Abstract
The neurexin-1 gene (NRXN1) has been shown to play a fundamental role in synaptogenesis and synaptic maintenance, as well as Ca2+ channel and NMDA receptor recruitment. A recent study reported that NRXN1 is associated with nicotine dependence (ND); this, together with the intriguing physiological functions of the gene, motivated us to investigate the involvement of NRXN1 with ND in independent samples. In this study, we analyzed 21 single nucleotide polymorphisms (SNPs) within NRXN1 for association with ND, which was assessed by smoking quantity (SQ), the heaviness of smoking index (HSI) and the Fagerström test for ND (FTND). Individual SNP and haplotype association tests were carried out in a sample consisting of 2037 individuals from 602 nuclear families of African-American (AA) or European-American (EA) origin. Individual SNP analysis revealed significant associations of rs2193225 with SQ, HSI and FTND (P = 0.00014–0.0010) in the EA sample and with SQ (P = 0.0019) in the pooled sample under the dominant model and rs6721498 with SQ, HSI and FTND in the AA (P = 0.000090–0.0000086) and pooled (P = 0.0010–0.00099) samples under the additive model, following correction for multiple testing. Haplotype analysis revealed six major haplotypes in the AA sample (minimum P-value = 0.000079), one major haplotype in the EA sample (P = 0.0062) and five major haplotypes in the pooled sample (minimum P-value = 0.00083), which showed significant association with all three ND measures; all of these contained one specific allele from one of the two aforementioned SNPs. Based on our findings that NRXN1 has significant association with ND in two independent samples, recent findings that NRXN1 plays an important role in synaptic development, and the previous report of association, we conclude that this gene represents a strong candidate for involvement in the etiology of ND.
INTRODUCTION
Despite the continued efforts of organizations and individuals, nicotine dependence (ND) continues to take a devastating toll on world health, primarily by way of cigarette smoking. In light of numerous epidemiological studies that indicate the importance of genetic factors in determining individual liability for ND (1,2), this continued smoking highlights the need for an improved genetic and molecular understanding of addiction. Identifying genetic factors that influence ND susceptibility has the potential to enhance individual pharmacological and preventative treatment options, and also to combat the difficulties that many dependent individuals find impossible to overcome. If future understanding of ND can achieve these goals, the loss of ∼435 000 American lives and more than $157 billion could be avoided annually (3,4). There is currently a substantial effort dedicated to reaching these goals (5).
Nicotine exerts its effect on the central nervous system by binding nicotinic acetylcholine receptors (6), and has been shown to increase dopamine levels in the nuclear accumbens (7). These molecular mechanisms are thought to underlie the rewarding and eventually addictive actions of nicotine. While the most obvious direct effects of nicotine result in changes in neurotransmitter release, there are likely complex multi-step pathways that are affected downstream from these actions such as neural circuit formation, which play a key role in ND development. Identifying genes involved in synaptic development and neural circuit formation has the potential to elucidate these pathways.
The neurexins are a trigenic family of proteins that function as cell adhesion molecules and play a key role in synaptic formation and maintenance (8,9). The three genes, NRXN1 (2q16.3), NRXN2 (11q13.2) and NRXN3 (11q13.2), produce both long α-neurexins and a truncated β-neurexins, which are regulated independently by their own promoters and are all predominantly expressed in neurons (10,11). Interestingly, neurexins undergo extensive alternative splicing, generating a great diversity of more than 2000 potential isoforms (10,11). These different isoforms of neurexins may display differential abilities to bind their extracellular ligands in the brain, which include neuroligins, neurexophilins and dystroglycans (8,9). First identified as receptors for the spider venom component α-latrotoxin to mediate neurotransmitter release (12), neurexins are becoming more appreciated for their functional significance in binding to neuroligins to create a key link in synapse formation and maintenance (8,9).
A recent high-density genome-wide association study revealed that NRXN1 is significantly associated with ND in a European sample (13). Additionally, another neurexin gene, NRXN3, was found to be associated with polysubstance (14). Subsequently, it was reported that NRXN3 is significantly associated with alcohol dependence and that polymorphisms in NRXN3 might affect expression of NRXN3 isoforms (15). These association results, taken together with the intriguing role of neurexins in synaptic organization and cell–cell recognition, make neurexin genes strong candidates for further genetic study on ND.
In this study, using a family-based analysis, we analyze single nucleotide polymorphisms and haplotypes within NRXN1, with a primary goal of providing an independent replication of the association of NRXN1 with ND reported in a case–control-based genome-wide association study (13). Owing to the massive size (1.1 Mb) of the gene, 21 SNPs distributed throughout the gene including the three polymorphisms that were significantly associated with ND in previous report (13), mostly intronic, were analyzed. Association analysis was performed on an independent set of sample consisting of 2037 individuals from 602 nuclear families of either African-American (AA) or European-American (EA) ancestry, using three selected ND measures: smoking quantity (SQ), the heaviness of smoking index (HSI) and the Fagerström test for ND (FTND).
RESULTS
Association analysis of individual SNPs
It has been documented that ethnic-specific characteristics of SNPs exist among ethnic groups (16,17) and genetic differences underlying nicotine metabolism exist between different ethnic populations (18). These characteristics and differences are highlighted in this study, as significant asymmetry is seen for individual association patterns in SNPs when comparing the AA and EA samples. Examining differences between the two samples also revealed significant intersample variation in allele frequencies for several SNPs (Table 1). Therefore, all statistical analyses were performed separately on AA and EA samples to eliminate this potential source of heterogeneity. Data for the pooled sample are also given with the aim of illustrating the effect of merging the two datasets; we would expect association signals to become more strongly positive when findings in the different populations are combined, implying that our observed effect of them opposing each other reflects the effect of the same underlying functional variant.
Table 1.
