Abstract
Histone deacetylase (HDAC) inhibitors are gaining interest as cancer therapeutic agents. We tested the hypothesis that natural organoselenium compounds might be metabolized to HDAC inhibitors in human prostate cancer cells. Se-methyl-L-selenocysteine (MSC) and selenomethionine (SM) are amino acid components of selenium-enriched yeast. In a cell-free system, glutamine transaminase K (GTK) and L-amino acid oxidase (LAAO) convert MSC to the corresponding α-keto acid, β-methylselenopyruvate (MSP), and LAAO converts SM to its corresponding α-keto acid, α-keto-γ-methylselenobutyrate (KMSB). Although methionine (sulfur analogue of SM) is an excellent substrate for GTK, SM is poorly metabolized. Structurally, MSP and KMSB resemble the known HDAC inhibitor butyrate. We examined androgen-responsive LNCaP cells and androgen-independent LNCaP C4-2, PC-3, and DU145 cells and found these human prostate cancer cells exhibit endogenous GTK activities. In the corresponding cytosolic extracts, metabolism of MSC was accompanied by the concomitant formation of MSP. In MSP- and KMSB-treated prostate cancer cell lines, acetylated histone 3 was increased within 5 hr, and returned to essentially baseline levels by 24 hr, suggesting a rapid, transient induction of histone acetylation. In an in vitro HDAC activity assay, the selenoamino acids, MSC and SM, had no effect at concentrations up to 2.5 mM, whereas MSP and KMSB both inhibited HDAC activity. We conclude that, in addition to targeting redox-sensitive signaling proteins and transcription factors, α-keto acid metabolites of MSC and SM can alter HDAC activity and histone acetylation status. These findings provide a potential new paradigm by which naturally occurring organoselenium might prevent progression of human prostate cancer.
Keywords: Prostate cancer, Se-methyl-L-selenocysteine, selenomethionine, HDAC inhibition, glutamine transaminase K; L-amino acid oxidase
INTRODUCTION
Prostate cancer is the most common cancer in men in the United States. Although there is an unequal burden by race and ethnicity, the mortality rate is moderate. In 2008, over 186,000 American men will be diagnosed with prostate cancer and 28,000 men will die from this disease (1). Because most treatments can have significant side effects, development of chemopreventive agents and strategies for prevention of prostate cancer are highly desirable. Epidemiological studies and clinical intervention trials have demonstrated a protective role of selenium compounds against prostate cancer (2–7). A clinical trial by Clark and his colleagues, and follow-up studies, provided support for the protective role of selenium-containing compounds against progression of human prostate cancer (6,7). Recently, however, the selenium and vitamin E cancer trial (SELECT) was halted due to an apparent lack of efficacy for supplemental vitamin E and organoselenium, either alone or in combination (8,9).
An important caveat to translational studies is to understand the precise chemical nature of both inorganic and organoselenium compound(s) since toxicity and therapeutic efficacy depend greatly on the chemical form and not the amount of elemental selenium. The SELECT study used purified selenomethionine, whereas some earlier diet studies used selenium-enriched yeast, which like seleniferous plants such as garlic, onions and broccoli, also contains the naturally-occurring organoselenium compound, Se-methyl-L-selenocysteine (MSC) (10,11). The anticancer potential of MSC has been reported in human breast cancer cells (12–15), mouse mammary tumor cell lines (16–17) and a mammary tumor model in rats (18). Recent laboratory studies (19) revealed a possible use of MSC for prevention of prostate cancer by showing that MSC can inhibit growth of human prostate cancer cells in a xenograft mouse model.
The chemopreventive efficacy of MSC and other organoselenium compounds has been suggested to result from in situ generation of methylselenol (CH3SeH) by β-lyases (20). Methylselenol can also be generated by intracellular reduction of dimethyldiselenide and methylseleninic acid by endogenous glutathione. Thus, methylselenol has been suggested to play a critical role in chemoprevention and the anticancer properties associated with selenium supplementation through its ability to alter cell signaling pathways and induce cellular apoptosis (16).
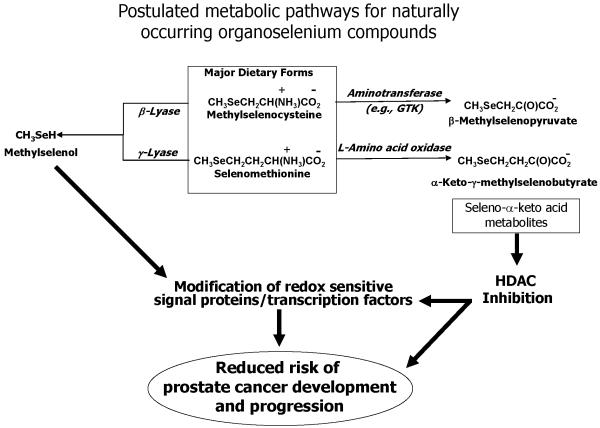
In order to generate methylselenol in situ using naturally-occurring organoselenium compounds, MSC and/or SM must undergo a β- and a γ-elimination reaction, respectively (21). β-Lyases and γ-lyases are pyridoxal 5'-phosphate (PLP)-dependent enzymes. Curiously, many aminotransferases can also catalyze β-elimination reactions particularly with cysteine S-conjugates that possess a good electronegative leaving group. For these enzymes, aminotransferase reactions often compete with elimination reactions. A full transamination reaction requires the concomitant presence of an α-keto acid substrate or a steady supply of PLP since pyridoxamine 5'-phosphate, the coenzymic product in a half transamination reaction, unlike PLP cannot catalyze the β-lyase reaction (22,23). Selenocysteine Se-conjugates, in particular MSC, have been shown to be 5 to 10-fold better aminotransferase substrates of rat kidney glutamine transaminase K (GTK) than of the corresponding cysteine S-conjugates (24). GTK is an aminotransferase that is widely distributed in rat tissues and the rat enzyme has broad specificity toward glutamine, methionine and other sulfur and selenium amino acids, aromatic amino acids and the corresponding α-keto acids (25). Human GTK has also been shown to have broad substrate specificity (26). Of interest to the current work, MSC was found to be both an aminotransferase and β-lyase substrate of human GTK (26). Because of the weaker C-Se bond compared to the C-S bond and/or the more facile abstraction of the β proton, selenocysteine Se-conjugates exhibit greater aminotransferase and β-lyase reactivity than the corresponding cysteine S-conjugates (27). These characteristics of MSC led us to question whether methylselenol was the only critical metabolite to explain the chemopreventive activity of organoselenium compounds in a physiological environment. Accordingly, under appropriate enzymatic conditions of regenerating PLP or in the presence of α-keto acid substrates, MSC should undergo a transamination reaction that competes with β-elimination. Thus, in addition to in situ formation of methylselenol through a β-elimination reaction, the deaminated product of MSC, β-methylselenopyruvate (MSP), should also be formed (Figure 1).
Figure 1.
Proposed metabolic pathways for naturally occurring organoselenium compounds. Dietary seleno amino acids can undergo either β-/γ-elimination reactions or transamination/oxidative deamination reactions. The α-keto acid metabolites from the latter reaction have been shown to exhibit HDAC inhibitor properties in human prostate cancer cells.
