Abstract
Studies using Nicotiana benthamiana protoplasts have determined that repression of upstream transcription by AL1 protein enhances AL2 and AL3 expression in Tomato golden mosaic virus (TGMV). Mutations resulting in the inability of TGMV AL1 protein to associate with its cognate binding site, result in a decrease in both AL2 and AL3 expression. Reduced expression correlates with an increase in transcription from the AL62 start site, and decreased transcription from downstream initiation sites (AL1935 and AL1629) present within the AL1 coding region. The results demonstrate that, in a tobacco protoplast system, repression of AL62 transcription, regulated through binding of AL1 protein to sequences in the origin of replication, is required prior to AL2 and AL3 gene expression from the AL1935 and AL1629 viral transcripts. This provides a mechanism to regulate expression of AL2, which is involved in suppression of host defense responses and is required for late gene expression.
Keywords: Geminivirus, AL1, autoregulation, binding, ori
Introduction
Geminiviruses are a family of single-stranded DNA viruses that replicate in the nuclei of infected plant cells, generating double-stranded DNA replicative (RF) intermediates (Stenger et al., 1991), as templates for viral transcription and further rounds of replication. These processes rely on cellular polymerases, making geminiviruses a valuable model system for the study of transcription and replication in plants (Bisaro, 1996; Hanley-Bowdoin et al., 1999).
Tomato golden mosaic virus (TGMV) belongs to the genus Begomovirus, with two DNA components (A and B), both of which are required for infectivity (Hamilton et al., 1983). DNA A encodes functions required for replication (AL1 and AL3) and encapsidation (AL2 and AR1) of the virus (Rogers et al., 1986; Sunter et al., 1987). Genes on the B component (BR1 and BL1) encode functions for viral movement (Brough et al., 1988; Jeffrey et al. 1996). A 5′ intergenic region (IR) of ~230 bp (conserved in both components of TGMV) separates divergent coding regions, and contains elements mediating bi-directional transcription (Fontes et al., 1994; Hanley-Bowdoin et al., 1990; Petty et al., 1988; Sunter et al., 1989; Sunter et al., 1993). The viral sense transcription unit of TGMV comprises a single RNA species spanning a single gene (AR1, coat protein; BR1, movement protein). The complementary sense transcription unit is more complex, consisting of multiple overlapping RNAs with different 5′ ends, all of which are 3′ co-terminal (Fig. 1; Hanley-Bowdoin et al., 1988; Sunter and Bisaro, 1989). The only RNA capable of producing a functional AL1 protein initiates at nucleotide 62 (AL62), but this RNA also has the potential to code for the AL2 and AL3 proteins. Recent data indicates two smaller RNAs initiating at nucleotides 1935 and 1629 (AL1935, AL1629), can express AL3, but AL2 appears to be expressed only from AL1629 (Shung et al., 2006).
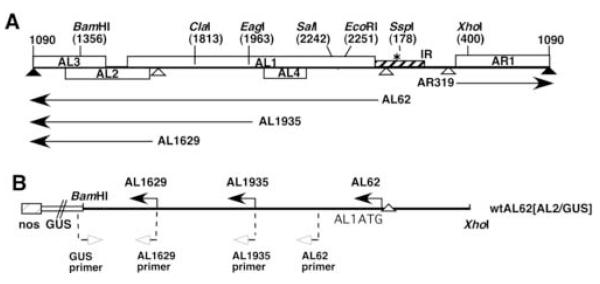
Fig. 1.
Genome organization of TGMV DNA A.
(A) The linear map illustrates the wild type TGMV DNA A genome component, indicating relevant restriction sites with nucleotide coordinates given in parentheses. Numbering is according to the TGMV sequence determined by Hamilton et al. (1984). Open boxes indicate coding regions and arrows designate relevant transcripts and direction of transcription, with initiation site indicated. The hatched box represents the ~230 bp intergenic region (IR), solid triangles represent polyadenylation signals, and open triangles indicate TATA boxes.
(B) A derivative of TGMV DNA A is illustrated, containing 5′ flanking sequences of the TGMV AL2 or AL3 coding region cloned as a translational fusion with the GUS reporter gene (open bar) using the BamHI site. The nopaline synthase (nos) 3′ polyadenylation signal, and relevant transcription initiation sites are indicated. The position of primers used in RT-PCR experiments is indicated with open arrows. Diagrams are not drawn to size.
TGMV conforms to the general strategy of DNA virus transcription where early gene products activate expression of viral late genes. For example, the TGMV AL1 protein negatively regulates expression from its own promoter by binding to sequences between the transcription start site for AL1 (AL62) and a consensus RNA polymerase II TATAA sequence within the IR (Eagle et al., 1994; Sunter et al., 1993). Repression of the AL1 promoter is not dependent on replication, as mutations in the AL1 protein that are deficient for DNA replication have no effect on repression (Eagle et al., 1994). In addition to AL1, the TGMV AL4 gene product is also capable of repressing the AL62 promoter, but mediates its effect through a unique sequence that does not overlap the AL1 binding site (Eagle and Hanley-Bowdoin, 1997). However, no role for AL4 during TGMV infection has been found, and the protein has not been detected in infected plants. The presence of sequence elements important for expression of AL1935 and AL1629 within the AL1 coding region (Shung et al., 2006) makes it likely that transcription from AL62 would be inhibitory to transcription from these downstream initiation sites. In this article, we study the effects of repression of AL62 transcription on downstream gene expression. Our results suggest that AL2 and AL3 gene expression are dependent on repression of AL62 transcription mediated through binding of AL1 to its cognate binding site. The results also indicate that AL4 plays some role in regulating AL2 or AL3 gene expression through repression. We discuss the possible role of repression in the life cycle of TGMV in particular, and geminiviruses in general.
Results
TGMV AL2 gene expression is dependent on the AL1 gene product
The presence of transcription initiation sites within the AL1 ORF (Fig. 1) suggests that upstream transcription could inhibit assembly of transcription complexes at these sites (Shung et al., 2006). Previous studies have demonstrated that the interaction between AL1 and its DNA binding site represses AL62 transcription (Eagle et al., 1994; Sunter et al., 1993). We therefore tested whether AL2 expression is dependent on this repression, using a 5′-truncated promoter linked to the β-glucuronidase (GUS) reporter in a translational fusion to the AL2 protein (Fig. 1B). This construct contains a deletion end-point at −1391, relative to the translation start site for AL2, and has been previously shown to direct AL2 expression (Shung et al., 2006). Two restriction sites were introduced into AL2[−1391]-GUS for cloning purposes (Table 1), generating wtAL62[AL2/GUS]. Neither change affects AL1 expression or function (Elmer et al., 1988; Hanley-Bowdoin et al., 1990), and AL2 expression is comparable to that observed for AL2[−1391]-GUS (data not shown).
Table 1.
