Abstract
Developmental biology has evolved from a descriptive science to one based on genetic principles and molecular mechanisms. While molecular biology and genetic technologies have been the primary drivers of this transformation, synthetic strategies have been increasingly utilized to interrogate the mechanisms of embryonic patterning with spatial and temporal precision. In this review, we survey how chemical tools and engineered proteins have been used to perturb developmental processes at the DNA, RNA, protein, and cellular levels. We discuss the design principles, experimental capabilities, and limitations of each method, as well as future challenges for the chemical and developmental biology communities.
INTRODUCTION
The transformation of a single, totipotent cell into a multicellular organism requires a complex ensemble of molecular programs that are executed with spatiotemporal fidelity. Understanding how these chemical mechanisms give rise to form and function is a major objective of modern embryology, which has evolved from a descriptive science based on tissue manipulations and morphological observations to one that integrates genomic technologies and quantitative, molecular models. For example, vertebrate limb development along the proximal-distal axis requires the “apical ectodermal ridge,” epithelial tissue overlying the limb bud tip, and it is now known that fibroblast growth factors (FGFs) expressed by these cells are key mediators of this growth (Fallon et al., 1994; Niswander et al., 1993). Digit identity is similarly dictated by a posterior “zone of polarizing activity” that is now associated with limb bud cells that secrete the morphogen Sonic Hedgehog (Shh) (Niswander et al., 1994), and dorsal-ventral patterning of the limb is regulated by the dorsal ectoderm, which produces another morphogen called Wnt7a (Parr and McMahon, 1995). Genetic analyses and computational models have further established that these signaling pathways do not act independently; rather, they interact through feedback mechanisms to coordinate vertebrate limb patterning in space and time (Mackem and Lewandoski, 2009; Zeller et al., 2009).
The integration of molecular principles into embryological theory can be primarily attributed to advances in molecular biology and genomic technologies. Mutagenesis screens of model organisms such as worms (Caenorhabditis elegans), fruit flies (Drosophila melanogaster), zebrafish (Danio rerio), and mice (Mus musculus) have identified thousands of genetic loci that are required for embryonic patterning, many of which have now been mapped by positional cloning. Methods for targeted gene disruption in whole organisms, such as knockouts generated by homologous recombination or non-homologous end-joining, have allowed biologists to study embryonic patterning genes in a more deliberate manner, and collectively these studies have revealed several evolutionarily conserved genes and their downstream effectors—FGFs, Hedgehogs (Hhs), Wnts, bone morphogenetic proteins (BMPs), and Notch receptors, to name a few—that are used iteratively and combinatorially to control cell behavior and fate during embryogenesis. The completion of multiple genome-sequencing efforts has further revolutionized the study of organismal development by providing a comprehensive list of patterning molecules and their cis-regulatory elements, and whole-genome approaches are now commonly applied in developmental biology research. These latter technologies include the use of DNA microarrays or massively parallel sequencing to identify genes that are expressed in specific embryonic contexts, as well as the coupling of these methods with chromatin immunoprecipitation to uncover trans-regulatory mechanisms. High-throughput strategies are also being used to conduct genome-wide phenotypic screens in cell-based models and even whole organisms, typically through RNA interference (RNAi) methods.
While genetic methodologies have dominated developmental biology for the past three decades, it has become increasingly clear that further advances in the field will require fresh ideas and new approaches. Like many biological disciplines, embryology is becoming a quantitative science that strives to explain complex phenotypes in terms of molecular components, thermodynamic equilibria, and kinetic rates (Lewis, 2008). In addition to the vertebrate limb development studies described above, Drosophila body segmentation has been predictively modeled according to the anterior-posterior distribution of specific transcriptional activators and repressors and their cooperative affinities for binding sites within segmentation gene enhancers (Segal et al., 2008). Dorsal-ventral patterning of the frog embryo (Xenopus laevis and Xenopus tropicalis) has also been quantitatively described as opposing concentration gradients of BMPs and their extracellular antagonists that are scaled relative to embryo size through a molecular shuttling mechanism (Ben-Zvi et al., 2008). Establishing, testing, and refining new models will require an ability to perturb developmental signaling mechanisms with a spatial and temporal precision that is difficult to achieve through genetic manipulations alone. Conventional knockout technologies have limited temporal control, RNAi usually requires hours to sufficiently deplete targeted transcripts, and post-translational processes are inherently difficult to target through genetic methods. Approaches that utilize chemical concepts and synthetic elements, however, are less constrained by Nature’s molecular architecture. As a result, the developmental biologist’s toolbox now includes a growing number of chemistry-based technologies.
In this review, we provide an overview of how synthetic strategies have been used to study embryonic development in a variety of model organisms. We categorize these technologies according to their mechanisms of action, including targeted perturbations at the transcriptional, RNA, protein, and cellular levels. The use of small-molecule modulators in developmental biology has been reviewed elsewhere and will not be revisited here (Firestone and Chen, 2010; Yeh and Crews, 2003). Rather, we focus on rationally designed technologies, examine their underlying chemical and biological principles, and consider their advantages and limitations. We also discuss emerging chemical approaches that have not yet been applied in model organisms but could prove to be valuable tools for in vivo studies. We hope that this review will not only provide its readers with a synopsis of this nascent field but also inspire further exploration of embryonic development through the lens of chemical biology.
CHEMICAL CONTROL OF GENE TRANSCRIPTION
Dynamic, stereotypic gene expression is a primary driver of embryonic patterning, and developmental biologists have therefore focused on technologies that enable in vivo transcriptional control. Commonly used gene inactivation strategies include knockouts through homologous recombination (Mansour et al., 1988), TILLING (Targeting Induced Local Lesions in Genomes) (McCallum et al., 2000), and RNAi-dependent gene silencing (Fire et al., 1998). The transcription of exogenous genes throughout the organism or in a cell-specific manner can also be readily achieved through the transient introduction or genomic integration of oligonucleotide constructs. While these approaches have yielded key insights into the genetic programs that give rise to tissue form, they are typically constitutive or dependent on endogenous trans-regulatory mechanisms. Genetic recombination technologies such as the Cre/loxP and FLP/FRT systems can provide greater conditionality in some cases (Dymecki, 1996; Gu et al., 1994), but even these methods are constrained by the promoters used to activate Cre or FLP recombinase expression. Small molecules, however, can alter embryonic gene transcription with precise spatial, temporal, and/or dose control. In particular, ligand-inducible transcription factors and recombinases have been used to achieve transcriptional regulation in vivo, combining the rapidity and tunability of chemical modulation with the specificity of genetic targeting.
Small molecule-dependent transcription factors
Like several chemical approaches, the development of synthetic transcriptional activators that are ligand-actuated has been largely inspired by Nature’s regulatory mechanisms. Signaling proteins that mediate hormonal responses were first co-opted as tools for transcriptional control, as it was found that the ligand-binding domains of the glucorticoid, estrogen, and ecdysone receptors (GR, ER, and EcR) can be fused to heterologous proteins to modulate their function (Figure 1a) (Christopherson et al., 1992; Eilers et al., 1989; Green and Chambon, 1987). As with endogenous hormone receptors, these chimeric transactivators are complexed with cytosolic proteins such as heat shock protein 90 (Hsp90), and ligand binding induces a protein conformational change that disrupts this cytosolic complex and allows the factor to translocate into the nucleus. Coupling these ligand-binding domains to DNA-binding and transactivating polypeptides therefore enables the small molecule-dependent transcription of genes driven by the corresponding response elements.
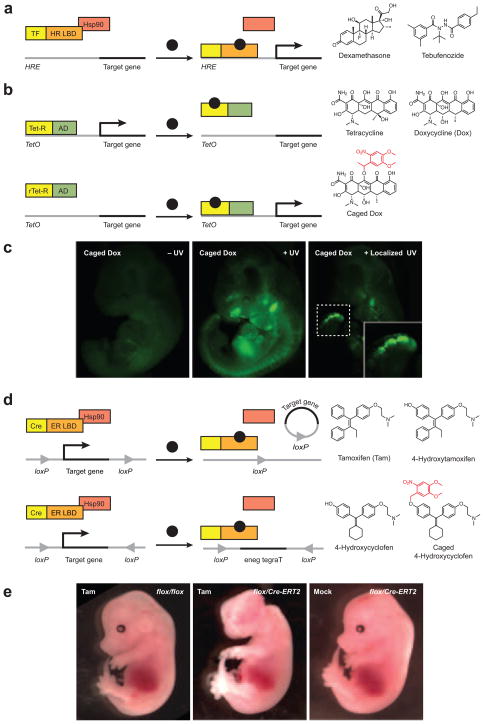
Figure 1. Synthetic control of gene transcription.
(a) Schematic representation of hormone receptor-based transactivator systems. Chemical structures for representative agonists are shown. TF, transcription factor; HR LBD, hormone receptor ligand-binding domain; HRE, hormone response element. (b) Schematic representation of tetracycline repressor-based transcription systems, including the “Tet-Off” and “Tet-On” configurations. Chemical structures of tetracycline and its synthetic derivatives are shown. Tet-R, Tet repressor; rTet-R, reverse Tet transactivator; AD, activation domain; TetO, Tet repressor operon. Although hormone receptor-and tetracycline repressor-based transactivators can each form protein dimers, monomeric forms of these proteins are depicted for schematic simplicity. (c) Control of GFP expression in mice embryos using the “Tet-On” system and caged doxycycline. Both global and localized (dashed box and inset) irradiation are shown. Adapted with permission (Cambridge et al., 2009; Copyright 2009, Nature America, Inc). (d) Schematic representation of tamoxifen-induced, Cre-dependent recombination, including excision and inversion reactions. The recombinase homodimer is depicted as a monomer for schematic simplicity, and chemical structures of tamoxifen and its derivatives are shown. ER LBD, estrogen receptor ligand-binding domain. (e) Tamoxifen-induced “self knockout” of the Otx2 gene in mouse embryos using the Cre-ERT2 system. Abnormal craniofacial and brain development is observed only in Otx2flox/Cre-ERT2 embryos treated with the ER agonist. Adapted with permission (Fossat et al., 2006; Copyright 2006, European Molecular Biology Organization).