Information for 21 SNPs within NRXN1 gene and a comparison of their minor allele frequencies in NCBI dbSNP database and in the AA and EA samples of the current study
| SNP number | dbSNP ID | Alleles | Position (Chr 2) | SNP location | MAF in dbSNP | TaqMan assay ID | MAF in this study | ||
|---|---|---|---|---|---|---|---|---|---|
| YRIa | CEUb | AA | EA | ||||||
| 1 | rs1995584 | A/G | 51116653 | 5′ end | 0.23 | 0.43 | C__11621223_10 | 0.33 | 0.44 |
| 2 | rs10490162 | A/G | 51101161 | Intron 1 | 0.08 | 0.09 | C__29665747_20 | 0.10 | 0.10 |
| 3 | rs12467557 | A/G | 51095774 | Intron 1 | 0.08 | 0.05 | C___2787998_10 | 0.08 | 0.05 |
| 4 | rs12623467 | C/T | 51078593 | Intron 1 | NA | 0.05 | C__31180525_10 | 0.13 | 0.06 |
| 5 | rs2052328 | A/C | 51004867 | Intron 2 | 0.43 | 0.13 | C___2787934_20 | 0.42 | 0.17 |
| 6 | rs2193225 | A/G | 50932986 | Intron 3 | 0.14 | 0.39 | C___2686011_20 | 0.22 | 0.45 |
| 7 | rs3850333 | C/T | 50856433 | Intron 3 | 0.47 | 0.38 | C___7547374_10 | 0.39 | 0.41 |
| 8 | rs858932 | C/G | 50783567 | Intron 3 | 0.43 | 0.44 | C___2857488_10 | 0.34 | 0.41 |
| 9 | rs11125321 | C/G | 50705521 | Intron 4 | 0.43 | 0.45 | C__31179632_10 | 0.41 | 0.41 |
| 10 | rs2351765 | A/C | 50647285 | Intron 7 | 0.35 | 0.27 | C___2683772_10 | 0.43 | 0.24 |
| 11 | rs6721498 | A/G | 50566516 | Intron 14 | 0.43 | 0.49 | C___2683870_10 | 0.48 | 0.50 |
| 12 | rs10490227 | A/G | 50513019 | Intron 16 | 0.30 | 0.10 | C___2683939_10 | 0.23 | 0.11 |
| 13 | rs10208208 | C/G | 50446727 | Intron 16 | 0.45 | 0.44 | C__30512948_10 | 0.15 | 0.03 |
| 14 | rs1622701 | C/G | 50365159 | Intron 16 | 0.42 | 0.43 | C___7547531_10 | 0.39 | 0.45 |
| 15 | rs601010 | C/T | 50302642 | Intron 17 | 0.45 | 0.43 | C___1736086_10 | 0.40 | 0.42 |
| 16 | rs10490239 | C/T | 50227124 | Intron 17 | 0.17 | 0.42 | C__29792310_10 | 0.24 | 0.35 |
| 17 | rs2123388 | G/T | 50166788 | Intron 18 | 0.23 | 0.23 | C__16130253_10 | 0.25 | 0.26 |
| 18 | rs10495995 | A/G | 50102417 | Intron 19 | 0.20 | 0.23 | C__30224587_20 | 0.21 | 0.33 |
| 19 | rs2882594 | C/T | 50060236 | Intron 19 | 0.42 | 0.43 | C__16142232_10 | 0.39 | 0.50 |
| 20 | rs1045881 | A/G | 50002476 | 3′-UTR | 0.18 | 0.10 | C___9551914_10 | 0.13 | 0.17 |
| 21 | rs971732 | G/T | 49995621 | 3′ End | 0.49 | 0.28 | C___2034714_20 | 0.44 | 0.38 |
NA, not available.
aSub-Saharan African sample from NCBI dbSNP database.
bEuropean sample from NCBI dbSNP database.
Individual SNP analysis yielded multiple significant associations. SNP rs6721498 showed significant association with the three ND measures (P = 0.0000086–0.00090; Table 2) in the AA sample under both dominant and additive models. The pooled sample showed significant association with all three ND measures under the additive model (P = 0.000099–0.0010) and with FTND under the dominant model (P = 0.0013). On the other hand, SNP rs2193225 showed significant association with all analyzed ND measures under the dominant model in the EA sample (P = 0.00014–0.0010; Table 2). The pooled sample also showed positive association with SQ (P = 0.0019) under the dominant model. All these above associations remained significant after correction for multiple testing. Additionally, five analyzed SNPs showed significant associations with one or more phenotypes in the AA (rs10490227, rs10208208 and rs1045881), EA (rs10490162 and rs10208208) and/or pooled samples (rs11125321 and rs10208208) under the additive or dominant model before correction, but failed to maintain significance after correction for multiple testing (Table 2).
Table 2.