MSP, as well as the α-keto acid product of selenomethionine (SM), namely α-keto-γ-methylselenobutyrate (KMSB) resemble butyrate, an inhibitor of histone deacetylase (HDAC). HDAC inhibitors are showing promise for the treatment of several human cancers, and several mechanisms have been proposed (28–31). To date, five classes of HDAC inhibitors have been identified: (1) short-chain fatty acids, for example, butyric acid; (2) hydroxamic acids, such as suberoylanilide hydroxamic acid (SAHA); (3) electrophilic ketones that include trifluoromethyl α-ketones and α-ketoamides; (4) aminobenzamides namely MS-275 and CI-994; and (5) natural cyclic peptides such as apicidin. The structural multiplicity of HDAC inhibitors reflects both the diversity of the substrates for HDAC and the heterogeneity of tumor cell phenotypes. An intriguing possibility of the oxidatively deaminated products of either MSC or SM is the presence of a selenium moiety capable of disrupting the charge relay system in the HDAC pocket by coordinating with the zinc cofactor (32, 33).
This study demonstrates previously unrecognized effects of MSC and SM as prodrug inhibitors of HDACs. We provide information, for the first time, that the α-keto acid metabolites of MSC and SM, in addition to methylselenol derived from β- and γ-lyase reactions, may be potential direct acting metabolites of organoselenium in the chemopreventive activity for prostate cancer. These studies provide a new understanding to mechanisms by which naturally-occurring organoselenium may decrease progression of human prostate cancer.
MATERIALS AND METHODS
Chemicals and Enzymes
Se-Methylseleno-L-cysteine (MSC), L-selenomethionine (SM), L-phenylalanine, ammediol [2-amino-2-methyl-1,3-propanediol], metaphosphoric acid (MPA), Trichostatin A, sodium butyrate and sodium α-keto-γ-methylthiobutyrate (KMB) were purchased from Sigma-Aldrich (St. Louis, MO). A synthetic organoselenium compound, p-XSC [1,4-phenylene bis (methylene)selenocyanate] was synthesized as reported previously (34–36) and was a gift from Dr. Karam El-Bayoumy, Penn State Milton S. Hershey Medical Center. Recombinant human glutamine transaminase K (rhGTK) (also referred to as human kynurenine aminotransferase I) was obtained by the method of Han et al. (37) and generously supplied (16 mg/mL, 18 U/mg in the standard phenylalanine-KMB assay; see below) by Dr. Jianyong Li, Department of Biochemistry, Virginia Tech., Blacksburg, VA. Crotalus adamanteus L-amino acid oxidase (LAAO) (7.3 U/mg) and bovine liver catalase (200,000 U/ml) were purchased from Sigma Chemical Company (St. Louis, MO).
Cell lines and culture conditions
Androgen responsive LNCaP and androgen-independent PC-3 and DU145 cells were obtained from the American Type Culture Collection (Manassas, VA) and the androgen-independent clone of LNCaP cells, LNCaP C4-2, was a generous gift from Dr. Warren D.W. Heston (The Lerner Research Institute, the Cleveland Clinic Foundation, OH). LNCaP, LNCaP C4-2, and PC-3 cells were cultured in phenol red-free RPMI 1640 medium (GIBCO/Invitrogen) supplemented with 5% (v/v) fetal bovine serum (FBS) (Mediatech, Cellgro, Manassas, VA) and 1× non-essential amino acid solution (Mediatech, Cellgro, Manassas, VA). DU145 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, GIBCO/Invitrogen) supplemented with 5% (v/v) FBS, 1× non-essential amino acid solution and 1 mM pyruvate. All four cell lines were seeded 48 to 72 hr before the experiments (1 × 106) into 100 mm culture dishes and cultured in a humidified incubator at 37 °C and 5% CO2. After treatments (10–50 μM of MSP or KMSB; 100–200 μM MSC or SM; 2.5 mM NaB or 20 μM TSA) for 5 and 24 hrs, cells were washed twice with ice-cold phosphate buffered saline (PBS) and harvested for Western blot analysis into RIPA (Radio Immuno Precipitation Assay) buffer (150 mM NaCl, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris, pH 8.0) including protease inhibitors, 200 μM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 100 μM leupeptin, 800 nM aprotinin and 1 μg/ml pepstatin A. Cells were also harvested into PBS and their nuclear and cytosolic fractions were separated using NE-PER nuclear and cytoplasmic extraction reagents (Pierce Chemical Co., Rockford, IL) for HDAC assay. All samples were stored at −80 °C until use.
HDAC activity assay
HDAC activity was determined in nuclear and cytosolic extracts using the fluorometric HDAC Activity Assay Kit (BioVision, CA) according to the manufacturer's instruction. This assay is based on the Fluor de Lys (Fluorogenic Histone Deacetylase Lysyl Substrate/Developer) Substrate and Developer combination. Incubations were performed at 37°C for 30 minutes with nuclear or cytosolic extracts from human prostate cancer cells (10 to 50 μg total protein) or with HeLa nuclear extract (supplied with the kit), and the reaction was initiated by the addition of HDAC substrate (Boc-Lys(Ac)-AMC). Both androgen responsive (LNCaP) and androgen independent (C4-2, PC3, and DU145) prostate cancer cells were used for HDAC activity measurements. After 30 min, lysine developer was added and the mixture was incubated at room temperature for 30 min. Fluorescence was measured in a SpectraMax Gemini XS Microplate (Molecular Devices Corporation, Sunnyvale, CA) using an excitation wavelength of 365 nm and an emission wavelength of 450 nm. HDAC activity was expressed as the relative ratio of HDAC activity in control (basal level) to that in cells treated with MSC, SM, and their α-keto acid products after calculating the relative fluorescence units per μg protein.
Protein determination and Western blot analyses for histone 3 and acetylated histone 3
Protein concentrations were determined using the BCA protein assay kit with bovine serum albumin (BSA) as a standard (Pierce Chemical Co., Rockford, IL). For Western blot analysis, 20 to 100 μg of total soluble proteins were separated by one-dimensional SDS-PAGE using 10 or 12% (w/v) gels under reducing conditions. Proteins were transferred to polyvinylidene fluoride (PVDF) membrane (Millipore) and the membrane was blocked for 1 hr with 10% non-fat milk solution (Carnation), probed with one of two primary antibody solutions for acetylated histone H3 and histone H3 (Santa Cruz Biotechnology, CA) for 2 hr at room temperature or overnight at 4 °C. The membrane was then probed with secondary antibody solution (goat anti-rabbit IgG or goat anti-mouse IgG, peroxidase conjugated, Pierce Chemical Co., Rockford, IL) for 1 hr at room temperature. Proteins were visualized by developing the membrane using chemiluminescence reagents (Pierce Chemical Co., Rockford, IL). Each membrane was stripped and re-probed with anti-β-actin antibody (Santa Cruz Biotechnology, CA). Equivalent sample loading was further confirmed using Ponceau S staining (Boston BioProducts, Boston, MA).
Enzyme assays
The GTK assay in whole cell extracts was performed according to the published procedure (26), measuring transamination between L-phenylalanine and KMB. The reaction mixture consisted of 20 mM L-phenylalanine, 5 mM KMB, 100 mM ammediol-HCl buffer (pH 9.0) and homogenates from prostate cancer cells. After incubation at 37 °C for 30 min, the reaction was stopped by adding 0.15 ml of 1 M NaOH and absorbance of phenylpyruvate-enol was measured at 320 nm wavelength (ε = 16,000 M−1 cm−1). The blank reaction lacked KMB. For determining substrate specificity of GTK for MSC and SM, a stock solution of rhGTK was prepared (57.6 mU/mg protein) in 15 mM potassium phosphate buffer (pH 6.8) containing 20% glycerol and stored at 4 °C until use. The reaction mixture consisted of 10 mM MSC or SM, 5 mM KMB, 100 mM sodium phosphate buffer (pH 7.4) and 3.2 μg purified rhGTK. The change in buffer and pH provides comparable GTK activity but under physiological conditions. In addition, when MSC and SM were used as amino acid substrates and KMB as co-substrate, concentrations of the corresponding transamination products namely, MSP, KMSB, and methionine, were measured by HPLC using CoulArray detection (see below).