Since their introduction during the late 1980s, chimeric hormone receptors have been widely used to regulate gene expression in cultured cells and embryos. For example, Kolm and Sive fused the myogenic transcription factor MyoD to the ligand-binding domains of either GR or ER, and frog embryos expressing these chimeric transactivators exhibited ectopic muscle cells upon treatment with dexamethasone or estradiol, respectively (Kolm and Sive, 1995). The Zivkovic laboratory generalized this approach by coupling the GR ligand-binding domain to the DNA and activation domains of the yeast transcription factor Gal4, enabling the dexamethasone-dependent expression of any UAS-driven transgene (de Graaf et al., 1998). Using the Gal4-GR transactivator, they chemically induced the expression of Xwnt1 and XactivinbB in zebrafish embryos, resulting in morphological phenotypes associated with the ectopic action of these morphogens. Due to potential crosstalk between these transactivator systems and endogenous GR or ER signaling, our laboratory subsequently developed a ligand-gated transcription factor containing the EcR ligand-binding domain, exploiting the orthogonality of this insect-specific hormone receptor to vertebrate signaling mechanisms (Esengil et al., 2007). By fusing this polypeptide to the Gal4 DNA-binding region and a minimal activation domain from herpes simplex virus protein VP16 and expressing this transactivator under the control of tissue-specific promoters, we were able to achieve heart and skeletal muscle-specific green fluorescent protein (GFP) expression in zebrafish embryos upon exposure to the synthetic EcR ligand tebufenozide.
The other commonly used technology for chemically inducible gene expression takes advantage of the tetracycline resistance mechanism in Escherichia coli (Thaker et al.). Bacterial resistance to this antibiotic involves the expression of a transmembrane efflux pump (TetA), which is regulated by a transcriptional repressor (TetR). In the absence of tetracycline (or its synthetic derivative doxycycline), TetR binds to operator sequences (tetO) in the tetA regulatory domain; the binding of tetracycline to TetR, however, promotes its dissociation from tetO, enabling tetA transcription and antibiotic clearance from the bacteria. Based on this system, Gossen and Bujard established two ligand-inducible gene transcription strategies that can be applied in mammals (Figure 1b). In the “Tet-Off” system, the DNA- and ligand-binding domains of TetR are fused with the VP16 activation domain to form a constitutive transcriptional activator (tTA) that is inhibited by tetracycline (Gossen and Bujard, 1992). The “Tet-On” system utilizes a “reverse” form of tTA (rtTA) that actually requires tetracycline for tetO binding (Gossen et al., 1995). Using these complementary methods, transgene expression can be inhibited or activated in a ligand-dependent manner, and it has been adapted for use in fruitflies (Bieschke et al., 1998), zebrafish (Huang et al., 2005), frogs (Ridgway et al., 2000), and mice (Furth et al., 1994; Kistner et al., 1996) with varying degrees of success. In one particularly elegant application of this technology, the Tilghman laboratory studied the role of endothelial receptor B (EDNRB) in murine embryogenesis by integrating either tTA or rtTA into one Ednrb locus and tetO responsive elements into the other allele (Shin et al., 1999). Administration of the tetracycline derivative doxycycline to EdnrbtTA/EdnrbtetO or EdnrbrtTA/EdnrbtetO embryos therefore suppressed or induced Ednrb transcription, respectively, while maintaining the tissue specificity of the endogenous Endrb gene. By varying the timing of doxycycline treatment, Tilghman and her co-workers were able to determine the developmental stages during which EDNRB signaling is required for the differentiation of neural crest cells into melanoblasts and enteric neurons.
Collectively, these examples demonstrate the temporal control of embryonic transcription that can be achieved through small-molecule ligands and synthetic receptors. They also illustrate the reliance of these systems on endogenous cis-regulatory elements for spatial control. While recapitulating native gene expression patterns is adequate for certain applications, promoter elements that will restrict transcription to targeted cell populations are not always available, and in some cases it would be advantageous to target tissues according to morphological cues alone. Synthetic chemistry has enabled developmental biologists to circumvent these limitations. In particular, the conjugation of caging groups to hormone or tetracycline receptor ligands provides an extra level of conditionality that is orthogonal to the timing of ligand addition and the spatiotemporal dynamics of transactivator expression. The precision of optical techniques makes photoactivatable ligands especially versatile reagents, and a number of light-activatable hormone receptor ligands have been reported (Link et al., 2004; Link et al., 2005). Although the control of in vivo gene transcription with these synthetic compounds has not yet been described, caged doxycycline has been successfully used to induce embryonic gene expression in a tissue-specific manner.
As a proof of principle, Cambridge and co-workers generated transgenic mice that ubiquitously express an rtTA-based transactivator and carry a tetO-dependent GFP reporter (Cambridge et al., 2009). Embryos surgically removed from these mice and cultured with the 1-(4,5-dimethoxy-2-nitrophenyl)ethyl ether of doxycycline (DMNPE-doxycycline) exhibited light-dependent GFP expression that could be induced throughout the organism or localized to a targeted area such as the dorsal root ganglia (Figure 1c). Transgenic Xenopus tadpoles that expressed rtTA and carried tetO-GFP reporter constructs responded to DMPNE-caged doxycycline in a qualitatively similar manner, suggesting the generality of this approach. However, the lower levels of GFP fluorescence observed in irradiated, DMPNE-doxycycline-treated tadpoles in comparison to those treated with unmodified doxycycline indicate that organismal penetrance of the caged ligand or its diffusion after photoactivation may be limiting factors in some experimental contexts.
Small molecule-dependent recombinases
Although hormone receptor ligand-binding domains are used endogenously to gate transcriptional activity, in principle they could be used to control the function of any protein that acts within the nucleus. Realizing this potential, the Chambon laboratory fused the bacteriophage P1 recombinase Cre to a mutated ER ligand-binding domain that responds to the synthetic inducer 4-hydroxytamoxifen but not endogenous estrogens (Cre-ERT; G525R human ER), thereby creating a small molecule-dependent genetic recombination system (Feil et al., 1996). Cells expressing the Cre-ERT chimera will undergo 4-hydroxytamoxifen-dependent excision or inversion of genomic DNA flanked by loxP sites, depending on whether these 34-base pair regions have the same or opposite orientations, respectively (Figure 1d). McMahon and co-workers provided the first demonstration that the Cre-ERT system can be used to effect genomic recombination in mouse embryos in utero, generating transgenic mice with Wnt1 enhancer-dependent Cre-ERT expression and a β-galactosidase reporter whose transcription requires the excision of loxP site-flanked cassette (Danielian et al., 1998). Injection of 4-hydroxytamoxifen into pregnant transgenic mice induced Cre-mediated recombination and β-galactosidase expression in the embryonic central nervous system, which coincides with the Wnt1 expression pattern.
The utility of this system for studying vertebrate development has been further established by the Lamonerie laboratory (Fossat et al., 2006). Mirroring Tilghman’s tTA/rtTA strategy for the Ednrb gene, they generated transgenic mice in which one allele of the Otx2 gene was replaced with cDNA encoding Cre-ERT2, another 4-hydroxytamoxifen-specific inducible recombinase (G400V/M543A/L544A mouse ER) (Feil et al., 1997). The other Otx2 allele was modified to have unidirectional loxP sites flanking the second exon (“floxed” Otx2). One major advantage of this “self-knockout” approach is that the Cre-ERT2 transgene will disrupt the remaining Otx2 allele only in cells that would normally express this homeobox transcription factor at the time of 4-hydroxytamoxifen exposure; cells that would express Otx2 at later developmental stages are spared. Accordingly, Otx2flox/Cre-ERT2 embryos in pregnant mice injected with tamoxifen prodrug 7.5 days post conception (E7.5) exhibited head phenotypes consistent with a loss of Otx2 function (Figure 1e). By varying the timing of tamoxifen treatment, Lamonerie and co-workers were able to determine that Otx2 is required for craniofacial development before E10.5 and midbrain patterning between E10.5 and E16.5. The Cre-ERT2/loxP system has also been validated in zebrafish embryos, and tamoxifen has been used to induce the excision and inversion of transgenes in this model organism (Hans et al., 2009).
In analogy to DMNPE-caged doxycycline, photoactivatable versions of tamoxifen could broaden the utility of Cre-ERT2 as a tool for embryological research. Toward this goal, Jullien and co-workers have synthesized and evaluated a number of caged tamoxifen derivatives (Sinha et al., 2010). Since ultraviolet light irradiation of 4-hydroxytamoxifen promotes its photoisomerization and decomposition, they established that 4-hydroxycyclofen, a photostable structural analog, can interact with the ERT2 ligand-binding domain with comparable potency (Figure 1d). 4,5-dimethoxy-2-nitrobenzyl (DMNB)-caged 4-hydroxycyclofen can therefore convey light-dependent nuclear translocation of proteins containing the ERT2 ligand-binding domain, as demonstrated in cultured cells and zebrafish embryos expressing a GFP-ERT2 chimera containing a nuclear localization sequence. However, whether DMNB-4-hydroxycyclofen will be an effective reagent for light-controlled genomic recombination in vivo remains uncertain. The success of this approach will depend on how the kinetics of 4-hydroxycyclofen efflux from irradiated cells compares to the rate of Cre-ERT2-mediated DNA recombination. Consistent with this potential limitation, Koh and co-workers have observed that even caged 4-hydroxytamoxifen derivatives that covalently modify the ER ligand-binding domain cannot effectively induce Cre-ERT-mediated recombination in cultured cells after photoactivation (Link et al., 2005).
In principle, these approaches could also be applied to the FLP/FRT recombination system, which has been widely used in fruit flies and to a more limited extent in frogs, zebrafish, and mice. The versatility of Cre/loxP-based technologies has also been expanded with the development of heterologous loxP sequences that efficiently recombine with themselves but not the wildtype loxP site (Siegel et al., 2001). More sophisticated genomic modifications can be achieved by combining these orthogonal recombination platforms, including serial inversion and excision steps to switch from the expression of one gene to another (known as the FLEx switch method) (Schnutgen et al., 2003). Nested, orthogonal recombination sites can also be used to allow sequential inversion steps (Boniface et al., 2009), potentially allowing genes to be switched on and off. Combining these strategies with ligand-regulated recombinases will allow developmental biologists to manipulate gene function in vivo with improved dexterity.
CHEMICAL CONTROL OF RNA FUNCTION
Although embryonic gene expression has been traditionally studied at the transcriptional level, tissue patterning also involves the regulation of RNA splicing, translation, and degradation. For example, splice variants of Delta-C, a Notch receptor ligand, have been found to have distinct signaling capabilities during embryogenesis, differentially affecting midline development and somitogenesis in zebrafish (Mara et al., 2008). Dozens of microRNAs (miRNAs) are expressed in a stereotypic manner to modulate RNA function in specific cell populations, and individual miRNAs have been implicated in the specification of muscle, lymphocytes, neurons, germ cells, and other cell types (Shi and Jin, 2009). Chemical technologies that enable the targeting of specific RNAs therefore constitute another effective means for studying the molecular mechanisms of embryonic patterning.
