Summary
H1 `linker' histones bind dynamically to nucleosomes and promote their compaction into chromatin fibers [1–4]. Developmental H1 isoforms are evolutionarily conserved, but their function, regulation, and post-translational modifications are poorly understood [5–7]. In Xenopus egg extracts, the embryonic linker histone H1M does not affect nuclear assembly or replication, but is required for proper chromosome architecture during mitosis [8, 9]. We report here that somatic H1 isoforms, which are more positively charged and feature multiple Cdk1 phosphorylation sites, cannot substitute for H1M at endogenous concentrations, instead causing chromatin compaction during interphase, and dissociating from chromosomes at the onset of mitosis. Mitotic Cdk1 phosphorylation is not responsible for this dissociation, and instead functions to enhance H1 binding in egg extracts and embryos. Nuclear import receptors RanBP7 and importin β bind tightly to somatic H1 but not H1M, and addition of a constitutively-active Ran mutant abolishes this interaction and enhances the ability of somatic H1 to rescue mitotic chromosome architecture. Our results reveal distinct regulatory mechanisms among linker histone isoforms and a specific role for H1M to compact chromosomes during egg meiotic arrest and early embryonic divisions. (183)
Keywords: histone H1, H1 isoforms, H1 phosphorylation, mitosis, Cdk1, RanGTP, importin beta, RanBP7
Results and Discussion
Somatic H1 Cannot Substitute for H1M at Endogenous Concentrations
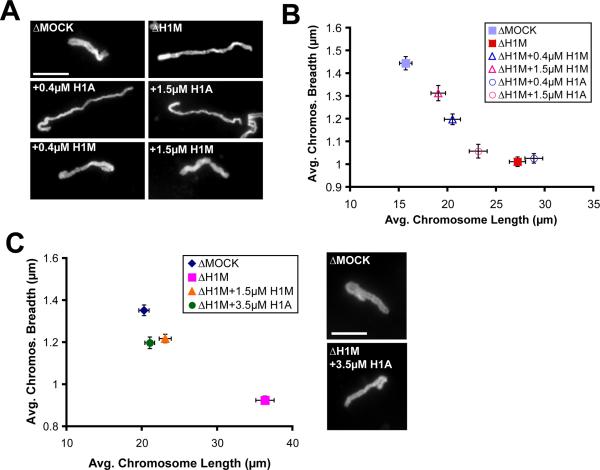
We showed previously that immunodepletion of H1M from egg extracts results in elongated mitotic chromosomes that align poorly in the spindle and are consistently about 2-fold longer and 30% thinner than controls in the same extract [10]. A strong rescue of chromosome morphology and H1M immunofluorescence required 1 μM recombinant H1M, which is equivalent to the endogenous concentration in the egg, while lower concentrations yielded partial rescues in a dose-dependent manner (Figure S1A–B and data not shown). To test the functional equivalence of developmental H1 isoforms, we generated recombinant, 6xHistidine-tagged H1M, H1A, or H10 proteins using X. tropicalis clones (to avoid allelic variants in the pseudotetraploid X. laevis). H1A typifies the replication-dependent linker histones present in most metazoan cells while H10 is specific to differentiated cell types [5–7]. Both are smaller and more basic than H1M, contain 4–5 consensus Cdk1 phosphorylation sites in their C-terminal domains, and behaved more similarly to one another than to H1M, and will therefore be referred to collectively as “somatic H1.” Surprisingly, unlike H1M, somatic H1 failed to rescue the elongated mitotic chromosome phenotype at concentrations of 1–2 μM (Figure 1A–B) and could restore mitotic chromosome morphology only at 3.5 μM (Figure 1C). Although we previously reported that H1 purified from calf thymus, which is 54% identical to somatic H1A, could rescue H1M-depleted mitotic chromosome compaction similarly to H1M [10], protein concentrations were approximate. In this study, precise protein concentrations were determined by Coomassie staining of a dilution series in comparison with known mass standards (data not shown), and equal amounts were added to H1M-depleted egg extracts, as confirmed by immunoblot blot analysis of the 6xHistidine tag (Figure S1C). Indeed, upon repeating the experiment we found that chromosome morphology was not restored with 1.3 μM calf thymus H1 but required concentrations above 3 μM, comparable to recombinant H1A (Figure S1D and data not shown). The quantification of dose-dependence and detailed morphometric analyses presented here were essential to distinguish differences among H1 proteins.