To synthesize the α-keto acid metabolites of MSC and SM, each of the organoselenium compounds was incubated for 2 hr with LAAO. The reaction mixture consisted of 5 mM MSC or SM, 100 mM potassium phosphate buffer (pH 7.4), 100 U catalase, and 0.1 U LAAO in a total volume of 0.5 mL. The reaction was conducted at 37 °C for 1 hr and deproteinized by either addition of 25% metaphosphoric acid (MPA) or passing the reaction mixture through a YM-10 Microcon Centrifugal Filter (10,000 nominal molecular weight cut off limit, Millipore Corporation, Danvers, MA) prior to measuring the corresponding α-keto acid products of MSC and SM by HPLC.
HPLC determination of MSC, SM, and their corresponding α-keto acids
The α-keto acids generated enzymatically from MSC and SM, namely MSP and KMSB, respectively, were analyzed by HPLC with electrochemical detection. When transamination reactions using rhGTK were performed with MSC and SM, the α-keto acid co-substrate was α-keto-γ-methylthiobutyrate (KMB). After the transamination reaction, the resulting amino acid formed from KMB is methionine, which was also detected by HPLC using CoulArray detection.
Since MSC, SM, MSP, KMSB, methionine and KMB are redox active compounds, they are easily determined by HPLC with CoulArray detection without the need for prior derivatization. After reaction with rhGTK or LAAO, the reaction was stopped by addition of 25% (w/v) MPA to yield a 5% (w/v) MPA solution. A Rheodyne injection valve with a 5-μl sample loop was used to manually introduce samples directly onto a Bio-Sil ODS-5S, 5-μm particle size, 4.0 × 250 mm, C18 column (Bio-Rad, Life Science Research Group, Hercules, CA). Samples were eluted with a mobile phase consisting of 50 mM NaH2PO4, 3% (v/v) acetonitrile, 1 mM octanesulfonic acid (pH 2.6) at a flow rate of 1 ml/min. PEEK™ (polyetheretherketone) tubing was used throughout the HPLC system, and 0.2 μ PEEK™ filters were placed pre- and post-column to protect both column and flow cells, respectively, from any particulate matter. The 8-channel CoulArray detectors (ESA, Inc., Chelmsford, MA) were set at 175, 250, 325, 400, 475, 550, 700 and 800 mV, respectively.
Mass Spectral Analysis
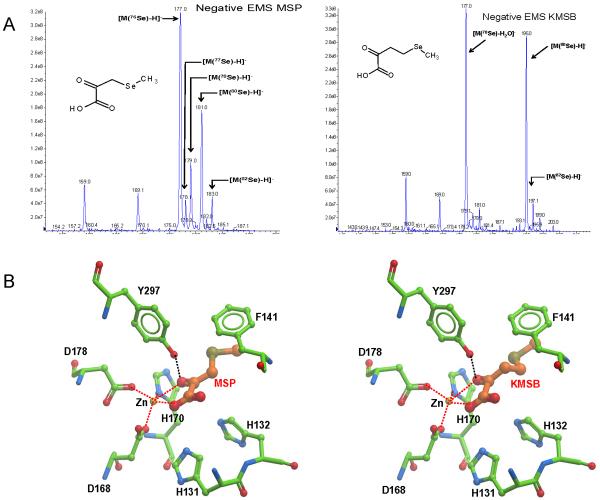
Negative electro spray ionization spectra were determined on an Applied Biosystems 4000 Q Trap (Triple quadrupole) mass spectrometer for compound MSP and MSKB. Prior to the mass infusion analysis, the synthetic mixture was desalted using a 1 mL, 30 mg of Oasis MAX SPE column from Waters. The cartridges were conditioned with 1 mL MeOH, and then equilibrated with 1 mL water, loaded with 50 μL of the synthetic mixture added with 200 uL H2O and 30 μL of 28% NH4OH. The column was washed with 1 mL H2O and eluted with 2 mL of 2% formic acid in MeOH. The eluant was evaporated to dryness with a speedvac and the sample reconstituted with 150 μL of 50/50 acetonitrile/0.1% formic acid. For compound MSP, the negative peak observed at m/z 178, 179, 181 and 183 reflect the isotopic distribution of Se (Figure 2) which is consistent with the natural abundance of selenium isotopes (Se74, Se76, Se78, Se77, Se79, Se80 and Se82) thus indicating the presence of a single selenium atom in a molecular formula of C4H6O3Se for MSP. Similarly, the peaks observed at m/z 193,195, 197 (C5H8O3Se) are the negative molecular ions of KMSB corresponding to Se78, Se80, Se82.
Figure 2.
Mass spectral analysis of α-keto acid metabolites of methylselenocysteine and selenomethionine and their molecular modeling docked into HDAC. A, Molecular ions ranging from 177 - 183 m/z were observed for MSP (left) and from 193 -197 m/z for KMSB (right). These peaks were consistent with isotope patterns for Se, Se-76 (9.36%), −77 (7.63%), −78 (23.8%), −80 (49.6%), and −82 (8.73%) and for molecular formulae of C4H6O3Se and C5H8O3Se, respectively. As a result of the collision energy, the peak at 177 for KMSB may represent a fragment ion containing a selenonium moiety following the removal of H2O. B, Docking of MSP (left) and KMSB (right) into Aquifex aeolicus HDAC homologue catalytic domain (ICM v3.5–1p). The receptor is represented by six grid potential maps accounting for hydrophobicity, van der Waals interactions, hydrogen-bonding and electrostatic potential. Both ligands are fully flexible in the field of the receptor. Each ligand was docked three times into the receptor and the most favorable orientation is show in the figure. The ligands (MSP and KMSB) are colored by atom type (carbon atoms, orange; oxygen, red, and selenium, pea green) and displayed as sticks. The zinc atom (Zn) and key HDAC residues are labeled in black lettering with the carbon atoms (light green) displayed as sticks. H-Bonds are represented by a black dashed line between tyrosine donor (Y297) and the selenoketo acid acceptor (SKA), respectively, and are defined as follows: Distance Y---SKA: 2.8 – 3.2 Å; Angle Y-H---SKA: 140–180°. Zinc coordination with the organoselenium derivatives, MSP and KMSB, and HDAC amino acid residues is represented by red dashed lines.
In silicomolecular modeling of MSP and KMSB docking into HDAC
The 3D coordinates of bacterial Aquifex aeolicus HDAC homologue catalytic domain were taken from the crystal structure data available in the Protein Data Bank (Pdb) 1C3R (Trichostatin A (TSA)/HDAC homologue) (38). The model was energetically refined in the internal coordinate space with Molsoft ICM v3.5-1p. The histidyl moieties at HDAC sites 131 and 132 were selected to be in the δ protonated state which was critical for demonstrating proper docking of hydroxamic acid–based inhibitors (39).
Energy calculation and optimization
The molecular system is described in terms of internal coordinate variables, using the electrostatically-driven Monte Carlo method with the Empirical Conformational Energy Program for Peptides/3 (ECEPP/3) force field (40), with distance-dependent dielectric constants for energy calculations as implemented in ICM (41). The method used in our studies is an extension of the Biased Probability Monte Carlo (BPMC) with minimization procedure for global energy optimization (42, 43). The BPMC global energy optimization method consisted of the following steps (i) a random conformation change of the free variables according to a pre-defined continuous probability distribution (42, 43); (ii) local energy minimization of analytical differentiable terms; (iii) calculation of the complete energy including non-differentiable terms such as entropy and solvation energy; iv) acceptance or rejection of the total energy based on the Metropolis criterion (44) and return to step (i).