One strategy has been to use ligand-inducible transcription to control the expression of exogenous short hairpin RNAs (shRNAs), which are processed by endogenous miRNA pathway machinery into short interfering RNAs (siRNAs) and consequently induce the degradation of targeted transcripts (Paddison et al., 2002). The tamoxifen/Cre-ERT platform has been used to selectively silence expression of the pluripotency factor Nanog in primordial germ cells (Yamaguchi et al., 2009) and to knockdown FGF receptor 2 (Fgfr2) expression in the developing limb (Coumoul et al., 2005). Reversible shRNA-mediated gene silencing has been achieved in mice through the doxycycline/Tet-on system (Seibler et al., 2007), and EcR-based transactivators have similarly employed in cultured cells (Gupta et al., 2004; Rangasamy et al., 2008). Since these methods are straightforward applications of the transcriptional technologies described above, we do not discuss them further here. Rather we highlight in this section approaches that utilize synthetic chemistry to directly modulate RNA function in embryos. We also discuss the application of ligand-gated riboswitches for inducible gene expression in vivo.
Caged RNA molecules
The microinjection of exogenous mRNAs and short interfering RNAs (siRNAs) into embryos is a common technique for studying gene function, especially at early developmental stages. In particular, mRNAs are frequently injected into early-stage ascidian (Ciona intestinalis), sea urchin (Lytechinus variegatus and other species), zebrafish, and frog embryos to assess gain-of-function phenotypes, and siRNAs are typically introduced into worms and fruitflies to induce loss-of-function phenotypes. Although these methods are simple and effective, their lack of conditionality limits their utility for studying genes that have pleiotropic functions during embryogenesis.
Since RNAs are not membrane-permeable, introducing mRNAs and siRNAs into embryos at later developmental time points presents a major technical hurdle. Developmental and chemical biologists have therefore pursued the caging of these molecules with light-sensitive groups, as in principle these latent RNAs could be microinjected at the one-cell stage and subsequently photoactivated in targeted cell populations. Blocking RNA function through chemical modifications, however, is associated with its own technical issues. For example, exogenous mRNAs are typically generated by in vitro transcription, necessitating the addition of caging groups to the full-length transcript rather in a site-specific manner.
The Okomoto group attempted to tackle this challenge by reacting in vitro transcribed mRNAs with 6-bromo-4-diazomethyl-7-hydroxycoumarin (diazo-BHC), thereby modifying the phosphodiester backbone with photocleavable groups (Figure 2a) (Ando et al., 2001). As a proof of principle, mRNAs encoding GFP and β-galactosidase reporters were caged with diazo-BHC and then injected into one-cell-stage zebrafish embryos. Subsequent exposure of the embryos to 365-nm light induced reporter expression as expected, yet the resulting levels of GFP fluorescence or β-galactosidase activity were approximately one-eighth that achieved with the microinjection of equivalent doses of unmodified mRNAs. In addition, the fold-change in reporter expression induced by photolysis was only four- to five-fold, a dynamic range that will be insufficient for many experimental applications. This low efficacy undoubtedly reflects the requirement of multiple caging groups to sufficiently abrogate mRNA translation (approximately 30 modified phosphodiesters per kilobase of RNA) and the low efficiency by which they can all be removed. Another limiting factor is the chemical instability of the caged mRNAs, as each BHC phosphotriester is susceptible to intramolecular attack by the ribose 2′-OH and subsequent hydrolysis. Similar shortcomings were observed by the Friedman group when they sought to cage siRNAs with 1-(1-diazoethyl)-4,5-dimethoxy-2-nitrobenzene (diazo-DMNB) (Shah et al., 2005), indicating that these issues are inherent to the general approach.
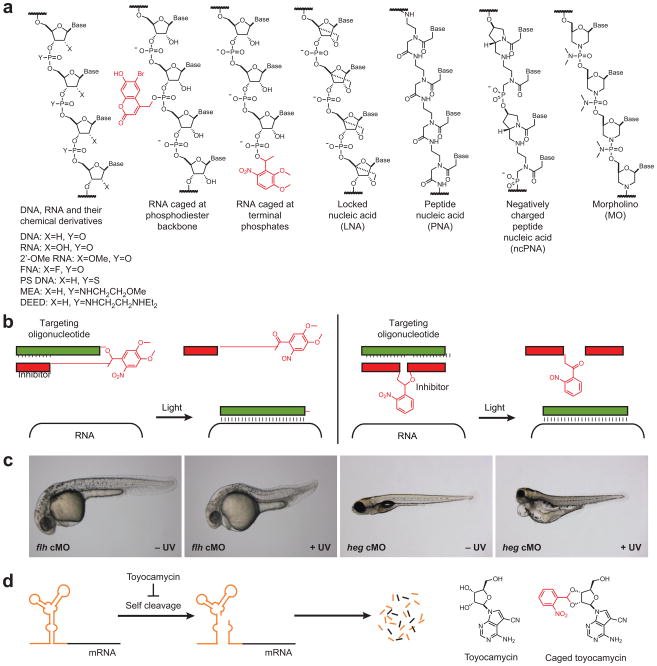
Figure 2. Synthetic control of RNA function.
(a) Chemical structures for natural oligonucleotides, their modified analogs, and non-natural oligonucleotides. Caging groups are shown in red. (b) Schematic representation of the caged MO hairpin (left) and PhotoMorphs™ technologies (right). (c) Light-controlled silencing of flh and heg in zebrafish embryos using caged MO hairpins. Adapted with permission (Ouyang et al., 2009; Copyright 2009, American Chemical Society). (d) Schematic representation of a small molecule-dependent riboswitch (orange) that regulates transcript stability. Chemical structures of toyocamycin and its caged derivative are shown.
The amenability of siRNAs to chemical synthesis does provide some possible ways forward. For example, siRNA analogs composed of 2′-deoxy-2′-fluoro nucleic acids (siFNAs) still promote the degradation of targeted transcripts, but their 2′-fluoro modification renders them less susceptible to hydrolysis (Figure 2a). siFNAs caged with diazo-DMNB consequently exhibit greater stability in vivo, and Monroe and co-workers have achieved light-inducible silencing of GFP expression in zebrafish embryos using these reagents (Blidner et al., 2008). The Friedman laboratory has recently reported an alternative approach, in which the terminal phosphates of blunt-ended siRNA precursors were selectively modified by diazo-DMNB to yield stable caged duplexes (Figure 2a) (Shah et al., 2009). Photoactivation of these oligonucleotides in cultured cells led to a six-fold decrease in gene expression, a dynamic range three times greater than that observed for siRNAs with caged phosphotriester linkages. Since siRNA- and siFNA-dependent toxicity has reported in zebrafish embryos, the promise of these new synthetic oligonucleotides as functional genomic tools may be best realized in worms, fruit flies, and other organisms that are more tractable to RNAi.
Non-natural, RNA-targeting oligonucleotides
Although RNAi is an effective method for loss-of-function studies in worms and fruit flies, it is less commonly used in other model organisms such as zebrafish and frogs. In addition to their possible cytotoxic effects, double-stranded RNAs have limited perdurance in vivo, effectively restricting their application to early developmental genes. Synthetic chemistry has yielded non-natural oligonucleotides that could potentially overcome some of these limitations, including 2′-OMe RNAs, phosphorothioate (PS) DNAs, methoxyethylamine (MEA) and N,N-diethylethylenediamine (DEED) phosphoramidates, locked nucleic acids (LNAs), peptide nucleic acids (PNAs), morpholinos (MOs), and chimeric oligomers composed of more than one nucleoside class (Figure 2a). Each oligonucleotide utilizes standard DNA bases while substituting the ribose backbone and/or phosphodiester linkages with other chemical substituents, resulting in enhanced in vivo stability. Depending on the specific structural modifications, hybridization of the oligomers to their complementary RNA targets either promotes RNase H-dependent transcript degradation or acts as a steric block at the site of hybridization.
Although there have been a few reports of gene silencing in frog embryos using PS DNAs (Woolf et al., 1990), MEA or DEED phosphoramidate/DNA hybrids (Dagle et al., 1990; Dagle et al., 2000; Veenstra et al., 2000), and LNA/PS DNA chimeras (Lennox et al., 2006), these RNase H-active antisense agents are not widely used by the developmental biology community. Similarly, PS DNAs and 2′-OMe RNAs are not generally used to study zebrafish embryogenesis, as they are both cytotoxic at doses sufficient to alter RNA function (Tomasini et al., 2009). RNAse H-inactive antisense oligonucleotides such as MOs have proven to be more effective reverse-genetic tools in vivo, and they have been used to interrogate gene function in the embryos of ascidians (Satou et al., 2001), sea urchins (Coffman et al., 2004), zebrafish (Nasevicius and Ekker, 2000), frogs (Heasman et al., 2000), chickens (Gallus gallus) (Kos et al., 2001), and mice (Coonrod et al., 2001). These oligomers are composed of a nuclease-resistant morpholine-phosphorodiamidate backbone, and typically 25-base oligonucleotides are injected into embryos at the one-cell stage, after which they can remain functional for days (Nasevicius and Ekker, 2000; Summerton and Weller, 1997). MOs that hybridize to the start site or its flanking 5′ untranslated region (UTR) can inhibit RNA translation, allowing them to block both maternal and zygotic RNA function (Summerton, 1999); those that target intron-exon junctions can yield misspliced zygotic transcripts that that include introns or skip exons, frequently introducing a reading-frame shift and a premature stop codon (Draper et al., 2001). MOs have also been used to target miRNAs or their precursors to investigate the functions of these non-coding RNAs (Flynt et al., 2007; Kloosterman et al., 2007). In fact, thousands of MOs have been used to study zebrafish development alone, targeting genes such as the morphogen wnt8a, the Hh pathway component suppressor of fused (sufu), and the mesodermal transcription factor no tail-a (ntla).
PNAs also inhibit RNA function through RNase H-independent steric blockade, but they have been applied in vivo to a much lesser degree than MOs. This limited use is due in part to the low water solubility of PNA oligomers, making it difficult to achieve the micromolar concentrations required to block gene expression in organisms. Farber and co-workers circumvented this technical hurdle by utilizing negatively charged PNAs (ncPNAs; also known commercially as gripNAs™), which have a backbone composed of trans-4-hydroxy-L-proline/phosphonate polyamide subunits (Urtishak et al., 2003). ncPNAs used for in vivo studies are typically shorter in length than MOs (18 bases) since they hybridize to RNA with stronger affinity. ncPNAs also exhibit greater sensitivity to base-pair mismatches than MOs. In practice, both RNase H-inactive antisense oligonucleotides can be effective gene-silencing tools in embryos, and in some cases one reagent is more efficacious than the other even when targeting the same RNA sequence. For example, the Farber laboratory observed that an ncPNA targeting the homeobox gene dharma (dha) start site was able to recapitulate dha mutant phenotypes, whereas a MO against the same region was ineffective. Yet only a handful of ncPNAs targeting embryonic transcripts have been reported in the scientific literature, and all of these genes are expressed early in development. It is therefore possible that the amide bond-containing ncPNAs are more labile in vivo than the phosphorodiamidate-based MOs.