Wild type and mutant TGMV sequences used in promoter-reporter assays.
| Nucleotide Coordinatesa |
Nucleotide Sequenceb |
Amino Acid Sequencec |
|
|---|---|---|---|
| WT AL1 binding site | 84-72 | 5′- CTACCTTACTACC-3′ | N/A |
| Mutant AL1 binding site | 84-72 | 5′- CTAggTTACTAgg-3′ | N/A |
| WT AL1 protein | 13-5 | 5′-1ATGCCATCG-3′ | MPS |
| Mutant AL1 protein | 13-5 | 5′- ATcCCATCG-3′ | N/A |
| WT AL1 protein | 2295-2287 | 5′-ACG103TACATC-3′ | TYI |
| Y103A mutant AL1 | 2295-2287 | 5′- ACGgcgATC-3′ | TAI |
| WT AL4 protein | 2447-2439 | 5′- ATGAAGATG-3′ | MKM |
| Mutant AL4 protein | 2447-2439 | 5′- AcGAAGAcG-3′ | N/A |
Nucleotide Coordinates are given according to the sequence of Hamilton et al. (1984).
The sequence is given in the 5′ to 3′ direction on the complementary strand. Viral sequences that have been mutated are indicated in lower case letters and the corresponding wild type sequence is underlined. Numbers within a sequence indicates the amino acid residue in the wild type protein.
The amino acid sequence of the predicted protein product is shown. N/A indicates no protein is predicted.
To analyze the effects of AL1 binding on AL2 expression, mutations (Table 1) were introduced into the AL1 translation initiation codon (mAL62[AL2/GUS]) or the AL1 DNA binding site (AL1bs−[AL2/GUS]). Mutation of the AL1 binding site has previously been shown to abolish binding of AL1 protein to DNA (Eagle et al., 1994). Constructs were transfected into protoplasts prepared from Nicotiana benthamiana suspension cells, extracts prepared three days post-transfection, and fluorometric GUS assays performed as described (Shung et al., 2006). A construct that generates replicating TGMV DNA A, lacking the GUS reporter gene (pTGA26), served as a background control. As shown (Fig. 2), significantly greater activity than background (Student’s t-test: P<0.05) was detected in extracts from protoplasts transfected with a construct containing wild type TGMV sequences (wtAL62[AL2/GUS] + pUC). A significant reduction in activity of three to four-fold (Student’s t-test: P<0.05), was detected with introduction of a mutation into either, the AL1 translation initiation codon (mAL62[AL2/GUS] + pUC), or into the AL1 DNA binding site (AL1bs−[AL2/GUS] + pUC). These results suggest that loss of AL1 protein, or loss of AL1 binding to viral DNA, impairs expression of AL2. Cloned DNA containing the mutation in the AL1 DNA binding site (AL1bs−[AL2/GUS]) can complement a mutation in the AL1 coding region (pTGA71; Brough et al., 1992b), which demonstrates that this DNA is capable of producing functional AL1 protein (data not shown). This therefore suggests that loss of AL2 expression is not due to the inability of the DNA template to produce AL1.
Fig. 2.
A functional AL1 binding site and AL1 protein are required for AL2 expression. The ability of promoter-reporter constructs containing mutations within the AL1 initiator codon (mAL62[AL2/GUS]) or AL1 binding site (AL1bs−[AL2/GUS]) to direct expression of AL2 was determined by comparing GUS activity to background fluorescence (pTGA26; ~3% activity). Protoplasts (5 × 105 cells) were transfected with 10 μg of a promoter-reporter construct with either pUC or pTGA73 DNA, and GUS activity measured in extracts isolated three days post-transfection. Columns represent mean relative GUS activity as compared to wtAL62[AL2/GUS], which was arbitrarily assigned a value of 100. Error bars represent the standard error of the mean from at least three independent experiments.
The reduction in AL2 expression observed with mutant DNA templates was not a consequence of template replication. Restriction patterns using DpnI and MboI that differentially cleave TGMV DNA isolated from eukaryotic (protoplasts) and prokaryotic (E.coli) cells (Brough et al., 1992a), demonstrated that template DNA isolated from protoplasts retained bacterial methylation (data not shown).
Functional AL1 protein can complement the loss of AL2 gene expression in an AL1 translation initiation codon mutant but not an AL1 binding site mutant
To test if functional AL1 can restore AL2 expression, protoplasts were transfected with wild type or mutant DNA in the presence of cloned DNA capable of expressing AL1 from the Cauliflower mosaic virus (CaMV) 35S promoter (pTGA73; Sunter et al., 1993). An increase in GUS activity (Student’s t-test: P<0.05) of ~ 2.5-fold was detected in extracts from protoplasts co-transfected with DNA containing a mutation in the AL1 initiator codon (mAL62[AL2/GUS]) and pTGA73 (Fig. 2). This indicates that the addition of exogenous AL1 protein in trans can complement a mutation in the translation initiation codon of AL1. Activity did not reach wild-type levels, probably as a consequence of two-hit kinetics involved in co-transfection experiments. In contrast, the presence of cloned DNA capable of expressing AL1 did not increase activity in extracts from protoplasts co-transfected with DNA containing a mutation in the AL1 binding site (Fig. 2), as anticipated. Taken together the results demonstrate that the presence of a functional AL1 protein is required for AL2 gene expression when a wild-type DNA binding site for AL1 is present within the template DNA.
Mutation in the AL1 protein results in loss of AL2 gene expression
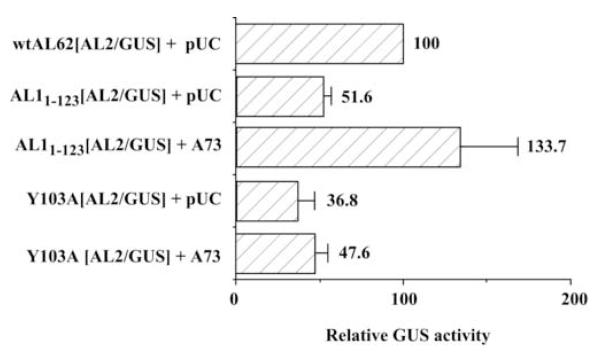
An alternative approach to prevent autoregulation of AL62 transcription was to introduce mutations within the AL1 protein itself. We therefore tested two additional constructs to assess the consequences of constitutive AL62 transcription on AL2 gene expression. A mutation in the SalI restriction site at nucleotide 2242 results in a truncated protein consisting of the N-terminal 123 amino acids of AL1. This mutation is known to abolish viral replication (Elmer et al., 1988). A second construct (Table 1) contains a single amino acid substitution at residue 103 (Y103A), which is known to abolish the ability of AL1 to bind DNA (Orozco and Hanley-Bowdoin, 1998). As can be seen (Fig. 3), GUS activity in extracts from protoplasts transfected with a DNA template containing the mutation at the SalI site (AL11-123[AL2/GUS]) is reduced ~ two-fold (Student’s t-test: P<0.05) relative to a wild type DNA construct (wtAL62[AL2/GUS]). However, the reduction in expression is not as dramatic as seen with the AL1 initiator codon mutant (Fig. 2). One possible explanation is that although transcriptional autoregulation by AL1 is primarily mediated through amino acids 1-93, amino acids 121-209 also make a contribution (Gladfelter et al., 1997). It is therefore possible that the truncated AL1 protein interacts with the binding site less efficiently than wild type protein.
Fig. 3.