Molecular docking
The Internal Coordinate ,Mechanics (ICM) structure-based virtual screening method was used (45) to simulate docking of each selenoketo acid. The receptor is represented by six grid potential maps accounting for hydrophobicity, van der Waals interactions hydrogen-bonding and electrostatic potential. The ligand is considered fully flexible in the field of the receptor. To ensure convergence of the BPMC based global energy minimization, each ligand was docked three times into the receptor, and the best scoring configuration for each compound was attained (Molsoft ICM v3.5-1p).
Statistical analyses
Results were statistically analyzed by a one-way ANOVA and Dunnett's comparison to test the significance of differences in measured variables between control and experimental groups in Minitab (Release 13.1). Statistically significant differences are denoted at P ≤ 0.05 or P ≤ 0.01.
RESULTS
Reactions of MSC and SM with LAAO
Based on previous findings that selenocysteine Se-conjugates are GTK and LAAO substrates (24, 26), we sought to prepare the corresponding α-keto acid derivatives of MSC and SM as depicted in Figure 1. Mass spectral analysis of the reaction products formed during 2 hr incubation with LAAO at 37 °C revealed α-keto acid products of MSC and SM, namely MSP and KMSB, respectively (Figure 2) both compounds exhibited patterns containing the naturally abundant isotopes of selenium. As MSC and SM were stoichiometrically metabolized into α-keto acid products via the action of LAAO (data not shown), we used this procedure to separate and quantify MSP and KMSB using HPLC with electrochemical detection.
Separation of reaction products using reverse-phase HPLC
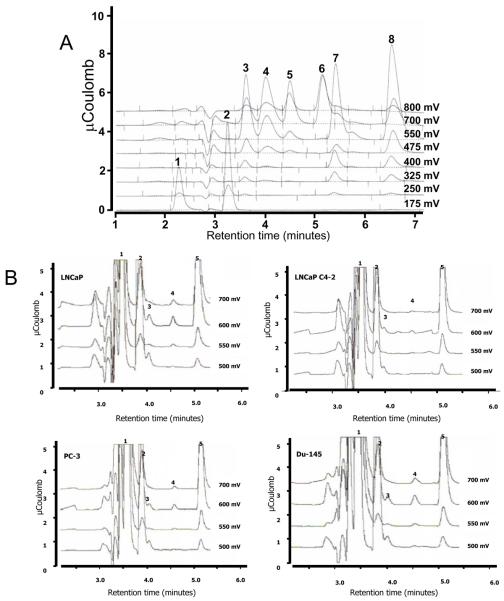
For the initial purification and characterization of the reaction products from rhGTK and prostate cancer cell lysates, reaction mixtures were precipitated with 5% MPA and were subjected to HPLC analysis using a C18 column. Compounds were eluted with phosphate buffer containing 3% acetonitrile. The elution pattern was monitored using coulometric detection. Figure 3 illustrates the peak elution times and oxidation patterns of a standard mixture containing MSC, MSP, methionine, KMB, SM, and KMSB and having retention times at 3.6, 4.0, 4.5, 5.2, 5.4 and 6.6 min, respectively. Ascorbic acid (retention time 3.2 min, 175 mV) was incorporated in the mixture as a redox protectant.
Figure 3.
A. HPLC coulometric analysis of selenium and sulfur amino acids and their keto acid metabolites. A, the peak elution times and patterns of oxidation of a standard mixture (50 nmol/mL each) of MSC (peak 3), MSP (peak 4), methionine (peak 5), KMB (peak 6), SM (peak 7), and KMSB (peak 8) are shown. Ascorbate (peak 2) (175 mVolts) is used in the mixture as a marker and a redox protectant; standard mixtures are prepared in 5% (w/v) metaphosphoric acid (peak 1). B, Generation of MSP (peak 3), the deaminated derivative of MSC (peak 1), was detected by HPLC analysis in prostate cancer cells as an action of transamination with α-keto-γ-methylthiobutyrate (KMB) (peak 5). Extracts (100 μg protein) from LNCaP, LNCaP C4-2, PC-3 and DU145 cells were incubated for 2 hr at 37°C and specific activities were 1.05, 0.56, 0.72, and 0.82 μmol of MSP formed. hr−1 . mg−1 protein, respectively. MSC (peak 1); glutathione (peak 2); MSP (peak 3); methionine (peak 4); KMB (peak 5).
Determination of GTK activity in human prostate cancer cells
Commandeur et al. (24) showed that rat kidney GTK catalyzes both a β-elimination reaction and a transamination reaction with MSC. The transamination product was not characterized in that study. Recently, Cooper et al. (26) showed that rhGTK catalyzes a similar reaction with MSC but not to any appreciable extent with SM. Here, we confirm that the α-keto acid product of MSC is MSP which structurally resembles the known HDAC inhibitor, butyrate. We hypothesized that prostate cancer cells possess GTK or other aminotransferases capable of generating MSP. Androgen responsive (LNCaP) and androgen independent (C4-2, PC-3, and DU145) cells were tested for GTK activity using the standard phenylalanine-KMB assay mixture. Table 1 demonstrates the presence of GTK in human prostate cancer cells and compares this activity with that present in rat kidney.
Table 1.
Specific activity of GTK in human prostate cancer cells.
| Cell line/tissue | Phenylpyruvate formed (nmol·hr-1·mg-1 protein) |
|---|---|
| LNCaP | 21.4 ± 2.1 |
| LNCaP C4-2 | 31.9 ± 3.6 |
| PC-3 | 23.3 ± 0.3 |
| DU145 | 30.9 ± 1.7 |
| Rat kidney | 641 ± 10 |
The reaction mixture consisted of 20 mM L-phenylalanine, 5 mM KMB, 100 mM ammediol-HCl buffer (pH 9.0) and homogenates from prostate cancer cells. After incubation at 37 °C for 30 min, the reaction was stopped by adding 0.15 ml of 1 M NaOH and absorbance of phenylpyruvate was measured at 320 nm wavelength (ε = 16,000 M-1 cm-1). The blank reaction lacked KMB. Rat kidney cytosol served as a positive control for GTK activity. Each value is mean ± SEM of triplicate determinations.
Subsequent measurements of human prostate cancer cell extracts using MSC rather than phenylalanine as the amino acid substrate and KMB as the α-keto acid co-substrate revealed formation of MSP (Figure 4). In this experiment, MSP was generated at least in part from endogenous activity of GTK. Specific activities for LNCaP, LNCaP C4-2, PC-3, and DU145 were 1.05, 0.56, 0.72, and 0.82 μmol of MSP formed · hr−1 · mg−1 protein, respectively.
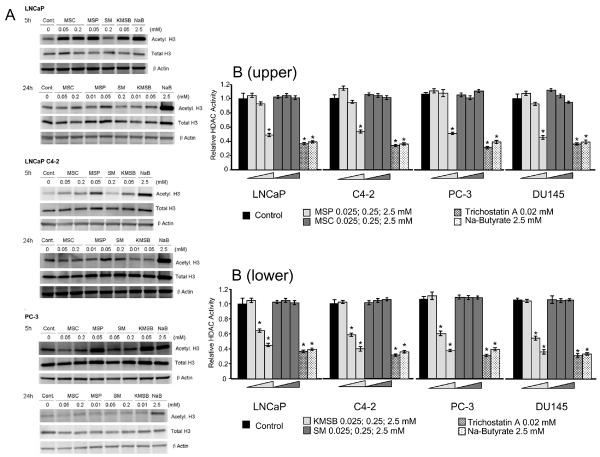
Figure 4.