P-values for individual SNP association with ND under the additive (at the top) and dominant (at the bottom) models in the EA, AA and pooled samples
| SNP ID | AA SQ | HSI | FTND | EA SQ | HSI | FTND | Pooled SQ | HSI | FTND |
|---|---|---|---|---|---|---|---|---|---|
| rs1995584 | – | – | – | 0.080 | 0.13 | 0.090 | – | – | – |
| 0.51 | 0.41 | 0.51 | – | 0.13 | – | 0.35 | 0.26 | 0.33 | |
| rs10490162 | 0.080 | 0.10 | 0.090 | 0.070 | 0.040 | 0.040 | – | – | – |
| – | – | – | – | – | – | 0.63 | 0.65 | 0.73 | |
| rs12467557 | 0.19 | – | 0.20 | – | – | 0.31 | 0.33 | – | 0.47 |
| – | 0.20 | 0.20 | 0.32 | 0.32 | – | – | 0.44 | 0.47 | |
| rs12623467 | – | – | – | 0.92 | 0.51 | – | – | – | – |
| 0.13 | 0.10 | 0.060 | 0.92 | – | 0.44 | 0.15 | 0.13 | 0.084 | |
| rs2052328 | – | – | – | 0.38 | 0.44 | 0.44 | – | – | – |
| 0.44 | 0.43 | 0.52 | – | – | – | 0.49 | 0.55 | 0.69 | |
| rs2193225 | – | – | – | 0.0050 | 0.017 | 0.032 | – | – | – |
| 0.71 | 0.46 | 0.48 | 0.00014 | 0.00058 | 0.0010 | 0.0019 | 0.0032 | 0.0052 | |
| rs3850333 | – | – | – | 0.27 | – | 0.33 | – | – | – |
| 0.17 | 0.42 | 0.33 | 0.27 | 0.26 | – | 0.15 | 0.34 | 0.25 | |
| rs858932 | – | – | – | 0.35 | – | – | 0.27 | 0.44 | – |
| 0.19 | 0.38 | 0.25 | – | 0.11 | 0.07 | – | – | 0.22 | |
| rs11125321 | 0.16 | – | – | 0.10 | 0.060 | 0.10 | 0.030 | 0.030 | 0.050 |
| – | 0.15 | 0.24 | – | – | – | – | – | – | |
| rs2351765 | – | – | – | – | 0.060 | 0.10 | – | 0.19 | 0.28 |
| 0.10 | 0.080 | 0.10 | 0.23 | – | – | 0.15 | – | – | |
| rs6721498 | 0.000090 | 0.000023 | 0.0000086 | – | – | – | 0.00099 | 0.0011 | 0.0010 |
| 0.00090 | 0.00033 | 0.00014 | 0.71 | 0.60 | 0.32 | 0.024 | 0.0030 | 0.0013 | |
| rs10490227 | 0.050 | – | – | 0.14 | – | – | – | – | – |
| 0.040 | 0.030 | 0.040 | – | 0.16 | 0.17 | 0.31 | 0.11 | 0.11 | |
| rs10208208 | – | – | – | 0.15 | 0.12 | 0.030 | – | – | 0.050 |
| 0.10 | 0.023 | 0.0066 | – | – | 0.040 | 0.050 | 0.010 | 0.0024 | |
| rs1622701 | – | 0.41 | 0.47 | 0.63 | – | – | – | 0.48 | – |
| 0.67 | – | – | 0.63 | 0.85 | 0.61 | 0.59 | – | 0.65 | |
| rs601010 | – | 0.60 | 0.90 | – | – | – | – | – | – |
| 0.57 | – | 0.90 | 0.62 | 0.43 | 0.19 | 0.64 | 0.73 | 0.69 | |
| rs10490239 | – | – | – | – | – | – | – | – | – |
| 0.43 | 0.53 | 0.60 | 0.81 | 0.6 | 0.46 | 0.44 | 0.62 | 0.65 | |
| rs2123388 | – | – | – | – | – | – | – | – | – |
| 0.13 | 0.29 | 0.55 | 0.10 | 0.11 | 0.55 | 0.58 | 0.74 | 0.80 | |
| rs10495995 | – | 0.23 | 0.19 | – | – | – | – | 0.47 | 0.46 |
| 0.88 | – | – | 0.53 | 0.33 | 0.28 | 0.90 | – | – | |
| rs2882594 | 0.81 | 0.47 | 0.34 | 0.18 | 0.070 | 0.13 | 0.36 | 0.11 | 0.10 |
| 0.81 | – | – | – | – | – | – | – | – | |
| rs1045881 | – | – | – | – | – | – | 0.27 | 0.25 | 0.19 |
| 0.040 | 0.040 | 0.033 | 0.17 | 0.084 | 0.11 | – | – | – | |
| rs971732 | – | 0.62 | – | – | – | – | 0.63 | 0.71 | 0.79 |
| 0.68 | – | 0.74 | 0.12 | 0.12 | 0.22 | – | – | – |
Adjusted P-value after correction for multiple testing at the 0.05 significance level is 0.00238. Only the lowest P-value given by any model is shown for non-significant associations. Age and gender were used as covariates for all calculations. Associations remained significant after correction for multiple testing are given in bold.
Haplotype analysis
The selected 21 SNPs distribute distantly over the large gene of NRXN1, which spans >1 Mb on human chromosome 2. Nonetheless, we employed the Haploview program (19) to calculate the pair-wise D′ values for all SNPs and to detect all haplotype blocks accruing to the criteria defined by Gabriel et al. (20). As shown in Figure 1, we found one block (rs1045881–rs971732, 6 kb) that was detected consistently in the AA, EA and pooled samples, and another block (rs12467557–rs12623467, 17 kb) that was detected only in the AA and pooled samples.
Figure 1.