Functional AL1 protein is required for AL2 expression. The ability of promoter-reporter constructs containing mutations within the AL1 coding region (AL1 1-123 and Y103A) to direct AL2 expression was determined by comparing GUS activity to background (pTGA26; ~7% activity). Protoplasts (5 × 105 cells) were transfected with 10 μg of a promoter-reporter construct with either pUC or pTGA73, and GUS activity measured in extracts isolated three days post-transfection. Columns represent mean relative GUS activity as compared to wtAL62[AL2/GUS], which was arbitrarily assigned a value of 100. Error bars represent the standard error of the mean from three independent experiments.
A single amino acid substitution (Y103A) results in a reduction in GUS activity (Student’s t-test: P<0.05) of ~ three-fold (Fig. 3), similar to that observed with the AL1 initiator codon mutant (Fig. 2). AL1 protein containing this mutation is unable to bind DNA (Orozco and Hanley-Bowdoin, 1998), or repress AL62 transcription. These results are therefore consistent with our finding that binding of AL1 to its cognate binding site is necessary for AL2 gene expression.
As observed earlier, when functional AL1 protein is provided in trans, GUS activity increases (Student’s t-test: P<0.05) from a DNA template containing the mutation at the SalI site (Fig. 3), probably as a consequence of AL1 binding, leading to repression of AL62 transcription. Surprisingly, the addition of exogenous AL1 did not significantly increase GUS activity (Student’s t-test; P<0.05) from a DNA template containing the Y103A mutation (Fig. 3). One possible explanation for this result is that AL1 protein containing the Y103A mutation could be acting as a dominant negative mutant as oligomerization of the AL1 protein is important for binding DNA (Orozco and Hanley-Bowdoin, 1998). The mutant AL1 protein contains the domain necessary for the formation of oligomers, and could therefore interact with the exogenous wild type AL1 protein to form inactive complexes that are unable to bind DNA. In fact, AL1 protein containing the Y103A mutation is capable of interacting with wild type AL1 in a yeast two-hybrid assay (data not shown).
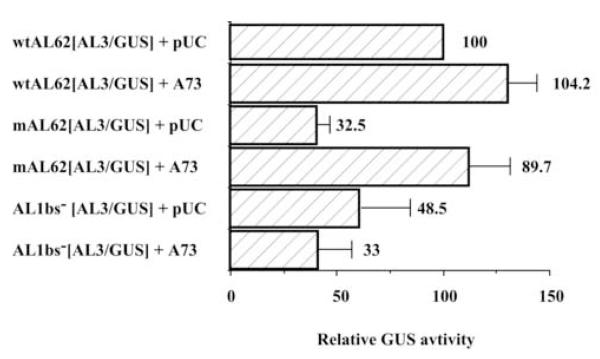
TGMV AL3 gene expression is also dependent on the TGMV AL1 gene product
Based on our hypothesis that repression of AL62 transcription is necessary for downstream gene expression, we performed similar experiments to those described above to test whether repression would affect TGMV AL3 gene expression. As shown (Fig. 4), AL3/GUS expression was reduced (Student’s t-test: P<0.05) ~ three-fold when a mutation was introduced into the AL1 initiator codon (mAL62[AL3/GUS]) and approximately two-fold when a mutation was introduced into the AL1 binding site (AL1bs−[AL3/GUS]). As observed for AL2/GUS templates, when functional AL1 protein is provided in trans, GUS activity increases (Student’s t-test: P<0.05) from a DNA template containing the mutation in the AL1 initiator codon, but not from a DNA template containing the mutation in AL1 binding site (Fig. 4).
Fig. 4.
A functional AL1 binding site and AL1 protein are required for AL3 expression. The ability of promoter-reporter constructs containing mutations within the AL1 initiator codon (mAL62[AL3/GUS]) or AL1 binding site (AL1bs−[AL3/GUS]) to direct expression of AL3 was determined by comparing GUS activity to background (pTGA26; <1% activity). Protoplasts (5 × 105 cells) were transfected with 10 μg of a promoter-reporter construct with either pUC or pTGA73 DNA, and significant differences in GUS activity (Student’s t-test: P<0.05) measured in extracts isolated three days post-transfection. Columns represent mean relative GUS activity as compared to wtAL62[AL3/GUS], which was arbitrarily assigned a value of 100. Error bars represent the standard error of the mean from at least three independent experiments.
Reduced AL62 promoter activity negates the requirement for repression by AL1 protein
To test the hypothesis that AL2 gene expression depends upon a reduction in transcription from the AL62 initiation site, sequences upstream of the SspI site in the conserved stem-loop region (Fig. 1A) were deleted (mAL1-SspΔ[AL2/GUS]). Deletion of these sequences has been shown to reduce AL62 promoter activity (Student’s t-test: P<0.05) by ~ three-fold relative to wild type (Eagle et al., 1994). As seen in Fig. 5, deletion of sequences upstream of the SspI site does not result in reduction of AL2 expression, as compared to wtAL62[AL2/GUS], and may in fact lead to a slight increase in AL2 expression, although this is not statistically significant (Student’s t-test: P<0.05). Interestingly, mutation of the AL1 translation initiation codon in this background (mAL1-SspΔ[AL2/GUS], has little effect on AL2 expression (Fig. 5). This is in contrast to the three to four-fold reduction in AL2 expression observed when this mutation is introduced into a wild type background (mAL1-[AL2/GUS]). Similar results were obtained for these mutations in an AL3/GUS background (data not shown). This suggests that reducing AL62 promoter activity removes the requirement for a functional AL1 protein to down-regulate AL62 transcription. In each case the addition of exogenous AL1 protein from pTGA73 had no effect on AL2 expression (Fig. 5). The data is again consistent with our hypothesis that downstream gene expression is dependent on repression of AL62 transcription.
Fig. 5.
Reduced AL62 promoter activity negates the requirement for repression by AL1 protein. The ability of promoter-reporter constructs containing a deletion at the SspI site within the conserved stem-loop to direct AL2 expression was determined by comparing GUS activity to background (pTGA26; ~5% activity). Protoplasts (5 × 105 cells) were transfected with 10 μg of a promoter-reporter construct and significant differences in GUS activity (Student’s t-test: P<0.05) measured in extracts isolated three days post-transfection. Columns represent mean relative GUS activity as compared to wtAL62[AL2/GUS], which was arbitrarily assigned a value of 100. Error bars represent the standard error of the mean from six independent experiments.
