitutively active protein kinase C epsilon. Cellular Signalling. 2009;21:745–752. doi: 10.1016/j.cellsig.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 20.Ono Y, Fujii T, Ogita K, Kikkawa U, Igarashi K, Nishizuka Y. The structure, expression, and properties of additional members of the protein kinase C family. J. Biol. Chem. 1988;263:6927–6932. [PubMed] [Google Scholar]
- 21.Aziz MH, Manoharan HT, Church DR, Dreckschmidt NE, Zhong W, Oberley TD, Wilding G, Verma AK. Protein kinase Cepsilon interacts with signal transducers and activators of transcription 3 (Stat3), phosphorylates Stat3Ser727, and regulates its constitutive activation in prostate cancer. Cancer Res. 2007;67:8828–8838. doi: 10.1158/0008-5472.CAN-07-1604. [DOI] [PubMed] [Google Scholar]
- 22.Aziz MH, Manoharan HT, Sand JM, Verma AK. Protein kinase Cepsilon interacts with Stat3 and regulates its activation that is essential for the development of skin cancer. Mol. Carcinog. 2007;46:646–653. doi: 10.1002/mc.20356. [DOI] [PubMed] [Google Scholar]
- 23.Kirkland RA, Franklin JL. Prooxidant Effects of NGF Withdrawal and MEK Inhibition in Sympathetic Neurons. Antioxidants & Redox Signaling. 2003;5:635–639. doi: 10.1089/152308603770310301. [DOI] [PubMed] [Google Scholar]
- 24.Racz GZ, Szucs A, Szlavik V, Vag J, Burghardt B, Elliott AC, Varga G. Possible role of duration of PKC-induced ERK activation in the effects of agonists and phorbol esters on DNA synthesis in Panc-1 cells. J. Cell Biochem. 2006;98:1667–1680. doi: 10.1002/jcb.20913. [DOI] [PubMed] [Google Scholar]
- 25.Imler JL, Wasylyk B. AP1, a composite transcription factor implicated in abnormal growth control. Prog. Growth Factor Res. 1989;1:69–77. doi: 10.1016/0955-2235(89)90001-x. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence MC, Jivan A, Shao C, Duan L, Goad D, Zaganjor E, Osborne J, McGlynn K, Stippec S, Earnest S, Chen W, Cobb MH. The roles of MAPKs in disease. Cell Res. 18:436–442. doi: 10.1038/cr.2008.37. 0 AD. [DOI] [PubMed] [Google Scholar]
- 27.Akira S. Functional roles of STAT family proteins: lessons from knockout mice. Stem Cells. 1999;17:138–146. doi: 10.1002/stem.170138. [DOI] [PubMed] [Google Scholar]
- 28.Papadopoulos V, Amri H, Boujrad N, Cascio C, Culty M, Garnier M, Hardwick M, Li H, Vidic B, Brown AS, Reversa JL, Bernassau JM, Drieu K. Peripheral benzodiazepine receptor in cholesterol transport and steroidogenesis. Steroids. 1997;62:21–28. doi: 10.1016/s0039-128x(96)00154-7. [DOI] [PubMed] [Google Scholar]
- 29.Yoo JY, Wang W, Desiderio S, Nathans D. Synergistic Activity of STAT3 and c-Jun at a Specific Array of DNA Elements in the +¦2-Macroglobulin Promoter. Journal of Biological Chemistry. 2001;276:26421–26429. doi: 10.1074/jbc.M009935200. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Wrzeszczynska MH, Horvath CM, Darnell JE., Jr. Interacting Regions in Stat3 and c-Jun That Participate in Cooperative Transcriptional Activation. Mol. Cell. Biol. 1999;19:7138–7146. doi: 10.1128/mcb.19.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginsberg M, Czeko E, Muller P, Ren Z, Chen X, Darnell JE., Jr. Amino acid residues required for physical and cooperative transcriptional interaction of STAT3 and AP-1 proteins c-Jun and c-Fos. Mol. Cell Biol. 2007;27:6300–6308. doi: 10.1128/MCB.00613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh D, Ezashi T, Ostrowski MC, Roberts RM. A central role for Ets-2 in the transcriptional regulation and cyclic adenosine 5′-monophosphate responsiveness of the human chorionic gonadotropin-beta subunit gene. Mol. Endocrinol. 2003;17:11–26. doi: 10.1210/me.2002-0223. [DOI] [PubMed] [Google Scholar]
- 33.Plaza-Menacho I, van der ST, Hollema H, Gimm O, Buys CH, Magee AI, Isacke CM, Hofstra RM, Eggen BJ. Ras/ERK1/2-mediated STAT3 Ser727 phosphorylation by familial medullary thyroid carcinoma-associated RET mutants induces full activation of STAT3 and is required for c-fos promoter activation, cell mitogenicity, and transformation. J Biol. Chem. 2007;282:6415–6424. doi: 10.1074/jbc.M608952200. [DOI] [PubMed] [Google Scholar]