Figure 1. Somatic H1 Cannot Substitute for H1M at Endogenous Concentrations.
(A) Individual chromosomes assembled in mock-depleted (ΔMOCK) or H1M-depleted (ΔH1M) extracts, or H1M-depleted extracts supplemented with 0.4 μM or 1.5 μM H1M or H1A. H1M consistently rescued more efficiently than H1A, even at lower concentrations.
(B) Quantification of chromosome morphology from the experiment described in (A). Images of individual chromosomes (n > 30) were thresholded and their average fiber lengths and breadths calculated and plotted.
(C) 3.5 μM somatic H1 rescues H1M-depleted chromosome morphology similar to a lower concentration of H1M. Average lengths and breadths of individual chromosomes (n > 40) and representative chromosomes are shown. Since somatic H1 causes interphase compaction at this concentration, H1 proteins were added only at the onset of mitosis. Scale bars, 10 μm; error bars, standard error. See also Figure S1.
Somatic H1 Dissociates From Mitotic Chromosomes, but Compacts Interphase Chromatin
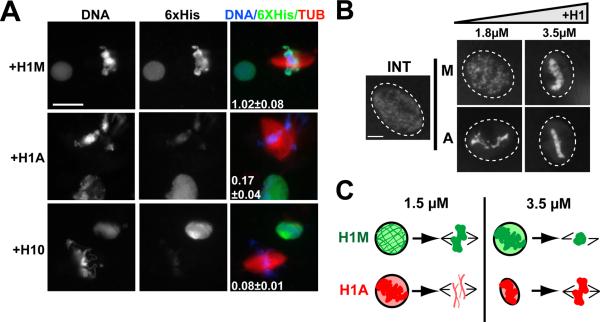
To determine whether the failure of somatic H1 to rescue reflected reduced mitotic chromosome binding, we performed immunofluorescence analysis of the recombinant 6xHistidine tag on samples of nuclei and mitotic chromosomes to compare its localization during interphase and mitosis (Figure 2A). Both somatic H1 and H1M localized to interphase nuclei, but a striking reduction in somatic H1 was observed on mitotic chromosomes, which could also be detected by comparing chromatin isolated from interphase and mitotic extracts by immunoblot (data not shown). We confirmed this dynamic localization pattern by fluorescence time-lapse analysis of individual nuclei, which showed that 1.5 μM Alexa488-labeled somatic H1, but not H1M, dissociated from chromatin shortly after nuclear envelope breakdown (Movies S1–2). This analysis indicated that somatic H1 isoforms do not bind well to chromatin during mitosis, but associate with interphase chromatin. In fact, the chromatin binding and compacting activity of somatic H1 during interphase was greater than that of H1M. Whereas interphase chromatin remained diffuse within the nucleus when 1.8 μM H1M was added, the same amount of somatic H1 hypercompacted chromatin, yielding bright foci connected by thin fibers of 0.81±0.11 μm width (Figure 2B). At the higher concentration of 3.5 μM, H1M also caused hypercompaction, which resembles an H1 overexpression phenotype reported in somatic cells [11]. In summary, somatic H1 compacts interphase chromatin at lower concentrations than H1M, but requires significantly higher concentrations to associate with and condense metaphase chromosomes (see schematic, Figure 2C).
Figure 2. Somatic H1 Dissociates from Mitotic Chromosomes.