MOs and ncPNAs do have certain limitations, however. Although synthetic oligonucleotides have proven to be valuable tools for studying developmental biology, phenotypes induced by these reagents must be interpreted with caution. Both reagents can have off-target effects that lead to embryonic defects, including p53-dependent cell death. Appropriate control experiments therefore must be conducted to confirm the specificity of these antisense reagents in vivo, including recapitulation of the phenotype with multiple MOs or ncPNAs against the same gene and the use of base pair-mismatch control oligonucleotides (Eisen and Smith, 2008). The Farber and Ekker labs have also demonstrated that MO and ncPNA off-target effects can be minimized by concurrently inhibiting p53 expression using a p53 MO, thereby unmasking the specific function of the gene of interest (Robu et al., 2007).
In addition, the constitutive activities of conventional MOs and ncPNAs renders them less effective for studying genes that are expressed in more than one tissue and/or at multiple developmental stages. Caged antisense oligonucleotides could therefore be more versatile probes of gene function, and those actuated by light would be particularly useful in optically transparent embryos such as those produced by ascidians, sea urchins, and zebrafish. Toward this goal, our laboratory caged MOs by using a DMNB-based photocleavable linker to tether the 25-base gene-targeting oligonucleotide to a complementary inhibitor composed of MO nucleosides (Figure 2b) (Shestopalov et al., 2007). Intramolecular Watson-Crick base-pairing produces a MO hairpin that cannot effectively hybridize to its RNA target, and linker photolysis with 365-nm light liberates the targeting MO to allow RNA function blockade. Using this approach, we have conditionally regulated ntla expression in zebrafish embryos with spatial and temporal precision, demonstrating that this T-box family transcription factor is required for notochord cell fate commitment, migration, and differentiation. We have also generated caged MOs against several other developmental patterning genes, including floating head (flh), heart of glass (heg), and ets varient gene 2 (etv2) (Figure 2c) (Ouyang et al., 2009).
A similar ncPNA caging strategy was concurrently reported by Dmochowski and coworkers, in this case using 18-base ncPNAs, complementary 2′-OMe RNA inhibitors, and a nitrobenzyl-based linker (Tang et al., 2007). By microinjecting caged ncPNAs targeting dha and the BMP antagonist chordin (chd) into zebrafish embryos and irradiating them globally with ultraviolet light, they were able to induce mutant phenotypes. Interpreting these developmental defects, however, is complicated by the release of 2′-OMe RNA upon linker photolysis, as these synthetic oligonucleotides have been shown to be toxic to zebrafish embryos (Tomasini et al., 2009).
More recently, the Mayer laboratory in collaboration with SuperNova Life Sciences developed an alternative caging strategy that employs MO/caged RNA duplexes called PhotoMorphs™ (Figure 2b) (Tomasini et al., 2009). Like the caged MOs and ncPNAs described above, these reagents utilize a single light-sensitive group, in this case a nitrobenzyl-based linker that joins two 12-base RNA oligonucleotides. The MO/caged RNA duplex resists strand exchange with endogenous transcripts, likely reflecting the fact that RNA-binding proteins and RNA tertiary structure restrict the accessibility of the latter. Photolysis of the caged RNA shifts the binding energetics in favor of MO/mRNA hybridization, and PhotoMorphs™ have been used to silence the expression of Ntla, E-cadherin, and Rheb in zebrafish embryos. MO activity, however, is not fully restored upon irradiation, as the caged RNA must be used in five- to ten-fold molar excess to compensate for its gradual degradation by endogenous nucleases. Unfortunately, other synthetic oligonucleotides tested by the Mayer laboratory were found to be toxic to zebrafish embryos, preventing their use as caged inhibitors. The PhotoMorph™ system is therefore conceptually similar to having multiple caging groups per oligonucleotide; more than one caged RNA must be photolyzed to activate each MO molecule, and the dynamic range of inducible activity is therefore limited.
Small molecule-dependent riboswitches
While synthetic oligonucleotides and their caged derivatives have proven to be valuable tools for the developmental biology community, their versatility is offset by their irreversible activities. Conventional MO and ncPNAs block RNA function in a constitutive manner, and caged versions reported to date cannot be turned off after they have been photoactivated. Since embryogenesis utilizes a limited number of signaling molecules in a dynamic, iterative manner, technologies that enable reversible RNA control would advance our understanding of patterning mechanisms. As with the hormone receptor family of transcriptional modulators, Nature has provided us with examples for how conditional RNA regulation can be achieved.
For instance, certain bacteria, fungi, and plant transcripts contain cis-regulatory elements that respond to endogenous metabolites, such as thiamine pyrophosphate, glycine, and glucosamine-6-phosphate. Upon metabolite-binding, these RNA sequences undergo conformational changes to adopt structures that either terminate transcription prematurely, prevent ribosomal docking, or generate self-cleaving ribozymes. Such mechanisms enable microbes and plants to modulate their expression of biosynthetic enzymes in response to changes in metabolite concentrations. The Mulligan laboratory pioneered the use of riboswitches to control mammalian gene expression by inserting a number of candidate ribozyme sequences into various regions of a β-galactoside reporter gene (Yen et al., 2004). Through this survey and subsequent optimization studies, they determined that a mutant form of the self-cleaving Sm1 ribozyme from the trematode Shistosoma mansoni can prevent reporter expression when inserted into the 5′ UTR (Figure 2d). By conducting a chemical library screen, they also discovered that the nucleoside analog toyocamycin inhibits Sm1 self-cleavage and rescues gene expression (Figure 2d) (Yen et al., 2006; Yen et al., 2004). Toyocamycin-dependent RNA translation could be achieved in both cultured cells and in mice infected with an adeno-associated virus encoding a ribozyme-firefly luciferase transgene, and in principle this technology could be used to control embryonic gene expression. Subsequent studies by the Mulligan group revealed that inhibition of RNA self-cleavage by toyocamycin actually requires its covalent incorporation into the transcript itself rather than simply RNA/toyocamycin binding (Yen et al., 2006). The kinetics by which transgene expression is lost upon toyocamycin removal will consequently depend upon the degradation rate of transcripts containing this nucleoside derivative. In principle, greater temporal control could therefore be achieved with small molecules that recapitulate the non-covalent modulation of riboswitch function by endogenous metabolites.
Deiters and co-workers have extended the capabilities of this approach by synthesizing a nitrobenzyl dioxolane derivative of toyocamycin that liberates the nucleoside upon 365-nm light exposure and intracellular hydrolysis (Figure 2d) (Young et al., 2009). Thus, light-activated gene expression could be achieved by combining these methodologies. It is also possible that orthogonal small molecule/riboswitch systems could be used in parallel to independently control the expression of multiple genes in a developing organism.
CHEMICAL CONTROL OF PROTEIN FUNCTION
The majority of technologies described in the preceding sections combine the specificity of genetically encoded components with the rapid kinetics of small-molecule binding or photochemical transformations. In most cases, however, the temporal precision by which these systems can perturb embryonic gene function is limited by endogenous processes. For techniques that activate gene expression, possible rate-determining steps include gene transcription, RNA splicing or translation, or post-translational modifications required for protein function. Methods for inhibiting oligonucleotide function are contingent on RNA and/or protein degradation rates. Technologies that act directly on signaling proteins could modulate patterning mechanisms with greater speed, in principle enabling the study of developmental processes that occur on the minute timescale. Indeed, the widespread application of small molecules that selectively target Hh, Wnt, BMP, or Notch pathway components exemplifies the power of this approach. Since implementing small-molecule modulators in a genome-wide manner has certain technical challenges—the serendipitous nature of compound discovery and the difficulty of establishing target specificity—we focus here on strategies for protein regulation that are potentially generalizable. Current technologies can be grouped into two categories: (1) methods control protein splicing or stability; and (2) those that are designed to regulate individual members of specific protein families.
Small molecule-dependent protein splicing
Rather than regulating protein function at the translational level, an alternative strategy would be to induce the assembly of active proteins from latent polypeptides. One mechanism that Nature has evolved for post-translational protein engineering is the use of intein domains that catalyze their self-excision from polypeptide precursors and the simultaneous ligation of flanking sequences (exteins) (Paulus, 2000). Most endogenous inteins mediate cis-splicing within single-chain polypeptides, but at least one intein subfamily that can couple two protein domains in trans has been discovered (Wu et al., 1998). In the latter reaction, the intein domain is split into two inactive fragments, each of which is linked to a portion of another protein. Reconstitution of the intein domain leads to its activation and trans-splicing of the tethered polypeptide chains.
These remarkable reactions have prompted chemical biologists to create synthetic congeners that enable small-molecule control of protein maturation. For example, the Muir laboratory developed an artificial split-intein system by separating the cis-splicing VMA intein of Saccharomyces cerevisiae into two halves and coupling them to individual, complementary fragments of a heterologous protein (Figure 3a) (Mootz and Muir, 2002). One split-intein fusion protein was then tagged with FK506-binding protein 12 (FKBP12) and the other with the FKBP12/rapamycin-binding domain of mammalian TOR (FRB). Co-expression of the split-intein system in cells and treatment with rapamycin leads to formation of the FKBP12/rapamycin/FRB complex, reconstitution of the VMA intein, and trans-splicing of the heterologous polypeptides. Using this technology, Muir and co-workers have achieved rapamycin-induced splicing of inactive firefly luciferase fragments in fruit flies, which can be tuned by varying the rapamycin dose or co-administering the FKBP12 ligand ascomycin to competitively inhibit FKBP12/rapamycin/FRB complex formation (Schwartz et al., 2007). In principle, this approach could be broadly applied to study embryonic patterning mechanisms through the use of rapamycin analog/FRB mutant pairs that are orthogonal to endogenous signaling proteins (see below).