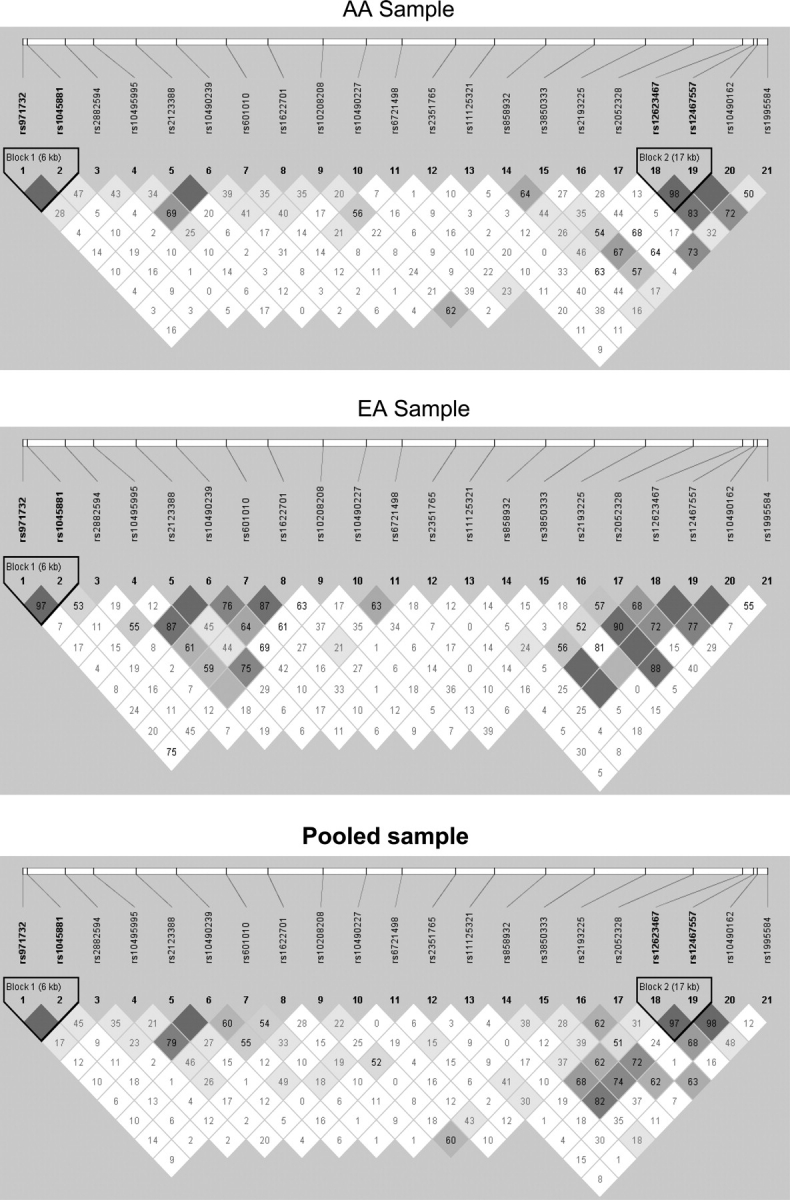
LD structures for 21 SNPs analyzed for NRXN1 in the AA, EA and pooled samples. The Haploview (v.3.2) program (17) was used to calculate all D′ values, and haplotype blocks were defined according to Gabriel et al. (18). The number in each box represents the D′ value for each SNP pair surrounding that box.
Since there were no large haplotype blocks containing more than two SNPs found in our AA, EA or pooled samples, we further performed haplotype association analysis for all possible consecutive three-SNP combinations of the 21 SNPs genotyped (i.e. ‘sliding window’ analysis). In the AA sample, each of the three possible consecutive three-SNP combinations containing rs6721498 (labeled as no. 11 in Table 3) was found to have haplotypes that were significantly associated with SQ, HSI and FTND measures. A total of six AA-sample haplotypes within this range (starting from rs11125321 to rs10208208, labeled as SNP nos. 9–13 in Tables 3–5) were found to have significant association with ND (minimum P-value range from 0.000079 to 0.0068). Of these six, haplotype G–A–C with a frequency of 35% (formed by rs2351765–rs6721498–rs10490227) showed the most significant association with SQ (minimum P = 0.00061; number of families = 194; global P-value = 0.0018) and HSI (minimum P = 0.000079; number of families = 195; global P-value = 0.00043) under the additive model, and with FTND under the dominant model (minimum P = 0.00064; number of families = 165; global P-value = 0.00007) (Table 3). The EA sample showed no significant association for any ND measures within this five-SNP window surrounding rs6721498 (minimum P = 0.22; Table 4). However, in the pooled sample, five of these six haplotypes showed significant associations with SQ (Z = −2.86 to 3.02; P = 0.0024–0.019), HSI (Z = −3.34–2.62; P = 0.0015–0.0097), and FTND (Z = −3.34–2.62; P = 0.00083–0.011; Table 5). Of these five haplotypes among the three samples, no single haplotype showed distinctly greater significance than any others, with haplotype G–A–C (formed by rs6721498–rs10490227–rs10208208) in the AA sample showing the strongest association with SQ (Z = 3.43; P = 0.00061; number of families = 194; global P-value = 0.0018) and with HSI (Z = 3.94; P = 0.000079; number of families = 195; global P-value = 0.00043) under the additive model, and with FTND (Z = 3.41; P = 0.00064; number of families = 165; global P-value = 0.000070) under the dominance model (Table 3).
Table 3.
Z-values, P-values and global P-values for associations of some major haplotypes with three ND measures in the AA sample
| Haplotype | % | SQ Number of families | Z(P-) value | Global P-value | HSI Number of families | Z- (P-) value | Global P-value | FTND Number of families | Z- (P-) value | Global P-value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | ||||||||||||||
| 5 | 6 | 7 | ||||||||||||
| C | G | T | 25 | 169 | −0.68 (0.49) | 0.61 | 170 | −0.58 (0.56) | 0.78 | 176 | −0.45 (0.65) | 0.72 | ||
| – | – | – | – | – | – | – | – | |||||||
| B | ||||||||||||||
| 9 | 10 | 11 | 12 | 13 | ||||||||||
| G | C | G | 9 | 87 | 3.19 (0.0015) | 0.0042 | 87 | 3.18 (0.0015) | 0.0036 | 89 | 3.25 (0.0011) | 0.0024 | ||
| 87 | 3.19 (0.0015) | 0.017 | 86 | 3.24 (0.0012) | 0.0089 | 89 | 3.25 (0.0011) | 0.0054 | ||||||
| C | G | A | 19 | 142 | 2.70 (0.0068) | 0.0055 | 142 | 2.91 (0.0025) | 0.00079 | 145 | 2.83 (0.0047) | 0.00042 | ||
| – | – | – | – | – | – | – | – | – | ||||||
| A | A | G | 7 | 65 | −2.62 (0.0087) | 0.0055 | 66 | −2.81 (0.0049) | 0.00079 | 68 | −2.85 (0.0045) | 0.00042 | ||
| 64 | −2.79 (0.0051) | 0.0057 | 65 | −3.11 (0.0018) | 0.00063 | 67 | −3.13 (0.0017) | 0.00020 | ||||||
| G | A | C | 35 | 194 | 3.43 (0.00061) | 0.0018 | 195 | 3.94 (0.000079) | 0.00043 | – | – | – | ||
| 157 | 2.89 (0.0038) | 0.00014 | 159 | 3.46 (0.00053) | 0.000025 | 165 | 3.41 (0.00064) | 0.000070 | ||||||
| A | A | C | 35 | – | – | – | – | – | – | – | – | – | ||
| 174 | −2.89 (0.0038) | 0.00014 | 204 | −2.30 (0.022) | 0.000025 | 182 | −3.11 (0.0018) | 0.000070 | ||||||
| A | G | C | 11 | 95 | −2.57 (0.013) | 0.0018 | 96 | −2.76 (0.0057) | 0.00043 | 99 | −2.94 (0.0031) | 0.00084 | ||
| 91 | −2.77 (0.0055) | 0.00014 | 92 | −3.25 (0.0011) | 0.000025 | 95 | −3.40 (0.00067) | 0.000070 | ||||||
(i) Test results for the additive model are given at the top (A); for the dominant model are given at the bottom (B); (ii) the adjusted P-value at the 0.05 significant level after Bonferroni correction for five (11–12–13), six (5–6–7, 10–11–12) and eight (9–10–11) major haplotypes is 0.010, 0.0083 and 0.0063, respectively, (iii) only the lowest P-value given by any model is shown for non-significant associations and (iv) age and gender were used as covariates for all calculations. Associations remained significant after correction for multiple testing are given in bold.