Mutation of the AL1 translation initiation codon or AL1 binding site results in decreased transcription of the AL1935 and AL1629 transcripts
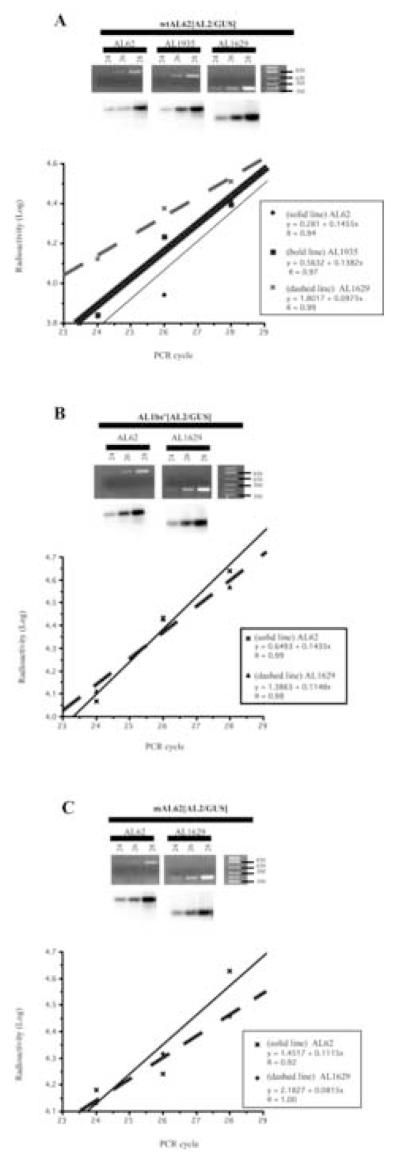
The experiments described above demonstrate that AL1 protein and a functional DNA binding site are required for AL2 and AL3 expression. To directly test whether this is a consequence of reduced AL62 transcription, we used RT-PCR to measure RNA levels corresponding to AL62, AL1935 and AL1629, the latter of which has been shown to be the only RNA capable of expressing AL2 (Shung et al., 2006). Constructs containing wild type sequences (wtAL62[AL2/GUS]), and mutations in the AL1 translation initiation codon (mAL62[AL2/GUS]), or AL1 DNA binding site (AL1bs−[AL2/GUS]), were cloned into pMON521. Cloned DNAs directed expression of AL2 in protoplasts, as determined by GUS assay, in a manner identical to that observed in the pUC-based vectors (data not shown). Agrobacterium cultures containing each construct were used to infuse N.benthamiana leaves (Sunter and Bisaro, 1989). Initial experiments indicated that three-days post-infusion was optimal for isolation of RNA (data not shown). We were unable to detect viral RNAs by Northern blotting (data not shown), even using poly (A)+ RNA, presumably due to the low level of expression from these templates as reported previously (Shung et al., 2006). We therefore used a semi-quantitative RT-PCR approach with nested primer pairs, to estimate the relative steady-state RNA levels of TGMV AL62, AL1935 and AL1629 RNAs, which overlap extensively (Fig. 1A). A common primer that anneals within the GUS reporter sequence was used in conjunction with a primer that would specifically amplify a fragment from RNA transcribed from initiation at either nt 62, nt 1935 or nt 1629 (Fig. 1B). Subsequent hybridization to a probe that would anneal to sequences contained within all three RNAs allowed us to obtain a relative ratio of complementary sense RNAs within any given sample. RNA amounts were first normalized by comparison to EF1α RNA levels using phosphorimager analysis (Shung et al., 2006). Linear regression analysis of the amount of RT-PCR amplified product versus the number of PCR cycles (Fig. 6) defines the linear range (Lee et al., 1996) and demonstrates that the primers amplify AL62, AL1935 and AL1629 cDNAs with similar efficiencies. The ratio of mRNAs in each sample was then determined by direct comparison of the levels of cDNA generated at a given cycle within the linear range of amplification. Products of the predicted size for RNA derived from AL62 (800 bp), AL1935 (700 bp) and AL1629 (400 bp) were detected in RNA isolated from leaves infused with all three constructs (Fig. 6). Phosphorimager analysis allows comparison of radioactive signals using the linear regression formula for each slope. As the primers for AL1629 would also amplify a product from AL62 and AL1935, this signal is comprised of cDNA product derived from all three RNAs. By subtracting the amount of signal detected for AL62, we can estimate that the residual signal is a consequence of amplification from both AL1935 and AL1629. Subtraction of the amount of signal detected for AL1935 from that detected for AL62 gives us an estimate of the signal resulting from amplification from AL1935 alone. Subtraction of this value from the value obtained for AL1629 and AL1935 provides an estimate of the signal derived from amplification of AL1629. The results obtained show that in leaves infused with a wild type DNA template, steady state TGMV complementary sense RNAs comprise an average of approximately 60% AL62, 6% AL1935 and 34% AL1629 (Fig. 6 and Table 2).. In the two experiments the level of AL1629 varied from 24 to 45% of the viral RNA detected which could reflect variation from the relatively asynchronous nature of the Agrobacterium infusion system. In both experiments the signal derived from AL1935 was difficult to detect, which is consistent with previous results that demonstrated transcription of AL1935 is low from these templates (Shung et al., 2006).
Fig. 6.
Expression of TGMV complementary sense RNAs from promoter-reporter constructs. The panels illustrate semi-quantitative RT-PCR analysis of TGMV AL62, AL1935 and AL1629 transcripts in leaves infused with wtAL62[AL2/GUS] (A), AL1bs−[AL2/GUS] (B) and mAL62[AL2/GUS] (C), after 24, 26, and 28 cycles of amplification. The top panel represents an ethidium bromide-stained gel of PCR products, and the bottom panel represents an autoradiogram of samples hybridized to a TGMV-specific probe. Marker DNA fragments are indicated in base pairs (bp). Semi-log plots of radioactivity (phosphorimager units) versus cycle number for the PCR fragments illustrated in A, B and C are shown. The linear regression equation for each plot is shown in the inset.
Table 2.
Semi-quantitative RT-PCR amplification of TGMV RNAs from infused leaf tissue.
| DNA template | ||||
|---|---|---|---|---|
| wtAL62[AL2/GUS] | AL1bs-[AL2/GUS] | mAL62[AL2/GUS] | ||
| AL1629 + GUSa | 1 | 13858 | 13849 | 13744 |
| 2 | 42364 | 26528 | N/D | |
| AL1935 + GUSb | 1 | 7586 | N/D | N/D |
| 2 | N/D | N/D | N/D | |
| AL62 + GUSc | 1 | 5929 (43%) | 12429 (90%) | 13434 (98%) |
| 2 | 30775 (76%) | 25604 (96%) | 50968 (100%) | |
| AL1935d | 1 | 1657 (12%) | N/D | N/D |
| 2 | N/D | N/D | N/D | |
| AL1629e | 1 | 6272 (45%) | 1420 (10%) | 310 (2%) |
| 2 | 11589 (24%) | 924 (4%) | 0 (0%) | |
| Meanf | AL62 | 60% (43-76%) | 93% (90-96%) | 99% (98-100%) |
| AL1935 | 6% (0-12%) | N/D | N/D | |
| AL1629 | 34% (24-45%) | 7% (4-10%) | 1% (0-2%) | |
Based on the linear regression equation for the plots shown in Figure 6, the signal intensity was determined at ether cycle 24 (Expt. 1) or cycle 28 (Expt. 2) by phosphorimager analysis, for each amplification product. The percentage of the total signal detected for AL62, AL1935 and AL1629 within each sample is given in parentheses. N/D = not detected.
Signal detected from RT-PCR products derived from AL62, AL1935 and AL1629.
Signal detected from RT-PCR products derived from AL62 and AL1935.
Signal detected from RT-PCR products derived from AL62 alone.
Signal detected from RT-PCR products derived from AL1935 alone was calculated by subtracting the signal for AL62 alone (AL62 + GUS) from the signal for AL62 and AL1935 combined (AL1935 + GUS).
Signal detected from RT-PCR products derived from AL1629 alone was calculated by subtracting the signal for AL62 and AL1935 combined (AL1935 + GUS) from the signal for AL62, AL1935 and AL1629 combined (AL1629 + GUS).