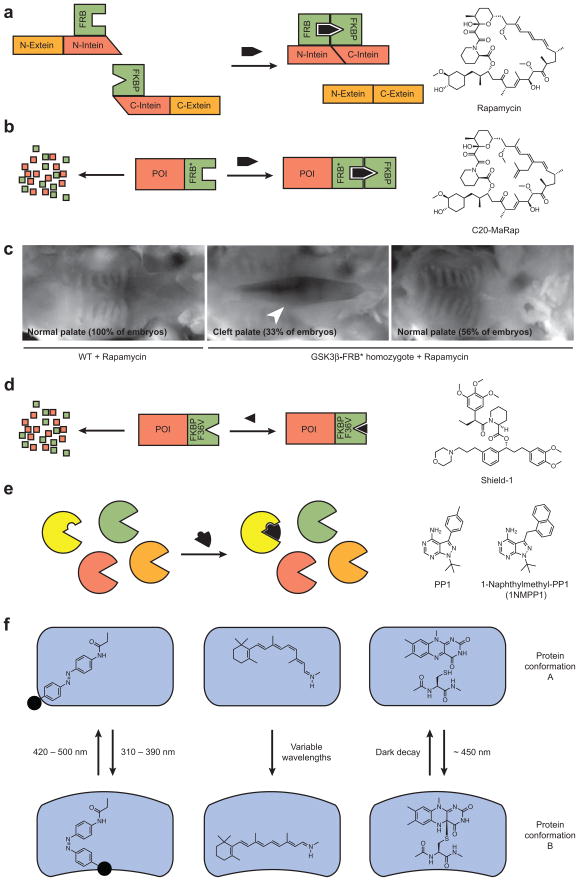
Figure 3. Synthetic control of protein function.
(a) Schematic representation of rapamycin-induced polypeptide splicing using fusion proteins composed of FRB, FKBP, and split inteins. (b) Schematic representation of ligand- and FKBP-dependent stabilization of FRB*-containing fusion proteins and the chemical structure of the FRB*-specific rapamycin analog C20-MaRap. POI, protein of interest. (c) Rapamycin-dependent rescue of GSK3β activity in homozygous GSK3β-FRB* knock-in mouse embryos. Rapamycin has no effect on wildtype embryos during the two-day drug regimen, but the majority of GSK3β-FRB* mice are fully rescued from cleft palate defects (white arrowhead) upon rapamycin treatment. Partial rescues were also observed (data not shown). Adapted with permission (Liu et al., 2007; Copyright 2007, Nature Publishing Group). (d) Schematic representation of ligand-dependent stabilization of FKBP mutant-containing fusion proteins and the chemical structure of Shield-1. (e) Schematic representation of the “bump-and-hole” strategy for achieving targeted protein inhibition with genome-wide specificity. Chemical structures of promiscuous (PP1) and mutant kinase-specific (naphthylmethyl-PP1) inhibitors are shown. (f) Schematic representation of light-regulated protein conformation and function using synthetic and endogenous cofactors. Azobenzene-, retinal-, and flavin mononucleotide-based approaches are shown, with the azobenzene-linked small-molecule modulator depicted as a solid black circle.
Liu and co-workers have reported a complementary method for conditional protein splicing that involves insertion of the ER ligand-binding domain into the RecA intein from Mycobacterium tuberculosis (Buskirk et al., 2004). By subjecting this synthetic construct to several cycles of mutagenesis and selection, they identified chimeric inteins that can mediate cis-splicing of flanking polypeptides in a 4-hydroxytamoxifen-dependent manner. The technology was subsequently used to regulate the Gli family of Hh pathway transcription factors in cultured cells (Yuen et al., 2006). Although the 4-hydroxytamoxifen-liganded intein fusions could generate only about one-third of the Gli transcriptional response observed with wildtype Gli1, this level of efficacy was sufficient to differentiate pluripotent mesenchymal cells into osteoblasts. Whether the intein-ER system in its current form will be applicable in vivo awaits further study.
Small molecule-dependent protein degradation
Rapamycin-mediated hetero-dimerization of FKBP12- and FRB-tagged proteins has also been exploited to attain conditional protein degradation. For instance, the Muir laboratory has developed a methodology that utilizes split ubiquitin for the rescue of function (SURF), in which the protein of interest is fused to a degron via the FRB domain and half of the ubiquitin polypeptide (Pratt et al., 2007). The targeted protein is therefore constitutively degraded by the proteosome, but when co-expressed in cells with another chimera composed of FKBP12 and the other half of ubiquitin, it is stabilized in a rapamycin-dependent manner. This is because the reconstituted ubiquitin that forms upon rapamycin binding is cleaved from the protein of interest by dedicated proteases, taking the degron along with it. The FKBP12/rapamycin/FRB system can be co-opted to achieve small molecule-inducible protein degradation as well, as demonstrated by Church and coworkers (Janse et al., 2004). In this case, the S. cerevisiae proteasome subunits Rpn10 or Pre10 were fused to the yeast ortholog of FKBP12, and the protein of interest was tagged with the yeast equivalent of the FRB domain. Rapamycin treatment of yeast expressing these chimeras then leads to the recruitment of the targeted protein to the proteosome, which is sufficient to promote its degradation. Finally, the Kanemaki laboratory has successfully transferred the auxin-based degron system from plants into multiple animal cell lines, allowing proteins tagged with auxin-induced degrons to be rapidly proteolyzed in a small molecule-dependent manner (Nishimura et al., 2009).
Although these systems can regulate protein degradation in cultured cells, their efficacy in vivo has not yet been established. To date, only two technologies for small molecule-dependent protein stability have been validated in whole organisms, and both utilize similar biochemical mechanisms. The first approach emerged serendipitously from collaborative efforts by the Crabtree and Wandless laboratories to develop rapamycin analog/FRB mutant pairs that would avoid crosstalk with endogenous TOR signaling (Stankunas et al., 2003). During the course of these studies, they observed that proteins fused to one FRB mutant containing three amino acid substitutions (FRB*; K2095P/T2098L/W2101F human TOR) were rapidly degraded by the proteosome, presumably because these mutations destabilize the FRB fold and consequently the entire chimeric protein. The addition of rapamycin or its C20-methyallyl derivative (MaRap) stabilized these fusion proteins by recruiting endogenous FKBP12 (Figure 3b).
The Crabtree and Longaker laboratories then generated mouse embryos homozygous for a glycogen synthase kinase 3β (GSK3β)-FRB* transgene knocked into the endogenous GSK3β locus (Liu et al., 2007). In the absence of rapamycin, these transgenic embryos phenocopied conventional GSK3β knockouts, exhibiting cleft palates and skeletal defects. Since rapamycin is teratogenic when administered to pregnant mice bearing wildtype embryos prior to the E10.5 stage but does not cause overt developmental abnormalities at later time points, they explored the phenotypic consequences of 48-hour drug treatments starting at E13.5. Through this temporal analysis, it was determined that GSK3β is required between E13.5 and E15 for palatogenesis and between E15.5 and E17 for skeletal development (Figure 3c). Unfortunately effective doses of the non-teratogenic compound MaRap could not be achieved in utero, so using this methodology to investigate protein functions at earlier developmental stages will require new rapamycin analogs/FRB mutant pairs.
The Wandless laboratory has further simplified this approach by creating a new small molecule-sensitive destabilization domain that does not require formation of a ternary complex (Banaszynski et al., 2006). In this “single-ligand, single-domain” system, a mutant, unstable form of human FKBP12 (F36V) is fused to proteins of interest, promoting their degradation in a FKBP12 ligand-dependent manner (Figure 3d). The availability of non-teratogenic FKBP12 ligands such as Shield-1 and the lack of endogenous binding partners help ensure that this method can be applied to whole organisms with uniform efficacy between cell types. Indeed, preliminary studies indicate that the Shield-1/FKBP12 mutant technology can be used to conditionally and reversibly control protein levels in zebrafish embryos without collateral developmental defects (T. J. Wandless, personal communication).
Small molecule-based targeting of kinases
One drawback of chemically modulated protein homeostasis is that functional perturbation of the targeted gene is not coincident with ligand binding; rather, synthesis or degradation of the mature protein is the rate-determining step. Direct activation or inhibition of protein function by small molecules can occur on faster timescales, but only a fraction of gene products have known chemical regulators. Nevertheless, certain gene families can now be systematically targeted by chemical inhibitors through structure-based protein engineering and ligand design. This strategy is exemplified by the Shokat laboratory’s development of kinase mutant-targeted inhibitors that achieve genome-wide specificity. By changing a large conserved residue in the ATP binding site (typically isoleucine, threonine, or phenylalanine) to either glycine or alanine, a “hole” can be introduced into the kinase without significantly altering its catalytic activity. Shokat and co-workers then designed kinase inhibitors with a complementary “bump” by adding naphthyl or naphthylmethyl groups to the Src family kinase inhibitor PP1 (Bishop et al., 1998). Kinase mutants with these “holes” can be inhibited by these “bump”-containing PP1 analogs while wildtype kinase activities are left intact (Figure 3e). Using this approach, individual tyrosine or serine/threonine kinases can be specifically and reversibly inhibited in whole organisms, even those that have closely related homologs. For example, Ginty and co-workers studied individual members of the Trk receptor kinase family, which respond to neurotrophins to regulate nervous system development (Chen et al., 2005). They knocked in naphthylmethyl-PP1-sensitive forms of each Trk receptor (TrkAF592A, TrkBF616A, and TrkCF617A) into the corresponding wildtype allele, and treated homozygous mutant mouse embryos in utero with the PP1 derivative. Drug treatment of the TrkAF592A, TrkBF616A, and TrkCF617A embryos led to loss of superior cervical ganglia, nodose ganglia, and parvalbumin-positive dorsal root ganglia neurons, respectively. The homozygous mutants developed normally, and wildtype embryos were insensitive to naphthylmethyl-PP1 treatment.
Optogenetic control of ion channels
In most cases, small molecules can be used to modulate gene function in physiologically relevant timescales. However, certain biological processes such as neuronal depolarization occur within milliseconds, and faster-acting technologies are necessary to study these systems. Modulating ion channel function during embryogenesis is an area of particular interest, as it is now widely appreciated that electrical activity influences multiple stages of neuronal development (Spitzer, 2006). Several synthetic strategies for controlling neuronal action potentials have been developed and applied in whole organisms, permitting the activation of specific neurons with millisecond precision. All of these methods involve light-gated channels that contain either synthetic or endogenous photoisomerizable cofactors. Although the application of these tools to investigate embryonic patterning has not yet been reported, the examples below illustrate their basic design principles and their utility for in vivo studies.