Table 5.
Z-, P- and global P-values for associations of some major haplotypes with three ND measures in the pooled sample
| Haplotype Number of families | % | SQ Number of families | Z- (P-) value | Global P-value | HSI Number of families | Z- (P-) value | Global P-value | FTND Number of families | Z- (P-) value | Global P-value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | ||||||||||||||
| 5 | 6 | 7 | ||||||||||||
| C | G | T | 21 | 216 | −1.77 (0.077) | 0.18 | 215 | −1.56 (0.12) | 0.39 | 225 | −1.51 (0.13) | 0.34 | ||
| – | – | – | – | – | – | – | – | – | ||||||
| B | ||||||||||||||
| 9 | 10 | 11 | 12 | 13 | ||||||||||
| G | C | G | 10 | 132 | 3.02 (0.0024) | 0.13 | 130 | 2.52 (0.0065) | 0.024 | 135 | 2.52 (0.011) | 0.035 | ||
| 129 | 2.89 (0.0038) | 0.22 | 127 | 2.59 (0.0054) | 0.049 | 132 | 2.59 (0.0095) | 0.088 | ||||||
| C | G | A | 24 | 215 | 1.95 (0.052) | 0.071 | 216 | 1.72 (0.049) | 0.023 | 223 | 1.72 (0.075) | 0.019 | ||
| – | – | – | – | – | – | – | – | – | ||||||
| A | A | G | 5 | 66 | −2.68 (0.0073) | 0.071 | 67 | −2.92 (0.0041) | 0.023 | 70 | −2.92 (0.0035) | 0.019 | ||
| 65 | −2.86 (0.0042) | 0.12 | 66 | −3.23 (0.0015) | 0.029 | 69 | −3.23 (0.0013) | 0.025 | ||||||
| G | A | C | 37 | 277 | 2.46 (0.014) | 0.0070 | 277 | 2.62 (0.0082) | 0.0054 | 290 | 2.62 (0.0088) | 0.0086 | ||
| 218 | 2.33 (0.019) | 0.0020 | – | – | – | – | – | – | ||||||
| A | A | C | 40 | – | – | – | – | – | – | – | – | – | ||
| 279 | −2.67 (0.0075) | 0.0020 | 283 | −2.73 (0.0097) | 0.00051 | 293 | −2.73 (0.0064) | 0.00071 | ||||||
| A | G | C | 8 | 101 | −2.34 (0.019) | 0.0070 | 102 | −3.34 (0.0081) | 0.0054 | 106 | −2.90 (0.0037) | 0.0086 | ||
| 97 | −2.63 (0.0084) | 0.0020 | 98 | −2.90 (0.0018) | 0.00051 | 102 | −3.34 (0.00083) | 0.00071 | ||||||
(i) Test results for the additive model are given at the top (A); test results for the dominant model are given at the bottom (B), (ii) the adjusted P-value at the 0.05 significant level after Bonferroni correction for four (11–13), five (10–12), six (5–7) and eight (9–11) major haplotypes is 0.013, 0.010, 0.0083 and 0.0063, respectively, (iii) only the lowest P-value given by any model is shown for non-significant associations and (iv) age and gender were used as covariates for all calculations. Associations remained significant after correction for multiple testing are given in bold.
Table 4.
Z-, P- and global P-values for associations of some major haplotypes with three ND measures in the EA sample
| Haplotype Number of families | % | SQ Number of families | Z- (P-) value | Global P-value | HSI Number of families | Z- (P-) value | Global P-value | FTND Number of families | Z- (P-) value | Global P-value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | ||||||||||||||
| 5 | 6 | 7 | ||||||||||||
| C | G | T | 13 | 47 | −2.74 (0.0062) | 0.0035 | 45 | −2.47 (0.013) | 0.047 | 50 | −2.55 (0.010) | 0.037 | ||
| B | ||||||||||||||
| 9 | 10 | 11 | 12 | 13 | ||||||||||
| G | C | G | 13 | 46 | 0.76 (0.45) | 0.58 | 45 | 0.33 (0.72) | 0.39 | 48 | −0.032 (0.97) | 0.35 | ||
| C | G | A | 34 | 62 | −0.51 (0.61) | 0.57 | 74 | −0.74 (0.46) | 0.87 | 78 | −0.82 (0.41) | 0.90 | ||
| A | A | G | NA | |||||||||||
| G | A | C | 40 | 80 | −0.28 (0.77) | 0.60 | 77 | −0.29 (0.77) | 0.48 | 83 | 0.32 (0.74) | 0.54 | ||
| A | A | C | 48 | 84 | −0.58 (0.56) | 82 | −1.22 (0.22) | 87 | −1.12 (0.26) | |||||
| A | G | C | NA | |||||||||||
NA, not enough informative families to calculate values.