(A) Anti-6XHistidine immunofluorescence of nuclei and mitotic spindles from H1M-depleted reactions supplemented with 1 μM of 6XHistidine-tagged H1M, H10, or H1A. Interphase and mitotic reactions were separately diluted into a fixation buffer, then mixed and spun onto a single coverslip for staining. The average H1:DNA (6XHistidine:Hoechst, ± standard error) fluorescence intensity of mitotic chromosome clusters (5–15 per condition) divided by that of interphase nuclei is shown for each condition in the merged image. Bar, 25 μm.
(B) Nuclei assembled in undepleted reactions supplemented with increasing concentrations of H1M (left) or H1A (right). Only H1A causes chromatin compaction at the lower dose (> 30 nuclei analyzed per condition). Bar, 10 μm.
(C) Schematic showing functional effects of two different concentrations of H1M and H1A during interphase and mitosis. See also Movies S1–2.
Somatic H1 Binding to Mitotic Chromosomes is Enhanced by Cdk1 Phosphorylation
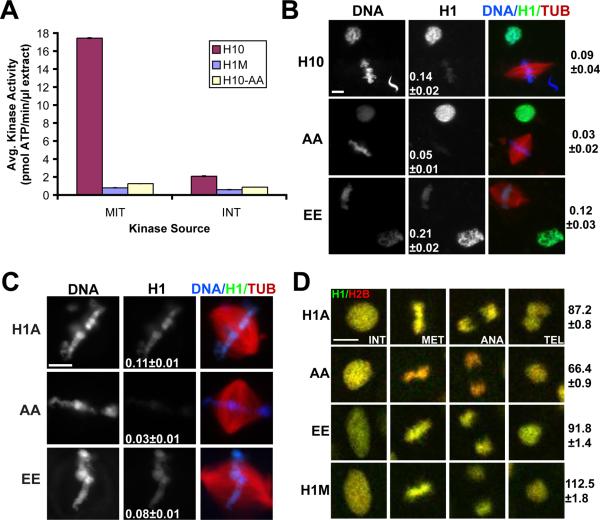
Cyclin-dependent kinase (Cdk1) phosphorylates somatic H1 isoforms, but not H1M, at multiple sites and has been proposed to reduce H1 binding to chromatin [12]. We therefore hypothesized that this post-translational modification could be responsible for the dissociation of somatic H1 from mitotic chromosomes in egg extract, which contains high Cdk1 activity. We generated a mutant somatic H1 protein in which the consensus Cdk1 phosphorylation serines (SPxK amino acid motif) were mutated to alanines, and confirmed that it was not phosphorylated in an extract kinase assay (Figure 3A). If Cdk1 phosphorylation causes H1 dissociation, this mutant should be retained on mitotic chromosomes. However, when added to metaphase reactions, the non-phosphorylatable mutant localized to chromosomes even less intensely than either wild-type H1 or a phosphomimetic mutant in which the serines were substituted with glutamic acid residues (Figure 3B). Reduced chromatin binding of non-phosphorylatable somatic H1 was specific to mitosis and did not depend on nuclear assembly or H1 import, since similar results were obtained upon addition of GFP-tagged proteins to metaphase cytostatic factor-arrested (CSF) egg extracts in which sperm nuclei were directly remodeled into mitotic chromatin without an intervening interphase (Figure 3C). Phosphorylation site mutations also did not improve somatic H1 function at concentrations of 1–2 μM, which still failed to rescue H1M-depleted chromosome morphology (data not shown). Thus, mitotic Cdk1 phosphorylation cannot be responsible for the dissociation of somatic H1 from mitotic chromosomes in Xenopus egg extracts.
Figure 3. Somatic H1 Chromosome Binding is Enhanced by Phosphorylation.
(A) H1 kinase assay from metaphase CSF-arrested (MET) or interphase-arrested (INT) egg extracts for H1M, H10, and alanine-substituted H10 (-AA).