The use of synthetic photoswitchable compounds to modulate ion channel activity state dates back nearly four decades, when Erlanger and co-workers found that electrophilic, azobenzene-containing quaternary amines were able to alkylate the nicotinic acetylcholine receptor (nAChR) and render it light-sensitive (Bartels et al., 1971). The trans isomer of the azobenzene-containing agonist locked the channel in an open state, and formation of the cis isomer upon exposure to 330-nm light caused channel inactivation (Figure 3f). Since this seminal work, several laboratories have applied this general strategy to other ion channels, including ligand-gated, voltage-gated, and mechanosensitive proteins. For example, the Trauner and Isacoff laboratories engineered an ionotropic glutamate receptor (iGluR6) to incorporate a modifiable cysteine proximal to the glutamate binding site (Volgraf et al., 2006). They then covalently attached a tether agonist composed of maleimide, azobenzene, and glutamate groups (MAG-1) to the iGluR6 mutant. The resulting synthetic receptor rendered cells permeable to ion currents in a light-dependent manner; irradiation with 380-nm light promoted cis isomerization of MAG-1 and current influx, and exposure to 500-nm light favored the trans MAG-1 isomer and ion channel closure. Photoswitching of the light-gated iGluR6 receptor (LiGluR6) occurred on the millisecond timescale, and the degree of channel activation could be modulated by changing the wavelength of light.
Even though the MAG-1/LiGluR6 technology requires channel adduct formation with an electrophilic reagent, Trauner and Isacoff have successfully used this approach to explore neuronal function in whole organisms (Wyart et al., 2009). They created a zebrafish line containing a UAS-dependent LiGluR6 transgene and then crossed it with several transgenic lines expressing Gal4 in different subsets of spinal neurons. The progeny were then treated with MAG-1 and stimulated with pulses of 390-nm light to depolarize the targeted cells, resulting in neuron-specific behaviors. For example, light activation of caudal Kolmer-Agduhr neurons induced tail oscillations that mimicked forward swimming, while bilaterial illumination of caudal Rohon-Beard neurons induced tail bends characteristic of the touch-escape response.
Light-gated channels found in Nature have been more widely applied to interrogate neuronal function, owing to their reliance on endogenous retinal as a cofactor rather than reactive synthetic ligands (Figure 3f) (Zhang et al., 2007a). Although several photo-responsive channels have been identified, channelrhodopsin-2 (ChR2) from the green algae Chlamydomonas reinhardtii and halorhodopsin from the halobacterium Natronomonas pharaonis (NpHR) are the two most commonly used proteins (Lanyi et al., 1990; Nagel et al., 2003). ChR2 is cation channel that allows Na+ ions to enter the cell in response to 470-nm light, and NpHR is a chloride pump that is activated by 580-nm light. Neuronal firing can therefore be induced and suppressed by ChR2 and NpHR, respectively, permitting the generation of specific action potential profiles with millisecond resolution. These light-gated channels have been used to modulate neuron function in worms (Nagel et al., 2005; Zhang et al., 2007b), fruit flies (Schroll et al., 2006), zebrafish (Baier and Scott, 2009; Douglass et al., 2008), mice (Bi et al., 2006; Zhang et al., 2007b), and even primates (Han et al., 2009), demonstrating their efficacy across neuronal subtypes and animal species. The Deisseroth laboratory’s development of photoactivatable rhodopsin-GPCR chimeras that actuate intracellular signaling pathways promises to extend this approach to other developmental processes (Airan et al., 2009).
Optogenetic control of small GTPases
Nature has created other photo-responsive systems that could be re-engineered into light-actuated probes of patterning mechanisms. For example, plants control phototropism and stomatal opening through light-sensing kinases called phototropins that contain tandem light-oxygen-voltage (LOV) domains (Christie, 2007). When exposed to blue light, the LOV domain reacts with its flavin mononucleotide (FMN) cofactor, generating a cysteinyl covalent adduct that is stable for seconds and then reverts to the non-covalent haloprotein (Figure 3f). In the Avena sative Phototropin1 protein, formation of this covalent intermediate is associated with a conformational change that disrupts interactions between one of its LOV domains and a C-terminal helical extension called Jα, causing the latter to unfold (Harper et al., 2003). By conjugating the LOV-Jα polypeptide to a constitutively active mutant of Rac1, a GTPase that regulates actin cytoskeletal dynamics, Hahn and coworkers were able to produce a photoactivatable Rac1 chimera (PA-Rac1) (Wu et al., 2009). In the absence of light, the LOV-Jα polypeptide prevented the binding of Rac1 to its downstream effector PAK, whereas exposure to 458-nm light induced unwinding of the Jα helix, restored Rac1/PAK interactions, and promoted lamellipodial protrusions and membrane ruffling. PA-Rac1 photoactivation is reversible, repeatable, and can be conducted in whole organisms, as demonstrated by the Huttenlocher laboratory (Yoo et al., 2010). In collaboration with the Hahn group, they created transgenic zebrafish that specifically expressed the PA-Rac1 fusion protein in neutrophils, and by activating PA-Rac1 with a spatiotemporally controlled beam of 458-nm light, they were able to direct neutrophil migration in the transgenic embryos.
Another LOV domain-containing polypeptide, the FKF1 protein that controls flowering in Arabidopsis thaliana, has been re-engineered to achieve light-activated protein-protein interactions in live cells. In this case, FKF1 associates with another flowering regulator called GIGANTEA (GI) in a light-dependent manner (Sawa et al., 2007), and the Dolmetsch laboratory has demonstrated that fusion proteins containing these polypeptides can photo-dimerize in a similar manner (Yazawa et al., 2009). For instance, cells co-expressing FKF1-Rac1 and farnesylated GI exhibit plasma membrane translocation of Rac1 activity upon exposure to 450-nm light, and focal illumination can induce the formation of lamellipodia. Dolmetsch and coworkers also achieved light-activated gene transcription by fusing FKF1 to the VP16 activation domain and GI to the Gal4 DNA binding domain. Unlike the Phototropin1-derived LOV-Jα system, the light-induced cysteinyl-flavin adduct in FKF1 persists for hours rather than seconds. Short illumination times can therefore induce long-lasting protein-protein interactions, making the FKF1/GI-based approach preferable for certain research applications.
CHEMICAL CONTROL OF CELL FUNCTION
As detailed in the sections above, our efforts to understand embryonic patterning at the molecular level is now matched with synthetic tools that enable perturbations of DNA, RNA, or protein function with spatiotemporal precision. It should be noted, however, that the surgical manipulations and ablations that characterized embryology prior to its molecular revolution are still valuable research tools. Genetic technologies that enable conditional and targeted cell ablation have also been developed, in particular methods that involve the tissue-specific expression of a diphtheria toxin-derived polypeptide (Breitman et al., 1987; Palmiter et al., 1987). Although this approach has provided important insights into metazoan development and physiology, its reliance on genomic regulatory elements limits its generality. The precise timing and efficacy of cell ablation are subject to the activities of these promoters, and due to the high toxicity of this diphtheria toxin fragment, even low levels of promoter activity can lead to unintended cell death and embryonic mispatterning. These problems can be mitigated to some extent by delivering full-length diphtheria toxin into transgenic organisms expressing the receptor for this polypeptide in targeted tissues (Saito et al., 2001); however, the proteolytic susceptibility of diphtheria toxin and its short biological half-life are other potential drawbacks.
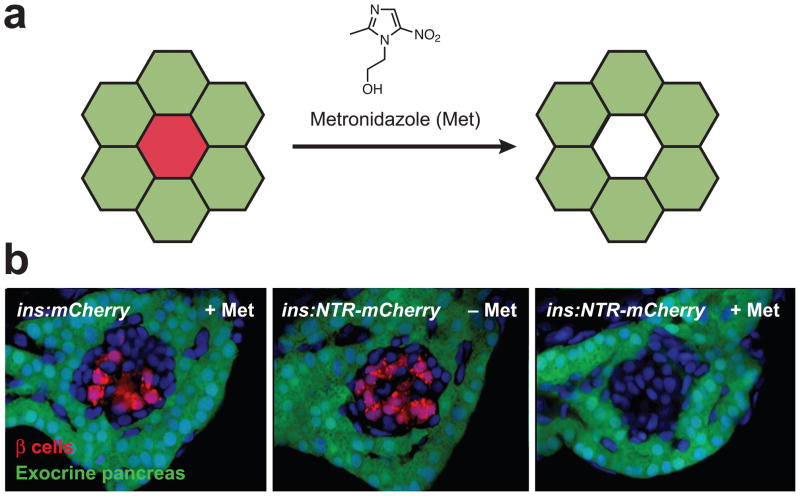
Cell ablation techniques that rely upon exogenous small molecules could provide greater experimental flexibility, particularly with respect to the timing, tunability, and sustainability of these perturbations. One such method takes advantage of nitro group-containing prodrugs, which are converted into potent cytotoxic agents by their nitroreductase-expressing bacteria targets (Roldan et al., 2008). The first demonstration of this approach was conducted by Clark and his co-workers, who generated transgenic mice that express E. coli nitroreductase in luminal cells of the mammary gland (Clark et al., 1997). Treatment of these mice with the prodrug CB1954 resulted in the rapid apoptosis of these cells, while neighboring myoepithelial cells were not affected. More recently, nitroreductase-mediated cell ablation has been executed in zebrafish by the Parsons and Stainier laboratories (Curado et al., 2007; Pisharath et al., 2007). In these studies, zebrafish lines that selectively express the bacterial enzyme in pancreatic β-cells, cardiomyocytes, or hepatocytes were generated, and the transgenic embryos were cultured in medium containing the prodrug metronidazole at various time points. Spatially and temporally controlled cell ablation was achieved in each case, and the targeted tissues were able to recover upon washout of the nitroreductase substrate (Figure 4). While tissue-dependent drug penetrance and ablation rates have been observed (Curado et al., 2007), the nitroreductase/metronidazole technique appears to be a generalizable strategy for studying tissue patterning and regeneration.
Figure 4. Chemically induced cell ablation.
(a) Schematic representation of metronidazole-dependent apoptosis of nitroreductase-expressing cells (red) without collateral damage to adjacent cells (green). (b) Metronidazole-induced ablation of pancreatic β cells (red) in transgenic zebrafish larvae with insulin promoter-driven nitroreductase-mCherry expression (ins:NTR-mCherry). Exocrine pancreas cells (green) were not affected, and no drug-dependent cell ablation was observed in ins:mCherry larvae. Adapted with permission (Pisharath et al., 2007; Copyright 2006, Elsevier Ireland Ltd.).
CONCLUDING REMARKS
The examples highlighted in this review illustrate how synthetic systems created by chemistry and protein engineering have changed the way we study embryogenesis and other organismal processes. As we try to understand increasingly complex systems, new approaches will be needed to interrogate patterning mechanisms with greater experimental finesse and spatiotemporal precision. For the immediate future, the developmental biology community would benefit from the extension of existing technologies. Just as the “bump-and-hole” approach has enabled the inhibition of individual kinases with genome-wide specificity, this method could be applied to other gene families, particularly those that share a common cofactor and/or structural motif. For instance, allele-specific inhibitors of methyltransferases or acetyltransferases could provide critical insights into the epigenetic mechanisms that regulate cell fate. Orthogonal riboswitches could enable simultaneous control of multiple transcripts, and parallel “single-ligand, single-domain” degradation systems could achieve the same versatility at the protein level. Both approaches would allow biologists to determine how developmental signaling molecules work in concert to effect specific morphological changes.