The mean value for the two experiments is given with the range in parentheses.
Comparison of signal intensities of RT-PCR products from leaves containing mutant DNA templates, suggests that AL1935 or AL1629 represent 10% or less of the total TGMV RNA detected, as signal intensities for AL62 and AL1629 are approximately equivalent (Fig. 6 and Table 2). This suggests that mutation of the AL1 initiator codon or AL1 binding site, leads to significant reduction in downstream transcription, consistent with results obtained for promoter-reporter analysis. Further support for this conclusion is provided when the relative levels of RT-PCR product derived from AL62 are directly compared. No product was detected when the reverse transcriptase was omitted indicating the absence of contaminating DNA template (data not shown).
AL4 contributes to AL2 and AL3 gene expression
Based on our hypothesis that AL1 binding to the ori is required for repression, we would predict that AL2 and AL3 expression should approach background in DNA templates containing mutations in the AL1 translation initiation codon (Fig. 2). However, we did not observe this, and one possible explanation is the presence of AL4 in the constructs used, which contributes to repression of AL1 through a unique DNA sequence within the common region between genome positions 136 and 326 (Eagle and Hanley-Bowdoin, 1997; Orozco and Hanley-Bowdoin, 1998). To therefore determine the effect of AL4-mediated repression on AL2 and AL3 gene expression, single base mutations were introduced into two ATG codons at the beginning of the AL4 coding region (Table 1). These changes occur in the third base of codons for the AL1 protein and do not change the amino acid specified for AL1. The mutation was introduced into both a wild type, and AL1 initiator codon mutant background, generating DNA templates with single (AL1+/AL4− and AL1−/AL4+) or double (AL1−/AL4−) mutations. In all constructs a wild type AL1 DNA binding sequence was maintained, along with the region responsible for AL4-mediated repression. As observed previously, a mutation in the AL1 initiator codon (AL1−/AL4+) results in a significant reduction (ANOVA: P<0.05) in AL2/GUS expression (Fig. 7A), but introduction of an AL4 mutation into this background (AL1−/AL4−) does not result in any further decrease in expression. In protoplasts transfected with a DNA template containing a mutation in the AL4 initiator codon, with a wild type AL1 protein (AL1+/AL4−), no significant increase in AL2/GUS activity was observed (Fig. 7). When each mutation was introduced into an AL3/GUS background (Fig. 7B), we observed slightly different results. As before, mutation of the AL1 initiator codon (AL1−/AL4+) results in a two to three-fold reduction in AL3/GUS expression (ANOVA: P<0.05). Mutation of the AL4 coding region (AL1+/AL4−) results in a 15-20% decrease in GUS activity, as compared to wild type, whereas a double mutation (AL1−/AL4− [AL2/GUS]) results in a four-fold decrease in activity as compared to wild type. To determine if this change affected AL1 function we performed assays to test autoregulation using a DNA template containing a transcriptional fusion between the AL62 promoter and GUS (Sunter et al., 1993). DNA capable of expressing the AL1 protein with the amino acid alteration in AL4 (AL1+/AL4−) had a small effect on repression of the AL62 promoter (data not shown). Our data is consistent with previous results demonstrating that AL4 has a relatively small effect on autoregulation of AL62 (Gröning et al., 1994; Eagle and Hanley-Bowdoin, 1997).This further demonstrates that AL4 repression plays a limited role in TGMV AL2 and AL3 gene expression.
Fig. 7.
TGMV AL4 plays a role in AL2 and AL3 expression. The ability of promoter-reporter constructs containing mutations within the AL4 coding region to direct AL2 (A) and AL3 (B) expression was determined by comparing GUS activity to background (pTGA26; ~4% activity). Protoplasts (5 × 105 cells) were transfected with 10 μg of a promoter-reporter construct, and significant differences in GUS activity (ANOVA (P<0.05) measured in extracts isolated three days post-transfection. Columns represent relative GUS activity as compared to a wild type (AL1+/AL4+) DNA template, which was arbitrarily assigned a value of 100, from either three (A) or two (B) independent experiments.
Discussion
Previous work has shown that AL1 autoregulates its own expression through binding to sequences within the intergenic region (Eagle et al., 1994; Sunter et al., 1993). The data presented here demonstrates that this autoregulation plays an important role in regulating expression of two downstream genes. Mutations within the AL1 binding site or within the AL1 protein itself, that destroy the ability of AL1 to bind DNA, result in a significant decrease in both AL2 and AL3 expression. Subsequent addition of exogenous AL1 in trans can restore expression close to wild type levels, providing a wild type AL1 DNA binding site is present.
Earlier results have shown that AL4 also plays a role in the autoregulation of AL1 expression through a unique sequence that does not overlap the AL1 binding site (Eagle and Hanley-Bowdoin, 1997; Gröning et al., 1994). Our results confirm those observations, and demonstrate that AL4 has a small, but measurable effect on AL2 and AL3 expression. In ACMV, the equivalent AL62 promoter is not sensitive to AL4 repression (Hong and Stanley, 1995), and therefore the role of AL4 in repression, and the requirement of AL4 for subsequent downstream gene expression, may not be conserved amongst all geminiviruses.
Transcription of two complementary sense RNAs, initiating at nucleotides 1935 (AL1935) and 1629 (AL1629), is directed by unique sequences located upstream of each transcription initiation site. One element(s) is located between 28 and 124 nucleotides upstream of the AL1935 transcription start site, and a second is located between 129 and 184 nucleotides upstream of the AL1629 transcription start site (Shung et al., 2006). Both sequences are located within the AL1 coding region, which would presumably be inaccessible to host transcription factors when transcription of AL62 is occurring. Analysis of TGMV RNAs transcribed from wild type and mutant DNA templates indicates that abolishing the ability of AL1 to autoregulate expression leads to a decrease in the level of expression of the two downstream transcripts (AL1935 and AL1629). This correlates with a decrease in AL2 expression, which is consistent with the observation that AL2 is only expressed from AL1629 (Shung et al., 2006). This interpretation is supported by results that demonstrate mutations within the AL1 initiator codon and AL1 DNA binding site, also lead to an increase in AL62 transcription. In addition, decreasing AL62 transcription directly through deletion of AL62 promoter sequences, can remove the requirement for a functional AL1 protein. Previous results from deletion analysis that demonstrate removal of sequences containing the intergenic region and the AL1 initiator codon have no effect on AL2 expression support our interpretation (Shung et al., 2006).
Although a similar fold reduction is observed for AL3 expression, a comparison of GUS activity directed by AL2/GUS and AL3/GUS constructs demonstrates that AL3 expression is two to four-fold higher than AL2 from a wild type DNA template (Shung et al., 2006). This is most likely due to the fact that AL3 can be expressed from all three complementary sense polycistronic mRNAs (Hanley-Bowdoin et al., 1989; Shung et al., 2006).