(B) Anti-6XHistidine immunofluorescence of reactions with H10 wild-type versus alanine (AA) or glutamic acid (EE) phosphorylation site mutants. Interphase or mitotic reactions were fixed separately, then mixed and processed together. Average H1:DNA (6XHistidine:Hoechst) fluorescence ratio is shown for mitotic chromosomes alone (center column) or mitotic chromosomes over interphase nuclei (right of figure).
(C) Sperm nuclei were remodeled directly into mitotic chromosomes in CSF egg extracts supplemented with 1 μM H1A-GFP or phosphorylation site mutants and the average H1:DNA (GFP:DAPI) fluorescence ratio was determined (center column).
(D) Time-lapse images of dividing cells in stage 13 embryos expressing H1-GFP (green) and H2B-RFP (red). The Cdk1 phosphorylation site alanine mutant (AA) binds to mitotic chromosomes with reduced affinity compared to wild-type H1A, H1A-EE, or H1M, causing chromatin to shift from yellow to red. H1-GFP signal was normalized to H2B-RFP levels for single cells at interphase and anaphase, and the anaphase:interphase fluorescence ratio was calculated for each individual cell and averaged (right of figure). Scale bars, 10 μm; error, standard error. 10–50 structures or cells were examined per condition. See also Figure S2 and Movies S3–5.
It was important to determine whether differences between H1 isoforms and mutants observed in egg extracts also hold true in somatic cells. We therefore investigated linker histone localization during mitosis in Xenopus embryos, where somatic H1 is normally expressed. H1A accumulated only after the midblastula transition (stage 8), consistent with previous reports [11, 13, 14], and appeared to localize brightly to mitotic chromosomes (Figure S2A–B). To directly compare mitotic chromosome binding of isoforms and phosphorylation site mutants, H1M- or H1A-GFP fusion proteins were exogenously co-expressed with histone H2B-RFP in neurula (stage 13–15) embryos and examined by confocal fluorescence time-lapse microscopy, allowing quantification of histone fluorescence intensity on chromosomes in single cells as they underwent mitosis (Figure 3D; Movies S3–5). Although GFP-tagged H1 proteins were similarly bright during interphase (Figure S2C and data not shown), we found that the intensity of H1A-GFP relative to H2B-RFP decreased transiently during mitosis to ~85 % of interphase levels (Movie S3), whereas H1M-GFP binding increased to ~110 % (Movie S4). Mutation of Cdk1 phosphorylation sites to alanines further decreased H1A-GFP binding to ~65 % of interphase levels (Movie S5), while mutation to glutamic acids was comparable to wild-type (Figure 3D). In addition, FRAP (Fluorescence Recovery After Photobleaching) of the alanine substitution mutant during mitosis was significantly faster than that of wild-type H1A-GFP or the phosphomimetic mutant, and fluorescence recovered to a slightly higher extent, suggesting that unphosphorylated H1 has a higher off-rate from chromatin and a greater mobile fraction (Figure S2D). These data suggest that relative differences between H1 isoforms at mitosis persist in somatic cells, but that somatic H1 dissociation occurs to a lesser degree, and is reversed by Cdk1 phosphorylation. Even though somatic H1 binding to mitotic chromosomes was lower in egg extracts than in embryos, the relative affinities of phosphorylation site mutants were similar (Figure 3B–D), indicating that H1 phosphorylation is a general mechanism that increases its binding to mitotic chromosomes.