There are also a number of technical challenges facing the chemical and developmental biology communities as we seek a more molecular, quantitative understanding of embryogenesis. The increasing demand for conditional reagents requires that new caging technologies be developed. Chromophores that are more sensitive to ultraviolet light or compatible with two-photon irradiation would minimize cell damage and facilitate the targeting of deeper tissues. Caging strategies for small molecules or oligomers that do not involve light-sensitive groups could also be valuable advances since not all tissues are readily targeted with focal or patterned light sources. In these cases, enzymatically triggered reagents could be used in conjunction with transgenic animals, in analogy to the nitroreductase-based cell ablation technology.
In addition, new methods for delivering and maintaining exogenous reagents in whole organisms are needed. In the case of caged small molecules, their efficacy in vivo hinges upon their rates of diffusion and the rates of the biological processes they actuate. Strategies for limiting the former after uncaging would therefore broaden the functionality of these compounds. The membrane impermeability of synthetic oligonucleotides poses a different challenge, as their delivery into non-injectable embryos or at later developmental stages is a major technical hurdle. Technologies that enable the uniform delivery of these compounds into embryos, ideally by simply adding them to the culture medium, would be a significant breakthrough.
Given the evolutionary diversity of metazoans, it is unlikely that a single synthetic strategy will be the “silver bullet” for deciphering the dynamic mechanisms that control embryogenesis. For optically transparent embryos that develop ex utero, technologies involving genetically encoded photoswitchable proteins hold particular promise. The direct optical control of protein function is unsurpassed for its immediate impact on developmental signaling, as well as its spatiotemporal precision. The reversibility of optogenetic systems is also unparalleled by methods that target DNA or RNA function, as the basal state can be restored seconds after the light source is removed. One can even envision a suite of engineered, light-activatable domains that survey a broad range of deactivation rates, allowing one to select protein activity lifetimes that match specific applications. Photoactivatable domains with different spectral properties could also be engineered to enable orthogonal control of two or more proteins, akin to the ChR2 and NpHR systems described above. Yet how far these approaches can be extended to other model organisms remains to be seen. Recent advances in fiber optic systems have enabled optogenetic manipulations in live mice (Barretto et al., 2009; Deisseroth et al., 2006), but applying these technologies in the womb will be experimentally challenging. In these cases, new non-optical, small molecule-actuated tools will likely have a broader impact on developmental biology research.
Just as molecular biology complemented the dissecting needle in the 1980s, small molecule- and synthetic protein-based technologies are now emerging as important embryological tools. Chemical biologists therefore have an opportunity to play leading roles in the post-genomic era of developmental biology. Trained in chemical principles and empowered by chemical synthesis, we are well positioned to investigate the thermodynamic and kinetic processes that underlie embryogenesis and to create new research modalities unfettered by Nature’s bonds. Through this unique perspective, chemists can help find new answers to the age-old question of how function begets form.
Acknowledgments
We gratefully acknowledge financial support from the NIH Director’s Pioneer Award (DP1 OD003792), the NIH/NIGMS (R01 GM072600), the March of Dimes Foundation (1-FY-08-433), and a Bio-X Stanford Interdisciplinary Graduate Fellowship in Human Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- Ando H, Furuta T, Tsien RY, Okamoto H. Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat Genet. 2001;28:317–325. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- Baier H, Scott EK. Genetic and optical targeting of neural circuits and behavior--zebrafish in the spotlight. Curr Opin Neurobiol. 2009;19:553–560. doi: 10.1016/j.conb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto RP, Messerschmidt B, Schnitzer MJ. In vivo fluorescence imaging with high-resolution microlenses. Nat Methods. 2009;6:511–512. doi: 10.1038/nmeth.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels E, Wassermann NH, Erlanger BF. Photochromic activators of the acetylcholine receptor. Proc Natl Acad Sci U S A. 1971;68:1820–1823. doi: 10.1073/pnas.68.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi D, Shilo BZ, Fainsod A, Barkai N. Scaling of the BMP activation gradient in Xenopus embryos. Nature. 2008;453:1205–1211. doi: 10.1038/nature07059. [DOI] [PubMed] [Google Scholar]
- Bi A, Cui J, Ma YP, Olshevskaya E, Pu M, Dizhoor AM, Pan ZH. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50:23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieschke ET, Wheeler JC, Tower J. Doxycycline-induced transgene expression during Drosophila development and aging. Mol Gen Genet. 1998;258:571–579. doi: 10.1007/s004380050770. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- Blidner RA, Svoboda KR, Hammer RP, Monroe WT. Photoinduced RNA interference using DMNPE-caged 2′-deoxy-2′-fluoro substituted nucleic acids in vitro and in vivo. Mol Biosyst. 2008;4:431–440. doi: 10.1039/b801532e. [DOI] [PubMed] [Google Scholar]
- Boniface EJ, Lu J, Victoroff T, Zhu M, Chen W. FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis. 2009;47:484–491. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman ML, Clapoff S, Rossant J, Tsui LC, Glode LM, Maxwell IH, Bernstein A. Genetic ablation: targeted expression of a toxin gene causes microphthalmia in transgenic mice. Science. 1987;238:1563–1565. doi: 10.1126/science.3685993. [DOI] [PubMed] [Google Scholar]
- Buskirk AR, Ong YC, Gartner ZJ, Liu DR. Directed evolution of ligand dependence: small-molecule-activated protein splicing. Proc Natl Acad Sci U S A. 2004;101:10505–10510. doi: 10.1073/pnas.0402762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambridge SB, Geissler D, Calegari F, Anastassiadis K, Hasan MT, Stewart AF, Huttner WB, Hagen V, Bonhoeffer T. Doxycycline-dependent photoactivated gene expression in eukaryotic systems. Nat Methods. 2009;6:527–531. doi: 10.1038/nmeth.1340. [DOI] [PubMed] [Google Scholar]
- Chen X, Ye H, Kuruvilla R, Ramanan N, Scangos KW, Zhang C, Johnson NM, England PM, Shokat KM, Ginty DD. A chemical-genetic approach to studying neurotrophin signaling. Neuron. 2005;46:13–21. doi: 10.1016/j.neuron.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Mark MR, Bajaj V, Godowski PJ. Ecdysteroid-dependent regulation of genes in mammalian cells by a Drosophila ecdysone receptor and chimeric transactivators. Proc Natl Acad Sci U S A. 1992;89:6314–6318. doi: 10.1073/pnas.89.14.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Iwobi M, Cui W, Crompton M, Harold G, Hobbs S, Kamalati T, Knox R, Neil C, Yull F, et al. Selective cell ablation in transgenic mice expression E. coli nitroreductase. Gene Ther. 1997;4:101–110. doi: 10.1038/sj.gt.3300367. [DOI] [PubMed] [Google Scholar]
- Coffman JA, Dickey-Sims C, Haug JS, McCarthy JJ, Robertson AJ. Evaluation of developmental phenotypes produced by morpholino antisense targeting of a sea urchin Runx gene. BMC Biol. 2004;2:6. doi: 10.1186/1741-7007-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coonrod SA, Bolling LC, Wright PW, Visconti PE, Herr JC. A morpholino phenocopy of the mouse mos mutation. Genesis. 2001;30:198–200. doi: 10.1002/gene.1065. [DOI] [PubMed] [Google Scholar]
- Coumoul X, Shukla V, Li C, Wang RH, Deng CX. Conditional knockdown of Fgfr2 in mice using Cre-LoxP induced RNA interference. Nucleic Acids Res. 2005;33:e102. doi: 10.1093/nar/gni100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- Dagle JM, Littig JL, Sutherland LB, Weeks DL. Targeted elimination of zygotic messages in Xenopus laevis embryos by modified oligonucleotides possessing terminal cationic linkages. Nucleic Acids Res. 2000;28:2153–2157. doi: 10.1093/nar/28.10.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagle JM, Walder JA, Weeks DL. Targeted degradation of mRNA in Xenopus oocytes and embryos directed by modified oligonucleotides: studies of An2 and cyclin in embryogenesis. Nucleic Acids Res. 1990;18:4751–4757. doi: 10.1093/nar/18.16.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- de Graaf M, Zivkovic D, Joore J. Hormone-inducible expression of secreted factors in zebrafish embryos. Dev Growth Differ. 1998;40:577–582. doi: 10.1046/j.1440-169x.1998.t01-3-00001.x. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AD, Kraves S, Deisseroth K, Schier AF, Engert F. Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr Biol. 2008;18:1133–1137. doi: 10.1016/j.cub.2008.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Picard D, Yamamoto KR, Bishop JM. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Esengil H, Chang V, Mich JK, Chen JK. Small-molecule regulation of zebrafish gene expression. Nat Chem Biol. 2007;3:154–155. doi: 10.1038/nchembio858. [DOI] [PubMed] [Google Scholar]
- Fallon JF, Lopez A, Ros MA, Savage MP, Olwin BB, Simandl BK. FGF-2: apical ectodermal ridge growth signal for chick limb development. Science. 1994;264:104–107. doi: 10.1126/science.7908145. [DOI] [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Firestone AJ, Chen JK. Controlling destiny through chemistry: small-molecule regulators of cell fate. ACS Chem Biol. 2010;5:15–34. doi: 10.1021/cb900249y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N, Chatelain G, Brun G, Lamonerie T. Temporal and spatial delineation of mouse Otx2 functions by conditional self-knockout. EMBO Rep. 2006;7:824–830. doi: 10.1038/sj.embor.7400751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth PA, St Onge L, Boger H, Gruss P, Gossen M, Kistner A, Bujard H, Hennighausen L. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc Natl Acad Sci U S A. 1994;91:9302–9306. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Green S, Chambon P. Oestradiol induction of a glucocorticoid-responsive gene by a chimaeric receptor. Nature. 1987;325:75–78. doi: 10.1038/325075a0. [DOI] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- Gupta S, Schoer RA, Egan JE, Hannon GJ, Mittal V. Inducible, reversible, and stable RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Qian X, Bernstein JG, Zhou HH, Franzesi GT, Stern P, Bronson RT, Graybiel AM, Desimone R, Boyden ES. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS One. 2009;4:e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Jou TS, Ho YL, Lee WH, Jeng YT, Hsieh FJ, Tsai HJ. Conditional expression of a myocardium-specific transgene in zebrafish transgenic lines. Dev Dyn. 2005;233:1294–1303. doi: 10.1002/dvdy.20485. [DOI] [PubMed] [Google Scholar]