Our observations that a reduction in AL62 transcription is required for subsequent downstream gene expression can be reconciled both, with a model for geminivirus infection, and studies of other eukaryotic viruses. Geminiviruses replicate in nuclei of terminally differentiated cells where replication and cell division has ended (Rushing et al., 1987), and express proteins that interact with the host to induce cells to re-enter cell cycle. TGMV AL1 and AL3 interact with a plant homolog of retinoblastoma (Rb), and AL1 induces expression of proliferating cell nuclear antigen, the processivity factor of host DNA polymerase δ, in non-dividing plant cells (Ach et al., 1997; Nagar et al., 1995; Settlage et al., 1996; Settlage et al., 2001). This is similar to the small DNA tumor viruses, SV40 and adenovirus, where E1A and large T antigen deregulate the host cell cycle through interaction with the Rb and p53 pathways respectively (Bargonetti et al., 1992; de Stanchina et al., 1998; Dobbelstein et al., 1992; Whyte et al., 1988). During the early infection cycle, AL1 and AL3 proteins could be translated from AL62 to aid in deregulation of the host cell cycle by interacting with cell cycle components (Settlage et al., 2001). As the infection cycle progresses transcription of AL62 is negatively regulated by AL1 protein through binding to sequences within the origin of replication. This would lead to loss of interference of downstream regulatory elements within the AL1 coding region, activating transcription of AL1935 and AL1629 mRNAs to express AL2 and AL3 protein. Synthesis of AL2 from AL1629 (Shung et al., 2006) would lead to inactivation of SNF1 and ADK as part of the viral response to host defense mechanisms (Hao et al., 2003; Wang et al., 2003), followed by activation of the coat protein and BR1 gene promoters late in infection (Sunter and Bisaro, 1991; 1992).
It is currently unclear why geminiviruses would express AL3 from multiple transcripts. One possibility is that large amounts of AL3 are required for its multiple functions in replication and interaction with cell cycle components. Alternatively, AL3 could be required at several stages of the viral life-cycle requiring expression from different RNAs transcribed at different times. It may be more critical to regulate the level of AL2 protein, which is involved in inactivating defense responses and regulation of late gene expression. Nonetheless, independent regulation of these two genes would appear to be crucial to ensure successful completion of the viral life cycle.
Materials and Methods
General DNA techniques
The map locations and restriction endonuclease sites cited here refer to the published DNA sequence of TGMV (Hamilton et al., 1984). All restriction endonucleases and DNA modifying enzymes were used as recommended by the manufacturers. General DNA and RNA manipulations, and polymerase chain reaction were performed essentially as described by Ausubel et al. (2001) unless otherwise stated. All sequence alterations were confirmed by DNA sequencing (Plant-Microbe Genomics Facility, The Ohio State University).
Promoter-reporter constructs
A construct capable of generating a replicating TGMV genome component (pTGA26) has been previously described (Sunter et al., 1990). A series of constructs were generated that contained a translational fusion between the GUS reporter gene and either the N-terminal 83 amino acids of the AL2 (AL2/GUS), or the N-terminal 36 amino acids of the AL3 (AL3/GUS), coding region (Shung et al., 2006). Cloned DNA containing nucleotide changes that create a BglII site upstream of the AL1 coding region and an NdeI site as part of the initiator ATG for AL1 (pTGA60) has been described previously (Sunter et al., 1993). The 366 bp BglII to EcoRI fragment of pMON434 (Sunter et al., 1993) was used to replace the 3606 bp BglII to EcoRI fragment of pTGA60 to yield pGS204. Using pGS204 as template, a 400 bp fragment was amplified by PCR using the pUC reverse primer and a mutagenic primer (5′-AGATCTTAATTACAAAAgATATcCCATCGC-3′) to introduce single base changes (bold, lower case) into the AL1 initiator codon. The amplified DNA fragment was restricted with BglII (underlined) and EcoRI, and used to replace the equivalent fragment of pGS204, to yield pGS208. Cloned pGS204 and pGS208 DNA was then cleaved by EcoRI and the 3038 bp EcoRI fragment of AL2[−1391]-GUS (Shung et al., 2006) introduced, generating wtAL62[AL2/GUS]) and mAL62[AL2/GUS] respectively. TGMV DNA templates containing mutations in the AL1 binding site (pNSB246; Eagle et al., 1994) or amino acid 103 (Y103A) of the AL1 protein (pNSB683; Orozco and Hanley-Bowdoin, 1998) were kindly provided by Dr. Linda Hanley-Bowdoin at North Carolina State University. Cloned pNSB246 DNA was restricted with BglII and NdeI and the resulting 196 bp fragment used to replace the corresponding wild type DNA fragment of wtAL62[AL2/GUS], to generate AL1bs−[AL2/GUS]. The AL1 coding region from pNSB683 was amplified by PCR and the resulting DNA fragment restricted with BglII and EagI. The 654 bp containing the Y103A mutation was used to replace the corresponding wild type DNA sequence of wtAL62[AL2/GUS] to generate AL1Y103A[AL2/GUS]. Cloned DNA containing a deletion end-point at the SspI restriction site (Fig. 1A) was generated by restriction of wtAL62[AL2/GUS] with SspI and SalI, and the resulting 485 bp fragment used to replace the 1145 bp HindIII to SalI DNA fragment of wtAL62[AL2/GUS], to generate AL1-SspΔ[AL2/GUS]. The same procedure was used with mAL62[AL2/GUS] DNA to generate mAL1-SspΔ[AL2/GUS], containing the AL1 initiator codon mutation. To generate a frame-shift mutation within the AL1 coding region, wtAL62[AL2/GUS] DNA was restricted with SalI, treated with Klenow and re-ligated. The resulting DNA, AL11-123[AL2/GUS], is capable of producing a truncated AL1 protein of 123 amino acids and an AL2/GUS fusion protein. Two single base changes (bold, lower case) were introduced into two potential AL4 initiator codons by PCR using AL4 5′ and AL4 3′ primers (5′-AATCTGCAGAGAGCTTCAcGAAGAcGGGCAACCTC-3′, and 5′-GCGCACGTGAATTGAGATCCAAATGC-3′) with AL2[−1391]-GUS as template. The amplified DNA fragment was restricted with PstI (underlined) and EcoRI and the resulting 200 bp fragment used to replace the wild type DNA fragment of wtAL62[AL2/GUS], to yield pGS46. The 3038 bp EcoRI fragment from wtAL62[AL2/GUS], containing the AL2/GUS translational fusion, was cloned into pGS46, to yield AL1+/AL4−[AL2/GUS]. The 931 bp HindIII-PstI fragment of pGS46 was replaced by the equivalent fragment of mAL62-[AL2-GUS] to generate a double mutant (AL1−/AL4−[AL2/GUS]). This DNA was restricted with EcoRI and the 3038 bp EcoRI fragment from mAL62[AL2-GUS], containing the AL2/GUS translational fusion and AL1 initiator mutation, was cloned into the EcoRI site, to yield AL1−/AL4−[AL2/GUS]. To generate AL3/GUS constructs, the cloned DNAs described above were cleaved with BamHI, end-filled with Klenow, and religated, resulting in a +1 frame shift.
Protoplast transfection and analysis
Protoplasts were isolated from an N.benthamiana suspension culture cell line and transfected with various DNAs as described (Sunter and Bisaro, 2003). After incubation in the dark for 3 days, protoplasts were harvested and fluorometric GUS assays performed using equivalent amounts of protein as described (Shung et al., 2006), and GUS activities compared by Student’s t-test or one-way Analysis of Variance (ANOVA).