Nuclear Import Receptors Bind and Inhibit Somatic H1 During Mitosis
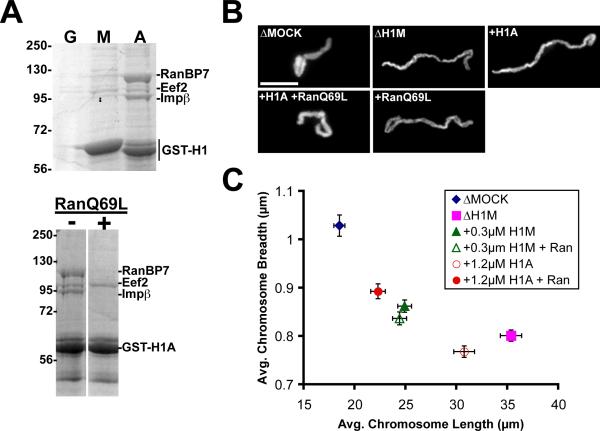
An important question is why somatic H1, but not H1M, dissociates from chromosomes at mitosis. Having ruled out somatic H1-specific phosphorylation by Cdk1, we performed pull-downs from egg extracts using recombinant GST-H1 fusion proteins to discover other potential mechanisms. Somatic H1, but not H1M, strongly associated with proteins of 90 kD and 110 kD, which mass spectrometry analysis revealed to be importin β and RanBP7 (Ran binding protein 7 or importin-7), respectively (Figure 4A). A 95 kD protein, Eef2 (eukaryotic translation elongation factor 2), frequently appeared in both H1M and somatic H1 pull-downs, but we have not determined whether this is a specific interaction. An importin β/RanBP7 heterodimer was previously reported to mediate nuclear import of somatic H1, releasing the H1 cargo upon binding to RanGTP in the nucleus [15, 16]. Consistent with this regulatory mechanism, we found that addition of 15 μM RanQ69L, a mutant locked in the GTP-bound state, disrupted binding of importin β/RanBP7 to somatic H1 in egg extracts (Figure 4A). To determine what regions of somatic H1 associate with importins, we performed pull downs with H1M/H10 chimeras in which the head of one isoform was fused to the tail of the other, and found that all combinations interacted with importin β/RanBP7, indicating that multiple regions of somatic H1 contribute to importin binding (Figure S3A). Mutation of somatic H1 Cdk1 phosphorylation sites did not have a detectable effect on its interaction with importin β/RanBP7 in pull-down assays (Figure S3B). Although H1M did not appear to bind tightly to these receptors, fluorescently-tagged H1M underwent active nuclear import, at rates that appeared to be slower than GFP-H1A (data not shown).
Figure 4. RanGTP Releases Somatic H1 from Nuclear Import Receptors and Promotes its Chromosome Binding.
(A) Top: Coomassie-stained gel of proteins pulled down with GST (G) or H1M- and H1A-GST fusion proteins (M, A) from metaphase-arrested extract. GST (22 kD) is not shown. Bottom: GST-H1A pull-downs supplemented with 15 μM RanQ69L (+) or buffer control (−).
(B) Representative mock- or H1M-depleted chromosomes assembled with or without recombinant H1A (1.2 μM) and RanQ69L (15 μM). A subpopulation of unrescued chromosomes was also observed in the +H1A +RanQ69L condition but is not shown.
(C) Average chromosome morphology (n > 30 per condition) comparing the effects of RanQ69L with either 1.2 μM H1A or 0.3 μM H1M. Doses were chosen that could not rescue chromosome morphology on their own. Error bars, standard error. See also Figure S3.
Our lab and others have previously shown that importin β functions during mitosis to bind and inhibit spindle assembly factors, raising the possibility that importin β/RanBP7 might similarly inhibit somatic H1. Importin β inhibition of spindle assembly factors is reduced in the immediate vicinity of mitotic chromosomes, where a gradient of RanGTP arises due to the presence of the chromatin-bound RanGEF, RCC1 [17, 18]. To test whether importin β/RanBP7 regulates the chromatin binding and activity of somatic H1, we added RanQ69L to extract reactions at the onset of mitosis. Ectopic generation of microtubules indicated that mitotic cargoes of importin β were released (data not shown; [17]). Morphometric analysis showed that addition of RanQ69L and somatic H1 together, but not individually, gave a substantial rescue of mitotic chromosome morphology. Significantly, this effect was specific to somatic H1, since RanQ69L did not enhance a partial rescue of chromosome structure by H1M (Figure 4B–C). Averaged over three experiments, adding somatic H1 or RanQ69L individually resulted in chromosomes that were 71±10 % or 79±9 % longer than mock-depletes respectively, while adding both factors yielded chromosomes only 22±7 % longer. These data suggest that the binding and compaction activity of somatic H1 on mitotic chromosomes is inhibited by importin β/RanBP7, and this inhibition is relieved in the presence of RanGTP. Although the rescue was substantial, it was not complete, indicating that other mechanisms may also regulate somatic H1.