A, Increased acetylated-histone-H3 expression in LNCaP, LNCaPC4–2 and PC-3 cell lysates. Cells were treated with MSC (50 and 200 μM), SM (50 and 200 μM), MSP (10 and 50 μM), or KMSB (10 and 50 μM) for 5 and 24-hrs. Sodium butyrate was used as positive control. Each lane was loaded with 25 μg of total soluble protein. HDAC inhibitory effects of MSP and KMSB occur as early as 5 hr post treatment in AR- and AI-human prostate cancer cells. B, HDAC inhibition by MSP and KMSB in nuclear fractions in human prostate cancer cells. Upper panel shows that the relative HDAC total activity in vitro exhibits a dose dependent decrease when nuclear fractions of prostate cancer cells are treated with 0.025, 0.25, and 2.5 mM MSP (wedge symbol). A 47–52% inhibition was observed at 2.5 mM MSP and marginal inhibition at 0.25 mM. No inhibition was observed with 0.025, 0.25 and 2.5 mM MSC (wedge symbol). Lower panel shows that KMSB at 0.025 mM exhibits no effect but 0.25 and 2.5 mM (wedge symbol) exhibit significant HDAC inhibition. SM at 0.025, 0.25 and 2.5 mM (wedge symbol) had no effect. Statistically significant differences are denoted: * = p < 0.01. Known HDAC inhibitors were included for comparison, namely trichostatin A (20 μM) and sodium butyrate (2.5 mM). Each assay was carried out in triplicate.
Since GTK has an unusual “crown” of aromatic amino acid residues in the binding pocket (46) and can accommodate a variety of large amino acid substrates e.g., 5-S-L-cysteinyl-L-DOPA and 5-S-L-cysteinyldopamine (26), we also considered the possibility that the cysteinylated or glutathionylated derivative of the synthetic organoselenium compound, p-XSC, may also undergo transamination. However, neither rhGTK nor the prostate cancer cell extracts revealed formation of any corresponding keto acid product (data not shown).
MSC treatment increases acetylated histone-3 expression in prostate cancer cell lines
Naturally occurring (MSC, SM) and synthetic (p-XSC) organoselenium compounds were added to cell culture medium and the expressed level of acetylated histone 3, as well as non-acetylated histone 3 (as loading control), were examined by immunoblot analyses of whole cell lysates (Figure 5). Compared with controls, MSC-treated cells expressed increased levels of acetylated histone3 after 5 hr incubation in LNCaP and LNCaP C4-2 cells, but slightly lower levels in PC-3 cell lysates. The finding that LNCaP cells showed a more marked accumulation of acetylated histone 3 in the presence of MSC at both 0.05 and 0.2 mM concentrations after 5 hr incubation may be reflective of the higher specific activity of GTK for MSC in LNCaP compared to LNCaP C4-2 and PC3 cells (see Figure 4). However, the expression of acetylated histone 3 decreased after 24-hr incubation suggesting a rapid, transient response. By contrast, incubation of cells for up to 48 hr with SM did not show an increase in acetylated-histone 3 in any of the cell lines tested, further demonstrating that it is not an active substrate for transamination in human prostate cancer cells. Treatment of cells with the α-keto acid forms of MSC and SM, namely MSP and KMSB, resulted in elevation of acetylated histone 3 in all cell lines within 5 hr at 50 μM concentration, supporting our hypothesis that the increase level of acetylated histone 3 following MSC treatment is due to its metabolite, MSP (PC-3 cells did not survive with 50 μM MSP or KMSB concentrations after 24 hr incubation). In addition, direct incubation of cells with p-XSC for 24 hrs did not show accumulation of acetylated-histone H3 in prostate cancer cells (data not shown). Thus, neither the selenoamino acids, MSC and SM, nor the selenoketo acids, MSP and KMSB, affect total histone 3 levels and only the selenoketo acids increase the ratio of acetylated to total histone 3.
A metabolite of MSC, rather than MSC, inhibits HDAC activity in prostate cancer cells
As shown in Figure 6a, MSC had no direct HDAC inhibitory effect in nuclear fractions of the four prostate cancer cell lines, despite the observation in Figure 5 of increased acetylated-histone H3 expression in cells exposed to MSC. We hypothesized that a metabolite of MSC, rather than MSC itself, inhibits HDAC activity in human prostate cancer cells. To test our hypothesis, MSP was generated by incubating MSC with LAAO for 2 hr at 37 °C and used for HDAC activity assay. HPLC analysis verified that the increase in concentration of MSP closely matched the decrease in concentration of MSC during the incubation (result not shown).
As shown in Figure 6a, whereas MSC had no effect up to 2.5 mM, HDAC activity was decreased 47 – 52% when nuclear fractions of all four human prostate cancer cell lines were treated with 2.5 mM MSP. The inhibition by 2.5 mM MSP was comparable to that seen with known HDAC inhibitors, trichostatin A (20 μM) and sodium butyrate (2.5 mM). Using the in vitro conditions reported here, we did not observe HDAC inhibition with concentrations lower than 0.025 mM MSP. This finding in vitro differs from that observed under cell culture conditions (See Figure 5). We surmise the disparity may be due to competitive differences between MSP and the fluorogenic histone substrate coupled with developer selectivity and concentration ratio as required by manufacturer's instruction. Results similar to those with MSC were observed with SM, in that SM had no direct effect on HDAC activity up to 2.5 mM, whereas its α-keto acid product, KMSB exhibited significant HDAC inhibition at 0.25 and 2.5 mM (Figure 6b). These results corroborate the findings in Figure 5 and demonstrate accumulation of acetylated histone H3 when KMSB rather than SM was introduced directly to cells in culture.
Molecular modeling and docking calculations for seleno α-keto acids
The bacterial Aquifex aeolicus HDAC homologue catalytic domain was built up using the experimentally resolved crystal structure complexed with ligand TSA available in the Pdb 1C3R (39) and energetically minimized in the internal coordinates space. The model shares most of the residues involved in the Zn-atom coordination and in the biological water-mediated catalysis of acetylated lysines with human class I HDACs as well as HDACs from other species (38, 39). To validate the model, TSA first was docked iteratively into the binding pocket, and the most energetically favorable position was compared with the crystal structure (39). TSA docked with an orientation similar to that found experimentally, producing an all-atoms root-mean-square distance value of 0.55 (results not shown).
Next, MSP and KMSB were docked into the HDAC model and the most energetically favorable docking pose obtained was essentially the same for both α-keto acid derivatives (Figure 2B left and right). The orientation of the α-carbonyl group and the carboxylate moiety of MSP and KMSB in the pocket closely resembles that seen for the carbonyl group of known HDAC inhibitors, MS-344 and TSA (38) with respect to both Zn-atom coordination and the hydrogen bond to the hydroxyl group of Y297. One of the oxygen atoms of the terminal carboxylate group also stabilizes Zn-atom coordination in a fashion similar to that reported earlier for sulforaphane-cysteine (45; 48–50). The aliphatic chain of both MSP and MSKB that contains the Se-atom interacts with the phenylalanine group (F141) and this interaction is comparable to that seen in complexes with SAHA (51).
DISCUSSION
Although most former studies on selenium as a nutrient and chemopreventive agent have focused on its role as an essential component of several selenium-containing enzymes (glutathione peroxidases, thioredoxin reductase, iodothyronine 5'-deiodinase) and other selenoproteins (52), recent studies show that small molecular weight organoselenium derivatives may also have intrinsic value. Over the past 10 years, a number of naturally occurring and synthetic organoselenium compounds have been examined for their anticancer properties. It appears that, depending on the molecular form of selenium, selenium compounds possess several modes of action affecting cancer promotion and progression. First, when metabolized as an essential nutrient, the incorporation of selenium into selenoproteins as the 21st coded amino acid, selenocysteine, enables several endogenous selenoenzymes to counteract peroxidative reactions and function as redox catalyzers. Secondly, exogenously administered organoselenium compounds may expose mitotic cells to a pool of novel organoselenium metabolites that have the potential to interact with specific molecular targets necessary for mediating chemopreventive activity. The former chemopreventive mechanisms may be relevant to protecting cells against cancer initiation while the latter mechanism may thwart tumor cell progression by maintaining a non-proliferative intracellular environment.