(i) Only the test results for the additive model are given, in which P-values are lower than those in the dominant model; (ii) the adjusted P-value at the 0.05 significant level after Bonferroni correction for three (11–12–13), five (5–6–7, 10–11–12) and seven (9–10–11) major haplotypes is 0.017, 0.010 and 0.0071, respectively. Associations remained significant after correction for multiple testing are given in bold.
On the other hand, the analysis of haplotypes for the three-SNP combination of rs2052328, rs2193225 and rs3850333 (labeled as SNPs 5–7 in Tables 3–5) yielded similar but opposite results. One haplotype (C–G–T) showed significant association with HSI and FTND in the EA population prior to correction, and retained significant association with SQ (P = 0.0062; number of families = 47; global P-value = 0.0035; Table 4) after correction. No significant association of this haplotype with any ND measure was detected in the AA or pooled sample.
DISCUSSION
The fundamental roles that neurexins play in synapse development and maintenance (8,9), along with previous reports of neurexin genes association with addiction to nicotine and other substances (13–15), make the NRXN1 gene a plausible candidate for association with ND pathology. To test for this association independently, we selected 21 SNPs within NRXN1 and genotyped them in a set of samples containing 2037 individuals from 602 EA and AA nuclear families. Three measures for ND were used to characterize phenotypes, and associations for each of these phenotypes were performed for all SNPs as well as all consecutive three-SNP combinations. By using a family-based sample, along with separating populations by ethnicity, we were able to eliminate or minimize potential population stratification.
Association analysis of 21 SNPs within NRXN1 revealed significant individual SNP and haplotypes associations with ND. Individual SNP analysis revealed a significant association of rs2193225 with ND in the EA sample for all the ND measures under both additive and dominant models, but yielded no significant associations in the AA population. The opposite effect was observed for rs6721498, which showed significance for all phenotypes under both additive and dominant models in the AA sample, but not in the EA population. Haplotype analysis showed a similar pattern regarding the two populations. All consecutive three-SNP combinations containing rs6721498 showed significance for one or more haplotypes using all three phenotypes in the AA sample (Table 3), but none of these haplotypes approached significance in the EA population (Table 4). Similarly, the C–G–T haplotype in the three-SNP combination of rs2052328, rs2193225 and rs3850333 showed significant association with all three phenotypes prior to correction for multiple testing, and with SQ following correction in the EA sample, yet showed no significant association with any ND phenotypes in the AA population.
Our results provided an independent replication of the association of NRXN1 with ND at the gene level, which was previously reported by Bierut et al.(13) in a genome-wide association study. In that case–control study with FTND as the ND measure, three SNPs (rs10490162, rs12467557 and rs12623467 in intron 1) were found significantly associated with ND in a set of sample consisting of Americans and Australians of European descent (13). However, in our family-based association analysis, we found only one of the three SNPs (rs10490162) had marginal significant association with ND in our EA sample, which did not remain significance after correction for multiple testing. On the other hand, we detected rs2193225 in intron 3, which is not so far away from the three SNPs in intron 1 but, not tested by Bierut et al., had more significant association with ND. The potential reason for such a discrepancy of the three SNPs between the two studies may be more or less related to the origins of each sample used in these two studies. The participants of the Bierut et al.’s sample were from different geographical locations in the USA and from Australia, whereas the samples used in the current study were primarily from the Mid-South states of the USA. Additionally, several other factors, such as different experimental designs (case–control versus family based) and sample sizes, may also contribute such differences at individual SNP level. However, at the gene level, we do replicate the result reported by Bierut et al. in which there exist a significant association between NRXN1 and ND. Thus, we think the evidence of NRXN1 associated with ND in EA population from two independent studies merits further investigation in the susceptibility regions of introns 1 and 3. Genotyping more SNPs in these areas may help find the causative loci that contribute to ND in EAs.
As in the other genetic studies (16,17), the ethnic specificity is also evident in our study on association of NRXN1 with ND. In contrast to the ND susceptibility regions of introns 1 and 3 for the EA population, we observed an ND susceptibility region around SNP rs6721498 in intron 14 of NRXN1 specific for the AA sample. Since NRXN1 is a large gene and a relatively loose density of SNPs was genotyped in the current study, we think it will be necessary to genotype more SNPs around the region where SNP rs6721498 is located in order to find potential causative loci within the region that contribute to the observed association of the gene with ND in our samples. Given that no participants were of AA origin in the Bierut et al.’s study, our significant association results of NRXN1 with ND in our AA sample provide first evidence on the involvement of NRXN1 in the development of ND in this ethnic group.
Each neurexin gene has at least two promoters to produce α- and β-neurexin. Further, alternative splicing is able to generate multiple potential isoforms of neurexins (10,11). These different splicing variants of neurexin, along with splicing variants of its binding partner, neuroligin, have been largely suggested to play distinct roles in cell–cell recognition, synaptic organization and synaptic signaling (8,9). To date, no polymorphisms in NRXN1 have been found to change the amino acids, whereas more polymorphisms have been found in the introns of this large gene. Some of these intronic polymorphisms may contribute to determining alternative splicing of NRXN1 and altering expression of specific NRXN1 isoforms, which further determines the generation of synapse specificity and influences the formation of neural circuits that are keys to addiction behaviors. If so, finding such polymorphisms that are associated with ND may help determine their predisposing role of NRXN1 in ND. Recently, a polymorphism in NRXN3 associated with alcohol dependence was found to be associated with altering expression of NRXN3 isoforms (15), which gives a good implication.