Altogether, our comparison of H1 isoforms suggests a model for their function during the cell cycle in egg extracts (Figure S3C). H1M interacts only weakly with importin β/RanBP7 and binds efficiently to both interphase and mitotic chromatin. Somatic H1, which is more positively charged, interacts strongly with importin β/RanBP7. In interphase nuclei, RanGTP concentrations are very high, releasing somatic H1 from these receptors and promoting its chromatin binding. Following nuclear envelope breakdown, concentrations of somatic H1 and RanGTP surrounding chromatin are reduced by diffusion. Although a steep gradient of RanGTP persists around mitotic chromosomes in egg extracts [18], a significant pool of linker histone does not colocalize and stoichiometric concentrations of somatic H1 may not be released from the receptors, thereby reducing chromatin binding and compaction during mitosis relative to interphase. Phosphorylation of somatic H1 by Cdk1 on multiple sites, which has been a mystery for over thirty years, functions to strengthen H1 binding to mitotic chromosomes and offset dissociation. This makes sense given that linker histones function to compact chromatin, and this occurs maximally during mitosis. The effects of somatic H1 phosphorylation were evident by single-cell analysis in the embryo, where it is normally expressed, and do not appear to reduce the interaction with importin β/RanBP7. Whether and how importin β/RanBP7 and RanGTP contribute to somatic H1 regulation in the context of the embryo is an open question that is difficult to address given the general import role of these factors, and the fact that positively charged residues throughout somatic H1 contribute to importin β/RanBP7 binding. It is tempting to speculate that since the mitotic RanGTP gradient occupies a larger proportion of the cytoplasmic volume in somatic cells (Kalab et al., 2006), more H1 may be released from the importin β/RanBP7 heterodimer.
H1 isoforms and phosphorylation sites may have evolved to match different cellular conditions. In egg cytoplasm, H1M ensures that chromosomes cluster together at the metaphase plate to achieve the compact morphology required for efficient and accurate segregation. Consequently, this isoform is optimized for chromosome binding within the large cells of the early embryo. In contrast, somatic H1 may have evolved to perform interphase functions and to associate with chromosomes in cells with a higher nuclear:cytoplasmic volume ratio. In such cells, the mitotic chromosome-binding properties of H1M might be detrimental, and indeed we have observed that H1M overexpression is highly toxic to embryos (B. Freedman and R. Heald, unpublished data). Future work will investigate the contribution of phosphorylated somatic H1 to chromosome condensation in embryos, and the consequences of its overexpression or replacement by other isoforms and mutants.