Leaf infusions
Cloned DNA containing wild type sequences (wtAL62[AL2/GUS]), and mutations in the AL1 translation initiation codon (mAL62[AL2/GUS]), or the AL1 DNA binding site (AL1bs−[AL2/GUS]), were cloned into the binary plasmid vector pMON521 (Rogers et al., 1987). Binary plasmid constructs were mobilized into Agrobacterium strain GV3111SE, containing the disarmed Ti plasmid pTiB36SE, by triparental mating (Horsch and Klee, 1986). Agrobacterium cultures containing promoter-reporter constructs were delivered to N.benthamiana leaves by leaf infusion as described (Johansen and Carrington, 2001; Wang et al., 2005).
RNA isolation and analysis
Total RNA was isolated from N.benthamiana leaves using Plant RNA Purification Reagent according to the manufacturers instructions (Invitrogen, Carlsbad, CA), and poly (A)+RNA purified as described previously (Sunter and Bisaro, 1989). Total RNA was further purified using the RNeasy mini-elute clean up kit (Qiagen,Valencia CA) followed by DNase I treatment (Turbo DNaseI, Ambion, Austin TX) at 37°C for 1 hour. Following phenol/chloroform extraction and ethanol precipitation, RNA was resuspended in H2O and stored at −80°C.
Semi-quantitative RT-PCR
Up to 500 ng total RNA was used for detection of EF1α or TGMV viral RNAs in semi-quantitative reverse transcription (RT)-PCR reactions using SuperScript one-step RT-PCR mix with Platinum Taq according to the manufacturers instructions (Invitrogen, Carlsbad, CA). The primers used for amplification were AL62 (5′-TCCACTAAAGAACTGGACTTTCCATAATGCG-3′); GUS (5′-CCCACCAACGCTGATCAATTCCAC-3′), AL1935 (5′-GGCGATAGTCGGACGGGAAAGACTATGTGGGC-3′), AL1629 (5′GCGCCATGGACTCCACTAAAGAACTGGAC), EF1αF (5′-TGGTGTCCTCAAGCCTGGTATGGTTGT-3′), and EF1αR (5′-ACGCTTGAGATCCTTAACCGCAACATTCTT-3′. For amplification of EF1α, 25 pm of each primer was used, and for amplification of viral RNAs, the AL62, AL1935 and AL1629 primers were used at 25 pm with 150 pm of the GUS primer. The optimum number of cycles required for a near linear relationship was determined using a variable cycle number during PCR. Samples were normalized by comparison to EF1α (Shung et al., 2006) at an equivalent number of cycles. Products from RT-PCR reactions were electrophoresed through 1% TAE agarose gels, transferred to Protran® pure nitrocellulose membranes (Schleicher and Schuell, Keene, NH) and immobilized by UV cross-linking (UV-Strata linker 1800, Stratagene, La Jolla, CA). Specific cDNA products were detected by hybridization to 32P-labeled probes specific for either TGMV or EF1α, generated by random priming (DECA Prime II labeling kit. Ambion, Austin, TX). DNA levels were quantified by phosphorimager analysis (Molecular Imager FX, Bio-Rad, Hercules, CA).
Acknowledgements
This research was supported in part by a National Institutes of Health MBRS/SCORE Grant (GM-08194). We thank Janet Sunter for maintenance of the suspension cell line and for assistance with protoplast transfection experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambriski PC, Gruissem W. An alternatively-spliced, multigene family in maize encodes retinoblastoma-related proteins which can interact with a plant D-type cyclin and a geminivirus replication protein. Mol. Cell. Biol. 1997;17:5077–5086. doi: 10.1128/mcb.17.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Siedman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley and Sons; New York: 2001. [Google Scholar]
- Bargonetti J, Reynisdottir I, Friedman PN, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6(10):1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- Bisaro DM. Geminivirus replication. In: DePamphilis M, editor. DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1996. pp. 833–854. [Google Scholar]
- Brough CL, Hayes RJ, Morgan AJ, Coutts RHA, Buck KW. Effects of mutagenesis in vitro on the ability of cloned tomato golden mosaic virus DNA to infect Nicotiana benthamiana plants. J. Gen. Virol. 1988;69:503–514. [Google Scholar]
- Brough CL, Gardiner WE, Inamdar N, Zhang XY, Ehrlich M, Bisaro DM. DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 1992a;18:703–712. doi: 10.1007/BF00020012. [DOI] [PubMed] [Google Scholar]
- Brough CL, Sunter G, Gardiner WE, Bisaro DM. Kinetics of tomato golden mosaic virus DNA replication and coat protein promoter activity in Nicotiana tabacum protoplasts. Virology. 1992b;187:1–9. doi: 10.1016/0042-6822(92)90289-2. [DOI] [PubMed] [Google Scholar]
- de Stanchina E, McCurrach ME, Zindy F, Shieh S-Y, Ferbeyre G, Samuelson AV, Prives C, Roussel MF, Sherr CJ, Lowe SW. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 1998;12(15):2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M, Arthur AK, Dehde S, van Zee K, Dickmanns A, Fanning E. Intracistronic complementation reveals a new function of SV40 T antigen that co-operates with Rb and p53 binding to stimulate DNA synthesis in quiescent cells. Oncogene. 1992;7(5):837–847. [PubMed] [Google Scholar]
- Eagle PA, Hanley-Bowdoin L. Cis-elements that contribute to geminivirus transcriptional regulation and efficient DNA replication. J. Virol. 1997;71:6947–1170. doi: 10.1128/jvi.71.9.6947-6955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle PA, Orozco BM, Hanley-Bowdoin L. A DNA sequence required for geminivirus replication also mediates transcriptional regulation. Plant Cell. 1994;6:1157–1170. doi: 10.1105/tpc.6.8.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer JS, Brand L, Sunter G, Gardiner WE, Bisaro DM, Rogers SG. Genetic analysis of tomato golden mosaic virus II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 1988;16(14):7043–7060. doi: 10.1093/nar/16.14.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EPB, Eagle PA, Sipe PS, Luckow VA, Hanley-Bowdoin L. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 1994;269:8459–8465. [PubMed] [Google Scholar]
- Gladfelter HJ, Eagle PA, Fontes EPB, Batts L, Hanley-Bowdoin L. Two domains of the AL1 protein mediate geminivirus origin recognition. Virology. 1997;239:186–197. doi: 10.1006/viro.1997.8869. [DOI] [PubMed] [Google Scholar]
- Gröning BR, Hayes RJ, Buck KW. Simultaneous regulation of tomato golden mosaic virus coat protein and AL1 gene expression: expression of the AL4 gene may contribute to suppression of the AL1 gene. J. Gen. Virol. 1994;75:721–726. doi: 10.1099/0022-1317-75-4-721. [DOI] [PubMed] [Google Scholar]