The anticancer mechanism(s) of selenium-enriched diets have focused on two organoselenium nutrients, MSC and SM, and their potential for being converted to methylselenol via hypothesized β- and γ-elimination reactions, respectively. The anticancer activity of methylselenol is purported to manifest through the oxidation of thiol compounds on proteins and/or enzymes, thereby changing the redox environment of cells (53–55).
Several PLP-containing enzymes are known to catalyze β-elimination reactions, for example serine deaminase. In addition, cystathionine γ-lyase can catalyze a β-elimination reaction with certain cysteine S-conjugates and with cystine (56 and reference quoted therein). In these two cases, there is no competing aminotransferase reaction. By contrast, several aminotransferases have been shown to catalyze β-elimination reactions with cysteine S-conjugates that contain a good leaving group in the β position. The β-elimination often competes with the transamination reaction. Thus, as noted in the introduction, an α-keto acid substrate or PLP is required for the β-lyase reaction to proceed. As also noted, we recently showed that rhGTK can catalyze both transamination and β-lyase reactions with MSC (26). This result suggested that the transaminated product of MSC, in addition to the product of β-elimination, methylselenol, may play critical roles in chemoprevention.
The present study considered the possibility that the α-keto acid products of MSC and SM would exhibit HDAC inhibitory properties. We first determined whether human prostate cancer cells metabolize naturally-occurring organoselenium amino acids to α-keto acid products, whether these products might increase the levels of histone-H3 acetylation. The α-keto acid products of both MSC and SM were generated using LAAO and a convenient assay system was developed using reverse phase HPLC coupled with electrochemical detection to identify the seleno-α-keto acid products. Both α-keto acids, MSP as a metabolite of MSC and KMSB as a metabolite of SM, were verified as the products of the LAAO reaction using MS analysis and further analyzed for HDAC inhibitory properties using a fluorescent-based assay for measuring Class I and II HDAC activity. Accordingly, each α-keto acid product inhibited HDAC activity in a dose dependent manner in a HeLa cell-based assay that monitors histone H3 acetylation.
Both androgen responsive (LNCaP) and independent (LNCaP C4-2, PC-3, and DU-145) cell lines were used to test our hypotheses that α-keto acid products of MSC and SM function as HDAC inhibitors in a whole cell environment. First, we determined whether prostate cancer cells possess GTK activity and have the ability to transaminate MSC. Using the standard aminotransferase substrates for GTK, phenylalanine and KMB, all four cell lines exhibited GTK activity. Curiously, although methionine functions as an excellent transamination substrate for rhGTK, SM is less than 0.3% as reactive (26). This was further corroborated when whole cell lysates from each of the prostate cancer cell lines were incubated with MSC and SM and aliquots removed after two hours and analyzed by HPLC to detect their corresponding α-keto acid metabolites. Only MSP could be detected whereas SM was not metabolized to KMSB even after 24 h incubation. In order not to negate long-term effects of SM when administered orally to animals, we are investigating the possibility that dietary forms of SM (from general protein sources or its free form) may undergo hepatic trans-selenation reactions to form methylselenocysteine and thus contribute to the seleno α-keto acid product via this pathway (53).
Treatment of LNCaP, LNCaP C4-2, PC-3 and Du-145 prostate cancer cells with MSC led to time- and dose-dependent changes in histone acetylation status. Treatment of cells with MSC resulted in an increased accumulation of acetylated histone H3 in LNCaP cells as early as 5 hrs and in the androgen independent cell lines within 24 hrs post-incubation. Cells treated with SM did not show acetylated histone H3 accumulation even after 24 hrs incubation. When cells were directly treated with the seleno α-keto acid products, MSP and KMSB, acetylated histone H3 accumulated as early as 5 hours post-treatment. We are currently investigating the possibility that longer incubation times (> 48 hr) may be required to generate sufficient concentrations of KMSB necessary to elicit HDAC inhibition. Preliminary enzyme kinetic data suggest that the Ki of KMSB for HDAC3 inhibition may be around 30 μM. These findings are consistent with our previous finding that MSC is >100 times more effective as an aminotransferase substrate for rhGTK than is SM despite the difference of only one methylene group (26).
By contrast, treatment of cells directly with the α-keto acid products of MSC and SM result in increased accumulation of acetylated histone H3 in as early as 5 hours after incubation. Selenomethionine is not directly metabolized by human prostate cancer cells to KMSB. KMSB can be detected in liver homogenates incubated with SM suggesting that liver tissue may contain other aminotransferases or an L-AAO as well as cystathionine γ-lyase, although not detected in prostate cancer cells, which may catalyze deamination or γ-elimination products, respectively, from selenomethionine. A question of residence time, stability, and extent of plasma circulation will be important factors to consider in subsequent studies, including potential follow-up to the recently halted SELECT study.
At present, we cannot conclude from our in vitro studies whether sufficient plasma levels of the α-keto acid products of MSC and SM can be achieved in preclinical models of prostate cancer over time with daily consumption or whether unequivocal HDAC inhibition occurs in vivo when dietary sources of selenium in the range of 2 to 6 ppm are consumed. This caveat withstanding, most tissues with exception of erythrocytes contain aminotransferases potentially capable of metabolizing MSC or even SM (26), however pharmacokinetic studies need to be conducted to determine bioavailability and metabolic fates of these compounds using our HPLC electrochemical method. As mentioned earlier, preliminary studies using an in vitro assay of HDAC activity suggest that the IC50 may be in the range of 30–50 μM, a level that may be achievable following administration or consumption of the pre-formed α-keto acid products. These effects remain to be established in a preclinical model of prostate cancer.
In conclusion, we have identified two metabolites of naturally occurring organoselenium derivatives as novel HDAC inhibitors. We first developed an assay system for these α-keto acid metabolites using HPLC with electrochemical detection and identified one of these metabolites (MSP) within prostate cancer cells. The findings for MSP and KMSB with respect to HDAC inhibition and induction of acetylated histone H3 in prostate cancer cells extend our current understanding of how organoselenium may decrease progression of human prostate cancer. By altering posttranslational modifications of histones, HDAC inhibitors facilitate access of transcription factors to DNA, and enable the unsilencing of tumor suppressor genes in cancer cells (57, 58). This paradigm has been viewed from the perspective of developing novel drugs for cancer therapy, but is now expanding to include dietary chemopreventive agents (48–50, 59). The next goal will be to define protein targets of seleno-α-keto acids and the underlying mechanisms, which might help to re-focus efforts in the wake of the recently halted SELECT study.
ACKNOWLEDGEMENTS
We thank Heather Twerdahl and Lawson Kurtz for technical assistance. This work was supported in part by NIH grants CA111842 (to JTP & RS), ES8421 (to AJLC) and CA090890 and CA122959 (to RHD).