In summary, our results indicated that NRXN1 has significant association with ND in two independent AA and EA samples, which not only replicate the previous association report in Caucasians, but also extend the association in the AA population. These genetic associations along with the intriguing physiological role of neurexins make NRXN1 a strong candidate for further investigation into the molecular and genetic mechanisms underlying ND. In addition, we provide evidence for substantial differences between the two ethnic samples. Given that NRXN1 is such a large gene, we believe that our indicated susceptibility regions within NRXN1 for the AA and EA populations, separately, will help us pinpoint potential causative loci for the observed association of the gene with ND in two separate ethnic groups in the future.
MATERIALS AND METHODS
Participants and ND measures
Participants in this family-based study were either of AA or EA origin, recruited between 1999 and 2004 primarily from the mid-South states within the USA. Criteria for eligible proband smokers consisted of: being at least 21 years of age, having consumed an average of 20 cigarettes per day for 12 months prior to recruitment, having smoked for a minimum of 5 consecutive years and being free of any current psychiatric diagnosis, with the exception of alcohol dependence. As for the parents and siblings of an eligible proband smoker are concerned, we recruited as many as we can regardless of their smoking status. Data collected for individuals and families included demographic information, such as age, sex, race, biological relationships, weight, height, education and marital status. Data related to medical histories, currents and past smoking behavior and personality traits were also collected using various questionnaires that are available at the NIDA Genetics Consortium website (http://zork.wustl.edu/nida). All participants provided informed consent, and the study protocol and forms were approved by all participating institutional review boards.
In order to quantify ND for each smoker, three of the most commonly employed measures in published studies on ND were used. These include SQ, HSI (0–6 scale) and FTND (0–10 scale) (21). There is no current consensus as to which measure provides the best overall assessment of ND, and performing association analysis for each individual ND measure can give different insights that selecting one specific measure cannot. SQ provides a straightforward index for total consumption, whereas HSI adds another question that assesses smoking urgency, i.e. ‘How soon after you wake up do you smoke your first cigarette?’ FTND includes the HSI factors, but adds other behavioral indicators that relate smoking to specific circumstances. The use of multiple measures also allows for cross-referencing with other studies, regardless of their chosen measure. There is a fairly robust correlation between these measures, ranging from 0.88 to 0.94 for both samples.
The study included 2037 participants (1366 AAs and 671 EAs) coming from 602 nuclear families (402 AAs and 200 EAs). Average participant age was 39.4 ± 14.4 (SD) years for the AA sample and 40.5 ± 15.5 years for the EA sample. Average nuclear family size for AAs was 3.14 ± 0.75 and 3.17 ± 0.69 for EAs. The average HSI and FTND scores for AA smokers were 3.7 ± 1.4 and 6.25 ± 2.15 and 3.9 ± 1.4 and 6.33 ± 2.22 for EA smokers, respectively. The average number of cigarettes smoked per day by AA and EA smokers was 19.4 ± 13.33 and 19.5 ± 13.4, respectively. More detailed descriptions of demographic and clinical data for participants can be found in previous publications (22–24).
DNA extraction, SNP selection and SNP genotyping
DNA was extracted from participant blood samples using the QIAamp DNA Blood Maxi Kit, purchased from Qiagen Inc (Valencia, CA, USA). SNP selection was based on the minor allele frequency (>5%; as listed in the NCBI dbSNP database) and optimal coverage of the gene. Three SNPs (rs10490162, rs12467557 and rs12623467) that were significantly associated with ND in previous genome-wide association study (13) were intentionally included. Information regarding each SNP is given in Table 1.
Each SNP was genotyped using a TaqMan SNP Genotyping Assay purchased from Applied Biosystems (Foster City, CA, USA). Reactions were run in 384-well plates, with total reaction volumes of 7 µl. Each reaction contained an MGB probe, primers, 15 ng of template DNA and 2.5 µl TaqMan Universal PCR Mastermix. Polymerase chain reaction conditions were: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 25 s and 60° for 1 min. Allelic discrimination analysis was carried out with an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Each plate was quality-controlled with four no-template controls, as well as eight DNA-containing positive controls which were monitored for consistency.
Linkage Disequilibrium (LD) and association analysis
The program Haploview (19) was used to determine pair-wise LD between all SNPs using the option of determining blocks based on the criteria defined by Gabriel et al. (20). Haploview was also used to detect inconsistencies in Mendelian inheritance, which would suggest nonpaternity or genotyping errors. Thirty-six genotyping errors from a total of 41 832 assays (0.08%) were identified and deleted from further analysis.
The three phenotypic ND measures discussed earlier were individually used to assess the association of ND with all 21 SNPs using the PBAT program (25). Age and sex were used as covariates within the PBAT program due to evidence that they influence ND differently (1,26–29). The FBAT (30) program was used to measure the association between all possible haplotypes in consecutive three-SNP combinations and the three ND measures. For both individual SNP and haplotype analysis, both additive and dominant models were used. All associations designated as significant were corrected for multiple testing of number of SNPs analyzed in the study using the SNP spectral decomposition (SNPSpD) method (31) for individual SNP analysis (which, unlike Bonferroni correction, accounts for correlation between SNPs). The SNPSpD method was favored because Bonferroni correction tends to be too conservative and overcorrects for inflated false-positive rates, resulting in a reduction of power. Bonferroni correction was used for Haplotype analysis though, as the SNPSpD method cannot handle correction for haplotypes.
FUNDING
This project was supported by National Institutes of Health grant DA-12844 to M.D.L. and DA-12849 to J.G.