- Janse DM, Crosas B, Finley D, Church GM. Localization to the proteasome is sufficient for degradation. J Biol Chem. 2004;279:21415–21420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, Bujard H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci U S A. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171:267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- Lanyi JK, Duschl A, Hatfield GW, May K, Oesterhelt D. The primary structure of a halorhodopsin from Natronobacterium pharaonis. Structural, functional and evolutionary implications for bacterial rhodopsins and halorhodopsins. J Biol Chem. 1990;265:1253–1260. [PubMed] [Google Scholar]
- Lennox KA, Sabel JL, Johnson MJ, Moreira BG, Fletcher CA, Rose SD, Behlke MA, Laikhter AL, Walder JA, Dagle JM. Characterization of modified antisense oligonucleotides in Xenopus laevis embryos. Oligonucleotides. 2006;16:26–42. doi: 10.1089/oli.2006.16.26. [DOI] [PubMed] [Google Scholar]
- Lewis J. From signals to patterns: space, time, and mathematics in developmental biology. Science. 2008;322:399–403. doi: 10.1126/science.1166154. [DOI] [PubMed] [Google Scholar]
- Link KH, Cruz FG, Ye HF, O’Reilly K E, Dowdell S, Koh JT. Photo-caged agonists of the nuclear receptors RARgamma and TRbeta provide unique time-dependent gene expression profiles for light-activated gene patterning. Bioorg Med Chem. 2004;12:5949–5959. doi: 10.1016/j.bmc.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Link KH, Shi Y, Koh JT. Light activated recombination. J Am Chem Soc. 2005;127:13088–13089. doi: 10.1021/ja0531226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KJ, Arron JR, Stankunas K, Crabtree GR, Longaker MT. Chemical rescue of cleft palate and midline defects in conditional GSK-3beta mice. Nature. 2007;446:79–82. doi: 10.1038/nature05557. [DOI] [PubMed] [Google Scholar]
- Mackem S, Lewandoski M. Limb development takes a measured step toward systems analysis. Sci Signal. 2009;2:pe33. doi: 10.1126/scisignal.271pe33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Mara A, Schroeder J, Holley SA. Two deltaC splice-variants have distinct signaling abilities during somitogenesis and midline patterning. Dev Biol. 2008;318:126–132. doi: 10.1016/j.ydbio.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Targeted screening for induced mutations. Nat Biotechnol. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- Mootz HD, Muir TW. Protein splicing triggered by a small molecule. J Am Chem Soc. 2002;124:9044–9045. doi: 10.1021/ja026769o. [DOI] [PubMed] [Google Scholar]
- Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Niswander L, Jeffrey S, Martin GR, Tickle C. A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature. 1994;371:609–612. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- Niswander L, Tickle C, Vogel A, Booth I, Martin GR. FGF-4 replaces the apical ectodermal ridge and directs outgrowth and patterning of the limb. Cell. 1993;75:579–587. doi: 10.1016/0092-8674(93)90391-3. [DOI] [PubMed] [Google Scholar]
- Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CL, Li WH, Olson AJ, Chen JK. Versatile synthesis and rational design of caged morpholinos. J Am Chem Soc. 2009;131:13255–13269. doi: 10.1021/ja809933h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Behringer RR, Quaife CJ, Maxwell F, Maxwell IH, Brinster RL. Cell lineage ablation in transgenic mice by cell-specific expression of a toxin gene. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- Parr BA, McMahon AP. Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature. 1995;374:350–353. doi: 10.1038/374350a0. [DOI] [PubMed] [Google Scholar]
- Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt MR, Schwartz EC, Muir TW. Small-molecule-mediated rescue of protein function by an inducible proteolytic shunt. Proc Natl Acad Sci U S A. 2007;104:11209–11214. doi: 10.1073/pnas.0700816104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy D, Tremethick DJ, Greaves IK. Gene knockdown by ecdysone-based inducible RNAi in stable mammalian cell lines. Nat Protoc. 2008;3:79–88. doi: 10.1038/nprot.2007.456. [DOI] [PubMed] [Google Scholar]
- Ridgway P, Quivy JP, Almouzni G. Tetracycline-regulated gene expression switch in Xenopus laevis. Exp Cell Res. 2000;256:392–399. doi: 10.1006/excr.2000.4853. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan MD, Perez-Reinado E, Castillo F, Moreno-Vivian C. Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol Rev. 2008;32:474–500. doi: 10.1111/j.1574-6976.2008.00107.x. [DOI] [PubMed] [Google Scholar]
- Saito M, Iwawaki T, Taya C, Yonekawa H, Noda M, Inui Y, Mekada E, Kimata Y, Tsuru A, Kohno K. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol. 2001;19:746–750. doi: 10.1038/90795. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. Action of morpholinos in Ciona embryos. Genesis. 2001;30:103–106. doi: 10.1002/gene.1040. [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- Schroll C, Riemensperger T, Bucher D, Ehmer J, Voller T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Schwartz EC, Saez L, Young MW, Muir TW. Post-translational enzyme activation in an animal via optimized conditional protein splicing. Nat Chem Biol. 2007;3:50–54. doi: 10.1038/nchembio832. [DOI] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- Seibler J, Kleinridders A, Kuter-Luks B, Niehaves S, Bruning JC, Schwenk F. Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Res. 2007;35:e54. doi: 10.1093/nar/gkm122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Jain PK, Kala A, Karunakaran D, Friedman SH. Light-activated RNA interference using double-stranded siRNA precursors modified using a remarkable regiospecificity of diazo-based photolabile groups. Nucleic Acids Res. 2009;37:4508–4517. doi: 10.1093/nar/gkp415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Rangarajan S, Friedman SH. Light-activated RNA interference. Angew Chem Int Ed Engl. 2005;44:1328–1332. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- Shestopalov IA, Sinha S, Chen JK. Light-controlled gene silencing in zebrafish embryos. Nat Chem Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- Shi Y, Jin Y. MicroRNA in cell differentiation and development. Sci China C Life Sci. 2009;52:205–211. doi: 10.1007/s11427-009-0040-5. [DOI] [PubMed] [Google Scholar]
- Shin MK, Levorse JM, Ingram RS, Tilghman SM. The temporal requirement for endothelin receptor-B signalling during neural crest development. Nature. 1999;402:496–501. doi: 10.1038/990040. [DOI] [PubMed] [Google Scholar]
- Siegel RW, Jain R, Bradbury A. Using an in vivo phagemid system to identify non-compatible loxP sequences. FEBS Lett. 2001;505:467–473. doi: 10.1016/s0014-5793(01)02806-x. [DOI] [PubMed] [Google Scholar]
- Sinha DK, Neveu P, Gagey N, Aujard I, Benbrahim-Bouzidi C, Le Saux T, Rampon C, Gauron C, Goetz B, Dubruille S, et al. Photocontrol of Protein Activity in Cultured Cells and Zebrafish with One- and Two-Photon Illumination. Chembiochem. 2010;11:653–663. doi: 10.1002/cbic.201000008. [DOI] [PubMed] [Google Scholar]
- Spitzer NC. Electrical activity in early neuronal development. Nature. 2006;444:707–712. doi: 10.1038/nature05300. [DOI] [PubMed] [Google Scholar]
- Stankunas K, Bayle JH, Gestwicki JE, Lin YM, Wandless TJ, Crabtree GR. Conditional protein alleles using knockin mice and a chemical inducer of dimerization. Mol Cell. 2003;12:1615–1624. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- Tang X, Maegawa S, Weinberg ES, Dmochowski IJ. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J Am Chem Soc. 2007;129:11000–11001. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- Thaker M, Spanogiannopoulos P, Wright GD. The tetracycline resistome. Cell Mol Life Sci. 2010;67:419–431. doi: 10.1007/s00018-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini AJ, Schuler AD, Zebala JA, Mayer AN. PhotoMorphs: a novel light-activated reagent for controlling gene expression in zebrafish. Genesis. 2009;47:736–743. doi: 10.1002/dvg.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urtishak KA, Choob M, Tian X, Sternheim N, Talbot WS, Wickstrom E, Farber SA. Targeted gene knockdown in zebrafish using negatively charged peptide nucleic acid mimics. Dev Dyn. 2003;228:405–413. doi: 10.1002/dvdy.10394. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Weeks DL, Wolffe AP. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science. 2000;290:2312–2315. doi: 10.1126/science.290.5500.2312. [DOI] [PubMed] [Google Scholar]
- Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf TM, Jennings CG, Rebagliati M, Melton DA. The stability, toxicity and effectiveness of unmodified and phosphorothioate antisense oligodeoxynucleotides in Xenopus oocytes and embryos. Nucleic Acids Res. 1990;18:1763–1769. doi: 10.1093/nar/18.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Hu Z, Liu XQ. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyart C, Del Bene F, Warp E, Scott EK, Trauner D, Baier H, Isacoff EY. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature. 2009;461:407–410. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kurimoto K, Yabuta Y, Sasaki H, Nakatsuji N, Saitou M, Tada T. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development. 2009;136:4011–4020. doi: 10.1242/dev.041160. [DOI] [PubMed] [Google Scholar]
- Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- Yeh JR, Crews CM. Chemical genetics: adding to the developmental biology toolbox. Dev Cell. 2003;5:11–19. doi: 10.1016/s1534-5807(03)00200-4. [DOI] [PubMed] [Google Scholar]
- Yen L, Magnier M, Weissleder R, Stockwell BR, Mulligan RC. Identification of inhibitors of ribozyme self-cleavage in mammalian cells via high-throughput screening of chemical libraries. RNA. 2006;12:797–806. doi: 10.1261/rna.2300406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D’Amato RJ, Mulligan RC. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A. Differential Regulation of Protrusion and Polarity by PI(3)K during Neutrophil Motility in Live Zebrafish. Dev Cell. 2010;18:226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DD, Garner RA, Yoder JA, Deiters A. Light-activation of gene function in mammalian cells via ribozymes. Chem Commun (Camb) 2009:568–570. doi: 10.1039/b819375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen CM, Rodda SJ, Vokes SA, McMahon AP, Liu DR. Control of transcription factor activity and osteoblast differentiation in mammalian cells using an evolved small-molecule-dependent intein. J Am Chem Soc. 2006;128:8939–8946. doi: 10.1021/ja062980e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R, Lopez-Rios J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K. Circuit-breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007a;8:577–581. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007b;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]