Abbreviations
- GTK
glutamine transaminase K
- HDAC
histone deacetylase
- KMB
α-keto-γ-methylthiobutyrate
- KMSB
α-keto-γ-methylselenobutyrate
- LAAO
L-amino acid oxidase
- MPA
metaphosphoric acid
- MSC
Se-methyl-L-selenocysteine
- MSP
β-methylselenopyruvate
- NaB
sodium butyrate
- PBS
phosphate buffered saline
- PLP
pyridoxal 5′-phosphate
- PVDF
Polyvinylidene fluoride
- p-XSC
1,4-phenylene bis (methylene)selenocyanate
- rhGTK
recombinant human GTK
- SM
selenomethionine
- TSA
trichostatin A
REFERENCES
- 1.ACS Cancer Facts and Figures. 2008 [Google Scholar]
- 2.Willett WC, Polk BF, Morris JS, et al. Prediagnostic serum selenium and risk of cancer. Lancet. 1983;2:130–4. doi: 10.1016/s0140-6736(83)90116-2. [DOI] [PubMed] [Google Scholar]
- 3.Clark LC, Alberts DS. Selenium and cancer: risk or protection? J Natl Cancer Inst. 1995;87:497–505. doi: 10.1093/jnci/87.7.473. [DOI] [PubMed] [Google Scholar]
- 4.Nyman DW, Suzanne Stratton M, Kopplin MJ, Dalkin BL, Nagle RB, Jay Gandolfi A. Selenium and selenomethionine levels in prostate cancer patients. Cancer Detect Prev. 2004;28:8–16. doi: 10.1016/j.cdp.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Brooks JD, Metter EJ, Chan DW, et al. Plasma selenium level before diagnosis and the risk of prostate cancer development. J Urol. 2001;166:2034–8. [PubMed] [Google Scholar]
- 6.Clark LC, Combs GF, Jr, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, Gross EG, Krongrad A, Lesher JL, Jr, Park HK, Sanders BB, Jr, Smith CL, Taylor JR. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. JAMA. 1996;276:1957–63. [PubMed] [Google Scholar]
- 7.Duffield-Lillico AJ, Reid ME, Turnbull BW, Combs GF, Jr., Slate EH, Fischbach LA, Marshall JR, Clark LC. Baseline characteristics and the effect of selenium supplementation on cancer incidence in a randomized clinical trial: a summary report of the Nutritional Prevention of Cancer Trial. Cancer Epidemiol Biomarkers Prev. 2002;11:630–9. [PubMed] [Google Scholar]
- 8.Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, Hartline JA, Parsons JK, Bearden JD, 3rd, Crawford ED, Goodman GE, Claudio J, Winquist E, Cook ED, Karp DD, Walther P, Lieber MM, Kristal AR, Darke AK, Arnold KB, Ganz PA, Santella RM, Albanes D, Taylor PR, Probstfield JL, Jagpal TJ, Crowley JJ, Meyskens FL, Jr, Baker LH, Coltman CA., Jr. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsavachidou D, McDonnell TJ, Wen S, Wang X, Vakar-Lopez F, Pisters LL, Pettaway CA, Wood CG, Do KA, Thall PF, Stephens C, Efstathiou E, Taylor R, Menter DG, Troncoso P, Lippman SM, Logothetis CJ, Kim J. Selenium and vitamin E: cell type- and intervention-specific tissue effects in prostate cancer. J Natl Cancer Inst. 2009;101:306–20. doi: 10.1093/jnci/djn512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinto JT, Sinha R, Papp K, Facompre ND, Desai D, El-Bayoumy K. Differential effects of naturally occurring and synthetic organoselenium compounds on biomarkers in androgen responsive and androgen independent human prostate carcinoma cells. Int J Cancer. 2007;120:1410–7. doi: 10.1002/ijc.22500. [DOI] [PubMed] [Google Scholar]
- 11.Dong Y, Lisk D, Block E, Ip C. Characterization of the biological activity of γ-glutamyl-Se-methylselenocysteine: a novel, naturally occurring anticancer agent from garlic. Cancer Res. 2001;61:2923–8. [PubMed] [Google Scholar]
- 12.Lyi SM, Heller LI, Rutzke M, Welch RM, Kochian LV, Li L. Molecular and biochemical characterization of the selenocysteine Se-methyltransferase gene and Se-methylselenocysteine synthesis in broccoli. Plant Physiol. 2005;138:409–20. doi: 10.1104/pp.104.056549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson WD, Morrissey RL, Kapetanovic I, Crowell JA, McCormick DL. Subchronic oral toxicity studies of Se-methylselenocysteine, an organoselenium compound for breast cancer prevention. Food Chem Toxicol. 2008;46:1068–78. doi: 10.1016/j.fct.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Bayoumy K, Sinha R. Mechanisms of mammary cancer chemoprevention by organoselenium compounds. Mutat Res. 2004;551:181–97. doi: 10.1016/j.mrfmmm.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 15.Medina D, Thompson H, Ganther H, Ip C. Se-Methylselenocysteine: A new compound for chemoprevention of breast cancer. Nutrition and Cancer. 2001;40:12–17. doi: 10.1207/S15327914NC401_5. [DOI] [PubMed] [Google Scholar]
- 16.Ip C, Dong Y. Methylselenocysteine modulates proliferation and apoptosis biomarkers in premalignant lesions of the rat mammary gland. Anticancer Res. 2001;21:863–7. [PubMed] [Google Scholar]
- 17.Unni E, Koul D, Yung WK, Sinha R. Se-methylselenocysteine inhibits phosphatidylinositol 3-kinase activity of mouse mammary epithelial tumor cells in vitro. Breast Cancer Res. 2005;7:R699–707. doi: 10.1186/bcr1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinha R, Kiley SC, Lu JX, Thompson HJ, Moraes R, Jaken S, Medina D. Effects of methylselenocysteine on PKC activity, cdk2 phosphorylation and gad gene expression in synchronized mouse mammary epithelial tumor cells. Cancer Lett. 1999;146:135–45. doi: 10.1016/s0304-3835(99)00250-5. [DOI] [PubMed] [Google Scholar]
- 19.Ip C, Ganther HE. Comparison of selenium and sulfur analogs in cancer prevention. Carcinogenesis. 1992;13:1167–70. doi: 10.1093/carcin/13.7.1167. [DOI] [PubMed] [Google Scholar]
- 20.Lee SO, Yeon Chun J, Nadiminty N, Trump DL, Ip C, Dong Y, Gao AC. Monomethylated selenium inhibits growth of LNCaP human prostate cancer xenograft accompanied by a decrease in the expression of androgen receptor and prostate-specific antigen (PSA) Prostate. 2006;66:1070–5. doi: 10.1002/pros.20329. [DOI] [PubMed] [Google Scholar]
- 21.Spallholz JE, Shriver BJ, Reid TW. Dimethyldiselenide and methylseleninic acid generate superoxide in an in vitro chemiluminescence assay in the presence of glutathione: implications for the anticarcinogenic activity of L-selenomethionine and L-Se-methylselenocysteine. Nutr Cancer. 2001;40:34–41. doi: 10.1207/S15327914NC401_8. [DOI] [PubMed] [Google Scholar]
- 22.Cooper AJL, Pinto JT. Aminotransferase, L-amino acid oxidase and β-lyase reactions involving L-cysteine S-conjugates found in allium extracts. Relevance to biological activity? Biochem. Pharmacol. 2005;69:209–20. doi: 10.1016/j.bcp.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 23.Stevens JL, Robbins JD, Byrd RA. A purified cysteine conjugate β-lyase from rat kidney cytosol. Requirement for an α-keto acid or an amino acid oxidase for activity and identity with soluble glutamine transaminase K. J Biol Chem. 1986;261(33):5529–37. [PubMed] [Google Scholar]
- 24.Cooper AJL. Mechanisms of cysteine S-conjugate β-lyases. Adv Enzymol Relat Areas Mol Biol. 1998;72:199–238. doi: 10.1002/9780470123188.ch6. [DOI] [PubMed] [Google Scholar]
- 25.Commandeur JNM, Andreadou I, Rooseboom M, Out M, de Leur LJ, Groot E, Vermeulen NPE. Bioactivation of selenocysteine Se-conjugates by a highly purified rat renal cysteine conjugate beta-lyase/glutamine transaminase K. J Pharmacol Exp Ther. 2000;294:753–61. [PubMed] [Google Scholar]