Experimental Procedures
X. laevis Egg Extract Reactions, Fluorescence Microscopy, and Image Analysis
Demembranated sperm nuclei and CSF low-speed egg extracts were supplemented with X-rhodamine-labeled tubulin (50 μg/ml), immunodepleted, and reacted as described [9, 19]. Purified linker histone was added to extracts prior to sperm addition and was included with fresh CSF extract at mitosis, except for overexpression experiments when it was added only at mitosis. For Ran addition, interphase reactions containing somatic H1 were split and 15 μM RanQ69L or XB buffer was added at mitosis. Live movies were obtained by spotting 5 μl of extract reaction onto a glass slide and overlaying it with an 18 mm × 18 mm coverglass. Immunofluorescence of extract interphase and mitotic structures was performed as described [9, 10]. To visualize interphase and mitotic structures side-by-side, reactions were fixed separately and then mixed before overlaying onto cushion. Staining antibodies included mouse monoclonal anti-His (Clontech #631212) and custom rabbit polyclonals anti-H1M and anti-H1A (X. laevis, Covance Research Services), which were directly labeled with AlexaFluor Monoclonal Antibody Labeling Kit (Molecular Probes) or Cy3 NHS ester (GE Healthcare, #PA13101) for co-staining. For morphological analysis, structures processed for immunofluorescence were imaged using identical exposures on an epifluorescence microscope (Olympus, model BX51) with CCD camera (Hamamatsu, model C4742-98), shutter controller (Sutter Instrument Co., model Lambda 10-2), and Metamorph software (Universal Imaging Corp.) using 40× dry (Olympus, N.A. 0.75) and 100× oil (Olympus, N.A. 1.3) objectives. Chromosomes were identified as paired sister chromatids. For morphological quantification, no fewer than 25 representative images were manually scaled and thresholded, cut where necessary, and subjected to Integrated Morphometry Analysis or linescans (Metamorph). For colocalization analysis, regions were drawn around nuclei or metaphase plates in the DNA channel and the background-subtracted H1:DNA intensity ratio was calculated for each region (Metamorph).
Purification of Linker Histone Isoforms, Mutants, and Fusion Proteins
X. tropicalis H1M (TEgg048d08), H1A (TGas008a06), and H10 (IMAGE: 5307589) coding sequences (Geneservice) were cloned into pET20b (Novagen), pGEX-KG (for N-terminal GST fusions), modified pET30 (for C-terminal GFP fusions), or pCS107 (for mRNA). To accommodate the NcoI site, the second codon of H10 was changed from ACA (threonine) to GAT (serine). To generate phosphorylation site mutants, all consensus phosphorylation sites (S/TPxK) were mutated (Quikchange Multi kit, Stratagene) to replace serine residues with either alanines or glutamic acids. For tailswaps, H1 was aligned with the H15 conserved domain (cd00073) and domains were cloned sequentially. Proteins were expressed in RIPL BL21-CodonPlus E. Coli cells (Stratagene) and nickel-purified in PBS plus 500 mM NaCl. All protein concentration and purity was determined by running a dilution series against standards of known mass on an SDS-PAGE gel stained with Coomassie blue and confirmed by Bradford assay, manufacturer's estimate, and/or immunoblotting against the 6XHistidine tag. RanQ69L protein was prepared in XB as described [21]. mRNA was generated using the mMessage mMachine SP6 kit (Ambion) and 500 pg H1A-GFP or 100 pg H1M-GFP mRNA was injected together with 500 pg H2B-RFP mRNA into one-cell stage embryos. Similar expression levels of H1-GFP were confirmed by embryo immunoblot.
Pull-downs and H1 Kinase Assays
For pull-downs, 5–10 μM GST-tagged or biotinylated H1 protein was added to 100 μl mitotic egg extract plus either 15 μM RanQ69L or buffer. After 1 hour incubation at room temperature, reactions were mixed with 30 μl Glutathione Sepharose 4B (GE Healthcare) or Streptavidin Agarose (Thermo Scientific) pre-equilibrated in XB, transferred carefully to a new tube, and rotated for one hour at 4 ° C. The matrix was then pelleted at 500 g for five minutes, the extract supernatant was removed, and the matrix was washed once in cold XB, twice in cold XB plus 100 mM KCl, once more in cold XB, and finally resuspended in 15 μl SDS-PAGE sample buffer. H1 filter-paper kinase assays were performed using CSF-arrested or interphase-arrested low-speed extracts as described [22].