- Hamilton WDO, Bisaro DM, Coutts RHA, Buck KW. Demonstration of the bipartite nature of the genome of a single-stranded DNA plant virus by infection with the cloned DNA components. Nucleic Acids Res. 1983;11(21):7387–7396. doi: 10.1093/nar/11.21.7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WDO, Stein VE, Coutts RHA, Buck KW. Complete nucleotide sequence of the infectious cloned DNA components of tomato golden mosaic virus: Potential coding regions and regulatory sequences. EMBO J. 1984;3:2197–2205. doi: 10.1002/j.1460-2075.1984.tb02114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Elmer JS, Rogers SG. Transient expression of heterologous RNAs using tomato golden mosaic virus. Nucleic Acids Res. 1988;16:10511–10528. doi: 10.1093/nar/16.22.10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Elmer JS, Rogers SG. Functional expression of the leftward open reading frames of the A component of tomato golden mosaic virus in transgenic plants. Plant Cell. 1989;1:1057–1067. doi: 10.1105/tpc.1.11.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Elmer JS, Rogers SG. Expression of functional replication protein from tomato golden mosaic virus in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1446–1450. doi: 10.1073/pnas.87.4.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Settlage S, Orozco BM, Nagar S, Robertson D. Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 1999;18:71–106. [PubMed] [Google Scholar]
- Hao L, Wang H, Sunter G, Bisaro DM. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell. 2003;15(4):1034–1048. doi: 10.1105/tpc.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YG, Stanley J. Regulation of African cassava mosaic virus complementary-sense gene expression by N-terminal sequences of the replication-associated protein AC1. J. Gen. Virol. 1995;76:2415–2422. doi: 10.1099/0022-1317-76-10-2415. [DOI] [PubMed] [Google Scholar]
- Horsch RB, Klee HJ. Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: Role of the T-DNA borders in the transfer process. Proc. Natl. Acad. Sci. U.S.A. 1986;83:4428–4432. doi: 10.1073/pnas.83.12.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey JL, Pooma W, Petty ITD. Genetic requirements for local and systemic movement of tomato golden mosaic virus in infected plants. Virology. 1996;223:208–218. doi: 10.1006/viro.1996.0469. [DOI] [PubMed] [Google Scholar]
- Johansen LK, Carrington JC. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 2001;126(3):930–938. doi: 10.1104/pp.126.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Slunt HH, Martin LJ, Thinakaran G, Kim G, Gandy SE, Seeger M, Koo E, Price DL, Sisodia SS. Expression of Presenilin 1 and 2 (PS1 and PS2) in Human and Murine Tissues. J. Neurosci. 1996;16(23):7513–7525. doi: 10.1523/JNEUROSCI.16-23-07513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar S, Pedersen TJ, Carrick K, Hanley-Bowdoin L, Robertson D. A geminivirus induces expression of host DNA synthesis protein in terminally differentiated plant cells. Plant Cell. 1995;7:705–719. doi: 10.1105/tpc.7.6.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco BM, Hanley-Bowdoin L. Conserved Sequence and Structural Motifs Contribute to the DNA Binding and Cleavage Activities of a Geminivirus Replication Protein. J. Biol. Chem. 1998;273(38):24448–24456. doi: 10.1074/jbc.273.38.24448. [DOI] [PubMed] [Google Scholar]
- Petty ITD, Coutts RHA, Buck KW. Transcriptional mapping of the coat protein gene of tomato golden mosaic virus. J. Gen. Virol. 1988;69:1359–1365. [Google Scholar]
- Rogers SG, Bisaro DM, Horsch RB, Fraley RT, Hoffmann NL, Brand L, Elmer JS, Lloyd AM. Tomato golden mosaic virus A component DNA replicates autonomously in transgenic plants. Cell. 1986;45:593–600. doi: 10.1016/0092-8674(86)90291-6. [DOI] [PubMed] [Google Scholar]
- Rogers SG, Klee HJ, Horsch RB, Fraley RT. Improved vectors for plant transformation: Expression cassette vectors and new selectable markers. Methods in Enzymology. 1987;153:253–277. [Google Scholar]
- Rushing AE, Sunter G, Gardiner WE, Dute RR, Bisaro DM. Ultrastructural aspects of tomato golden mosaic virus infection in tobacco. Phytopathology. 1987;77(8):1231–1236. [Google Scholar]
- Settlage SB, Miller B, Hanley-Bowdoin L. Interactions between geminivirus replication proteins. J. Virol. 1996;70:6790–6795. doi: 10.1128/jvi.70.10.6790-6795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settlage SB, Miller AB, Gruissem W, Hanley-Bowdoin L. Dual interaction of a geminivirus replication accessory protein and a plant cell cycle regulator. Virology. 2001;279:570–576. doi: 10.1006/viro.2000.0719. [DOI] [PubMed] [Google Scholar]
- Shung C-Y, Sunter J, Sirasanagandla SS, Sunter G. Distinct viral sequence elements are necessary for expression of tomato golden mosaic virus complementary sense transcripts that direct AL2 and AL3 gene expression. Mol. Plant-Microbe Interact. 2006;19:1394–1405. doi: 10.1094/MPMI-19-1394. [DOI] [PubMed] [Google Scholar]
- Stenger DC, Revington GN, Stevenson MC, Bisaro DM. Replicational release of geminivirus genomes from tandemly repeated copies: Evidence for rolling circle replication of a plant viral DNA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:8029–8033. doi: 10.1073/pnas.88.18.8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter G, Bisaro DM. Transcription map of the B genome component of tomato golden mosaic virus and comparison with A component transcripts. Virology. 1989;173:647–655. doi: 10.1016/0042-6822(89)90577-1. [DOI] [PubMed] [Google Scholar]
- Sunter G, Bisaro DM. Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology. 1991;180:416–419. doi: 10.1016/0042-6822(91)90049-h. [DOI] [PubMed] [Google Scholar]
- Sunter G, Bisaro DM. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell. 1992;4:1321–1331. doi: 10.1105/tpc.4.10.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter G, Bisaro DM. Identification of a minimal sequence required for activation of the tomato golden mosaic virus coat protein promoter in protoplasts. Virology. 2003;305(2):452–462. doi: 10.1006/viro.2002.1757. [DOI] [PubMed] [Google Scholar]
- Sunter G, Gardiner WE, Bisaro DM. Identification of tomato golden mosaic virus-specific RNAs in infected plants. Virology. 1989;170:243–250. doi: 10.1016/0042-6822(89)90372-3. [DOI] [PubMed] [Google Scholar]
- Sunter G, Hartitz MD, Bisaro DM. Tomato golden mosaic virus leftward gene expression: Autoregulation of geminivirus replication protein. Virology. 1993;195:275–280. doi: 10.1006/viro.1993.1374. [DOI] [PubMed] [Google Scholar]
- Sunter G, Gardiner WE, Rushing AE, Rogers SG, Bisaro DM. Independent encapsidation of tomato golden mosaic virus A component DNA in transgenic plants. Plant Mol. Biol. 1987;8:477–484. doi: 10.1007/BF00017993. [DOI] [PubMed] [Google Scholar]
- Sunter G, Hartitz MD, Hormuzdi SG, Brough CL, Bisaro DM. Genetic analysis of tomato golden mosaic virus. ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology. 1990;179:69–77. doi: 10.1016/0042-6822(90)90275-v. [DOI] [PubMed] [Google Scholar]
- Wang H, Hao L, Shung C-Y, Sunter G, Bisaro DM. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell. 2003;15(12):3020–3032. doi: 10.1105/tpc.015180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 2005;79(12):7410–7418. doi: 10.1128/JVI.79.12.7410-7418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, Harlow E. Association between an oncogene and an antioncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]