ACKNOWLEDGEMENTS
We acknowledge the invaluable contributions of clinical information and blood by all participants in the genetic study, as well as the dedicated work of many research staff at different clinical sites.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Li M.D., Cheng R., Ma J.Z., Swan G.E. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan P.F., Kendler K.S. The genetic epidemiology of smoking. Nicotine Tob. Res. 1999;1(Suppl. 2):S51–S57. doi: 10.1080/14622299050011811. Discussion S69–S70. [DOI] [PubMed] [Google Scholar]
- 3.Mokdad A.H., Marks J.S., Stroup D.F., Gerberding J.L. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 4.Leistikow B.N. The human and financial costs of smoking. Clin. Chest Med. 2000;21:189–197. doi: 10.1016/s0272-5231(05)70017-4. x–xi. [DOI] [PubMed] [Google Scholar]
- 5.Li M.D., Ma J.Z., Beuten J. Progress in searching for susceptibility loci and genes for smoking-related behaviour. Clin. Genet. 2004;66:382–392. doi: 10.1111/j.1399-0004.2004.00302.x. [DOI] [PubMed] [Google Scholar]
- 6.Leonard S., Bertrand D. Neuronal nicotinic receptors: from structure to function. Nicotine Tob. Res. 2001;3:203–223. doi: 10.1080/14622200110050213. [DOI] [PubMed] [Google Scholar]
- 7.Pontieri F.E., Tanda G., Orzi F., Di Chiara G. Effects of nicotine on the nucleus accumbens and similarity to those of addictive drugs. Nature. 1996;382:255–257. doi: 10.1038/382255a0. [DOI] [PubMed] [Google Scholar]
- 8.Craig A.M., Kang Y. Neurexin–neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean C., Dresbach T. Neuroligins and neurexins: linking cell adhesion, synapse formation and cognitive function. Trends Neurosci. 2006;29:21–29. doi: 10.1016/j.tins.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Rowen L., Young J., Birditt B., Kaur A., Madan A., Philipps D.L., Qin S., Minx P., Wilson R.K., Hood L., et al. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics. 2002;79:587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- 11.Tabuchi K., Sudhof T.C. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- 12.Ushkaryov Y.A., Petrenko A.G., Geppert M., Sudhof T.C. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- 13.Bierut L.J., Madden P.A., Breslau N., Johnson E.O., Hatsukami D., Pomerleau O.F., Swan G.E., Rutter J., Bertelsen S., Fox L., et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q.R., Drgon T., Walther D., Johnson C., Poleskaya O., Hess J., Uhl G.R. Pooled association genome scanning: validation and use to identify addiction vulnerability loci in two samples. Proc. Natl Acad. Sci. USA. 2005;102:11864–11869. doi: 10.1073/pnas.0500329102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hishimoto A., Liu Q.R., Drgon T., Pletnikova O., Walther D., Zhu X.G., Troncoso J.C., Uhl G.R. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Hum. Mol. Genet. 2007;16:2880–2891. doi: 10.1093/hmg/ddm247. [DOI] [PubMed] [Google Scholar]
- 16.Bamshad M., Wooding S., Salisbury B.A., Stephens J.C. Deconstructing the relationship between genetics and race. Nat. Rev. Genet. 2004;5:598–609. doi: 10.1038/nrg1401. [DOI] [PubMed] [Google Scholar]
- 17.Burchard E.G., Ziv E., Coyle N., Gomez S.L., Tang H., Karter A.J., Mountain J.L., Perez-Stable E.J., Sheppard D., Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N. Engl. J. Med. 2003;348:1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 18.Benowitz N.L., Perez-Stable E.J., Fong I., Modin G., Herrera B., Jacob P., III Ethnic differences in N-glucuronidation of nicotine and cotinine. J. Pharmacol. Exp. Ther. 1999;291:1196–1203. [PubMed] [Google Scholar]
- 19.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 21.Heatherton T.F., Kozlowski L.T., Frecker R.C., Fagerstrom K.O. The Fagerstrom test for nicotine dependence: a revision of the Fagerstrom tolerance questionnaire. Br. J. Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 22.Li M.D., Beuten J., Ma J.Z., Payne T.J., Lou X.Y., Garcia V., Duenes A.S., Crews K.M., Elston R.C. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum. Mol. Genet. 2005;14:1211–1219. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- 23.Beuten J., Ma J.Z., Payne T.J., Dupont R.T., Quezada P., Huang W., Crews K.M., Li M.D. Significant association of BDNF haplotypes in European-American male smokers but not in European-American female or African-American smokers. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2005;139:73–80. doi: 10.1002/ajmg.b.30231. [DOI] [PubMed] [Google Scholar]
- 24.Ma J.Z., Beuten J., Payne T.J., Dupont R.T., Elston R.C., Li M.D. Haplotype analysis indicates an association between the DOPA decarboxylase (DDC) gene and nicotine dependence. Hum. Mol. Genet. 2005;14:1691–1698. doi: 10.1093/hmg/ddi177. [DOI] [PubMed] [Google Scholar]
- 25.Lange C., Silverman E.K., Xu X., Weiss S.T., Laird N.M. A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics. 2003;4:195–206. doi: 10.1093/biostatistics/4.2.195. [DOI] [PubMed] [Google Scholar]
- 26.Madden P.A., Heath A.C., Pedersen N.L., Kaprio J., Koskenvuo M.J., Martin N.G. The genetics of smoking persistence in men and women: a multicultural study. Behav. Genet. 1999;29:423–431. doi: 10.1023/a:1021674804714. [DOI] [PubMed] [Google Scholar]
- 27.Perkins K.A., Donny E., Caggiula A.R. Sex differences in nicotine effects and self-administration: review of human and animal evidence. Nicotine Tob. Res. 1999;1:301–315. doi: 10.1080/14622299050011431. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Stable E.J., Herrera B., Jacob P., III, Benowitz N.L. Nicotine metabolism and intake in black and white smokers. JAMA. 1998;280:152–156. doi: 10.1001/jama.280.2.152. [DOI] [PubMed] [Google Scholar]
- 29.Edwards K.L., Austin M.A., Jarvik G.P. Evidence for genetic influences on smoking in adult women twins. Clin. Genet. 1995;47:236–244. doi: 10.1111/j.1399-0004.1995.tb04303.x. [DOI] [PubMed] [Google Scholar]
- 30.Horvath S., Xu X., Lake S.L., Silverman E.K., Weiss S.T., Laird N.M. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet. Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 31.Nyholt D.R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]