Embryo Confocal Microscopy and FRAP
Embryos were fertilized, maintained, and staged as described [23, 24]. For live imaging, stage 13–15 embryos injected with H1-GFP and H2B-RFP mRNAs were autoscaled using the Find function and imaged every minute with a 40× oil objective on a Zeiss Aviovert200M confocal microscope running LSM software. Movies were thresholded, corrected for background, and the fluorescence ratio of H1-GFP:H2B-RFP was calculated for selected cells at timepoints during G2 and anaphase. For FRAP, two-color images were taken every half-second with a 63× objective. A 4 second photobleach at 100 % power for the Argon/488 laser was applied to a ~2 μm diameter circle on individual metaphase plates, and aligned stacks were analyzed using the FRAP Profiler plugin for ImageJ.
Highlights.
-Somatic H1 compacts interphase chromatin, but unlike H1M dissociates at mitosis
-Cdk1 phosphorylation of somatic H1 enhances its binding to mitotic chromosomes
-RanGTP releases somatic H1 from importin β/RanBP7 to promote chromatin binding
Supplementary Material
Acknowledgments
We thank S. Zhou for mass spectrometry, P. Kalab for purification of RanQ69L, E. Kieserman for help with confocal microscopy and reagents, K. Miller for preliminary experiments, and T. Maresca for advice and reagents. We thank members of the Heald and Harland labs for reagents and helpful discussions, and K. Weis and Z. Cande for comments on the manuscript. R. Heald is supported by the National Institute of Health (GM057839).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thoma F, Koller T. Influence of histone H1 on chromatin structure. Cell. 1977;12:101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- 2.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lever MA, Th'ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 4.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 5.Godde JS, Ura K. Cracking the enigmatic linker histone code. J Biochem. 2008;143:287–293. doi: 10.1093/jb/mvn013. [DOI] [PubMed] [Google Scholar]
- 6.Happel N, Doenecke D. Histone H1 and its isoforms: Contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Izzo A, Kamieniarz K, Schneider R. The histone H1 family: specific members, specific functions? Biol Chem. 2008;389:333–343. doi: 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- 8.Dasso M, Dimitrov S, Wolffe AP. Nuclear assembly is independent of linker histones. Proc Natl Acad Sci U S A. 1994;91:12477–12481. doi: 10.1073/pnas.91.26.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maresca TJ, Heald R. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol Biol. 2006;322:459–474. doi: 10.1007/978-1-59745-000-3_33. [DOI] [PubMed] [Google Scholar]
- 10.Maresca TJ, Freedman BS, Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dworkin-Rastl E, Kandolf H, Smith RC. The maternal histone H1 variant, H1M (B4 protein), is the predominant H1 histone in Xenopus pregastrula embryos. Dev Biol. 1994;161:425–439. doi: 10.1006/dbio.1994.1042. [DOI] [PubMed] [Google Scholar]
- 12.Baatout S, Derradji H. About histone H1 phosphorylation during mitosis. Cell Biochem Funct. 2006;24:93–94. doi: 10.1002/cbf.1293. [DOI] [PubMed] [Google Scholar]
- 13.Dimitrov S, Almouzni G, Dasso M, Wolffe AP. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Dev Biol. 1993;160:214–227. doi: 10.1006/dbio.1993.1299. [DOI] [PubMed] [Google Scholar]
- 14.Saeki H, Ohsumi K, Aihara H, Ito T, Hirose S, Ura K, Kaneda Y. Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci U S A. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakel S, Albig W, Kutay U, Bischoff FR, Schwamborn K, Doenecke D, Gorlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. Embo J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. Embo J. 2002;21:377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 18.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 19.Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 20.Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 21.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 22.Felix MA, Pines J, Hunt T, Karsenti E. A post-ribosomal supernatant from activated Xenopus eggs that displays post-translationally regulated oscillation of its cdc2+ mitotic kinase activity. Embo J. 1989;8:3059–3069. doi: 10.1002/j.1460-2075.1989.tb08457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2000. [Google Scholar]
- 24.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin): a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Garland Pub.; New York: 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.