- 26.Cooper AJL. The role of glutamine transaminase K (GTK) in sulfur and α-keto acid metabolism in the brain, and in the possible bioactivation of neurotoxicants. Neurochem Int. 2004;44:557–77. doi: 10.1016/j.neuint.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Cooper AJL, Pinto JT, Krasnikov BF, Niatsetskaya ZV, Han Q, Li J, Vauzour D, Spencer JP. Substrate specificity of human glutamine transaminase K as an aminotransferase and as a cysteine S-conjugate β-lyase. Arch Biochem Biophys. 2008;474:72–81. doi: 10.1016/j.abb.2008.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rooseboom M, Vermeulen NPE, Durgut F, Commandeur JNM. Comparative study on the bioactivation mechanisms and cytotoxicity of Te-phenyl-L-tellurocysteine, Se-phenyl-L-selenocysteine, and S-phenyl-L-cysteine. Chem Res Toxicol. 2002;15:1610–8. doi: 10.1021/tx020034f. [DOI] [PubMed] [Google Scholar]
- 29.Butler KV, Kozikowski AP. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr Pharm Des. 2008;14:505–28. doi: 10.2174/138161208783885353. [DOI] [PubMed] [Google Scholar]
- 30.Insinga A, Monestiroli S, Ronzoni S, et al. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med. 2005;11:71–6. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- 31.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–52. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 32.Ungerstedt JS, Sowa Y, Xu WS, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. PNAS. 2005;102:673–8. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buggy JJ, Sideris ML, Mak P, Lorimer DD, McIntosh B, Clark JM. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem J. 2000;350:199–205. [PMC free article] [PubMed] [Google Scholar]
- 34.Vanommeslaeghe K, Loverix S, Geerlings P, Tourwé D. DFT-based ranking of zinc-binding groups in histone deacetylase inhibitors. Bioorg Med Chem. 2005;13:6070–82. doi: 10.1016/j.bmc.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 35.El-Bayoumy K, Narayanan BA, Desai DH, et al. Elucidation of molecular targets of mammary cancer chemoprevention in the rat by organoselenium compounds using cDNA microarray. Carcinogenesis. 2003;24:1505–14. doi: 10.1093/carcin/bgg103. [DOI] [PubMed] [Google Scholar]
- 36.El-Bayoumy K, Chae YH, Upadhyaya P, Meschter C, Cohen LA, Reddy BS. Inhibition of 7,12-Dimethylbenz(a)anthracene-induced tumors and DNA adduct formation in the mammary glands of female Sprague-Dawley rats by the synthetic organoselenium compound 1,4-phenylenebis(methylene)selenocyanate. Cancer Res. 1992;52:2402–7. [PubMed] [Google Scholar]
- 37.Ip C, Lisk DJ, Ganther HE. Activities of structurally-related lipophilic selenium compounds as cancer chemopreventive agents. Anticancer Res. 1998;18:4019–25. [PubMed] [Google Scholar]
- 38.Han Q, Li J, Li J. pH dependence, substrate specificity and inhibition of human kynurenine aminotransferase I. Eur J Biochem. 2004;271:4804–14. doi: 10.1111/j.1432-1033.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 39.Somoza JR, Skene RJ, Katz BA, et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–34. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Finnin MS, Donigian JR, Cohen A, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–93. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 41.Nemethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini MG, Zagari A. Energy parameters in polypeptides. 10. Improved geometrical parameters and non bonded interactions for use in the ECEPP/3 algorithm, with application to proline-containing peptides. J Phys Chem. 1992;96:6472–84. [Google Scholar]
- 42.Abagyan R, Totrov M, Kuznetsov D. ICM-a new method for protein modeling and design-applications to docking and structure prediction from the distorted native conformation. J Comp Chem. 1994;15:488–506. [Google Scholar]
- 43.Abagyan R, Totrov M. Biased probability Monte-Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 2003;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 44.Abagyan R, Totrov M. Ab-initio folding of peptides by the optimal-bias Monte Carlo minimization procedure. J Comp Phys. 1999;151:402–21. [Google Scholar]
- 45.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equation of state calculations by fast computing machines. J Chem Phys. 1953;21:1087–92. [Google Scholar]
- 46.Totrov M, Abagyan R. Protein-Ligand Docking as an Energy Optimization Problem. In: Raffa RB, editor. Drug-Receptor Thermodynamics: Introduction and Experimental Application. John Wiley & Sons; New York: 2001. pp. 603–24. [Google Scholar]
- 47.Rossi F, Han Q, Li J, Li J, Rizzi M. Crystal structure of human kynurenine aminotransferase I. J Biol Chem. 2004;279:50214–20. doi: 10.1074/jbc.M409291200. [DOI] [PubMed] [Google Scholar]
- 48.Rooseboom M, Commandeur JNM, Floor GC, Rettie AE, Vermeulen NPE. Selenoxidation by flavin-containing monooxygenases as a novel pathway for beta-elimination of selenocysteine Se-conjugates. Chem Res Toxicol. 2001;14:127–34. doi: 10.1021/tx0001326. [DOI] [PubMed] [Google Scholar]
- 49.Myzak MC, Karplus PA, Chung F-L, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Canc Res. 2004;64:5767–74. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 50.Dashwood RH, Myzak MC, Ho E. Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis. 2006;27:344–9. doi: 10.1093/carcin/bgi253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17:363–9. doi: 10.1016/j.semcancer.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berman HM, Westbrook J, Feng Z, Iype L, Schneider B, Zardecki C. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;11:3565–76. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki KT, Tsuji Y, Ohta Y, Suzuki N. Preferential organ distribution of methylselenol source Se-methylselenocysteine relative to methylseleninic acid. Toxicol Appl Pharmacol. 2008;227:76–83. doi: 10.1016/j.taap.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Medina D, Thompson H, Ganther H, Ip C. Se-Methylselenocysteine: a new compound for chemoprevention of breast cancer. Nutr Cancer. 2001;40:12–7. doi: 10.1207/S15327914NC401_5. [DOI] [PubMed] [Google Scholar]
- 56.Ganther HE. Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis. 1999;20:1657–66. doi: 10.1093/carcin/20.9.1657. [DOI] [PubMed] [Google Scholar]
- 57.Cooper AJL, Pinto JT. Role of cysteine S-conjugate βlyases in the bioactivation of renal toxicants. In: Elfarra AA, editor. Biotechnology: Pharmaceutical Aspects. Springer; New York, New York: 2008. pp. 323–346. Advances in Bioactivation Research. [Google Scholar]
- 58.Esteller M, Fraga MF, Paz MF, Campo E, Colomer D, Novo FJ, Calasanz MJ, Galm O, Guo M, Benitez J, Herman JG. Cancer epigenetics and methylation. Science. 2002;297:1807–8. doi: 10.1126/science.297.5588.1807d. [DOI] [PubMed] [Google Scholar]
- 59.Esteller M, Almouzni G. How epigenetics integrates nuclear functions. Workshop on epigenetics and chromatin: transcriptional regulation and beyond. EMBO Rep. 2005;6:624–8. doi: 10.1038/sj.embor.7400456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nian H, Delage B, Pinto JT, Dashwood RH. Allyl mercaptan, a garlic-derived organosulfur compound, inhibits histone deacetylase and enhances Sp3 binding on the P21WAF1 promoter. Carcinogenesis. 2008;29:1816–24. doi: 10.1093/carcin/bgn165. [DOI] [PMC free article] [PubMed] [Google Scholar]