Abstract
Two non-synonymous single-nucleotide polymorphisms (SNPs), Ser217Leu and Ala541Thr, in the elaC homolog 2 (Escherichia coli) (ELAC2) gene have been related to prostate cancer risk in previous studies, though with inconsistent results. The association of ELAC2 haplotypes with prostate cancer risk has not yet been explored. We assessed whether sequence variants in ELAC2 were associated with the risk of total or aggressive prostate cancer. In a nested case–control design within the Health Professionals Follow-Up Study, we identified 659 participants with prostate cancer diagnosed after they provided a blood specimen in 1993 and before January 2000. Controls were 656 age-matched men without prostate cancer who had had a prostate-specific antigen test after providing a blood specimen. We genotyped eight tagging SNPs in ELAC2 to test for the association between sequence variances in ELAC2 and prostate cancer. No individual SNP (including Ser217Leu) was associated with the risk of prostate cancer. Ala541Thr is a rare SNP in this population. One common haplotype (hap4) was statistically significantly associated with an increased risk of prostate cancer [odds ratio (OR) = 1.39, 95% confidence interval = 1.05–1.85]. Two common promoter SNPs and three common haplotypes were statistically significantly associated with aggressive prostate cancer (carriers versus non-carriers—snp2: OR = 1.43, snp3: OR = 0.69, hap1: OR = 1.47, hap2: OR = 0.72, hap4: OR = 1.51; global P-value for all common haplotypes = 0.11). Common SNPs and haplotypes of ELAC2 were associated with risk of aggressive prostate cancer.
Introduction
elaC homolog 2 (Escherichia coli) (ELAC2) is located on chromosome 17p11, spans 27 kb and includes 24 exons. Identified as a prostate cancer susceptibility gene (1), it is expressed at high levels in rat testis and some mouse tissues (2). Although ELAC2 is expressed at low levels in mouse prostate (2), experimental studies showed that ELAC2 plays a role in germline proliferation, which is related to cell cycle and sterility (3), and thus may relate to prostate carcinogenesis. The amino acid sequence of ELAC2 is similar to that of PSO2 DNA interstrand cross-link repair proteins and the 73 kD subunit of messenger RNA 3′ end cleavage and polyadenylation specificity factor (CPSF73), which may in turn be related to blocking transcription, replication and segregation of DNA and regulation of messenger RNA modifications (1,4).
Two non-synonymous polymorphisms in ELAC2, Ala541Thr and Ser217Leu, have been well studied. The allele frequency of Thr541 was lower in Japanese subjects (0–1.1%) (5–7) than in African-Americans (21%) (8) or Caucasians (2.7–10.6%) (8–20). However, comparing Thr541 carriers with non-carriers, the risk of sporadic prostate cancer was higher among Japanese subjects [odds ratio (OR) = 3.4–5.1] (6,7) than among Caucasians (OR = 1.0–2.2) (8–20). Furthermore, Thr541 was not associated with hereditary prostate cancer (10,12).
In contrast, the Leu217 allele is more prevalent in Caucasians (27–46%) than in African Americans (23%) (8) or Japanese (0–3.3%) (5,6). Three (5,10,17) out of 14 studies found that the Leu217 allele was statistically significantly associated with prostate cancer [Leu217 carriers versus non-carriers—all cases versus controls: OR = 0.78 (10); non-aggressive cases versus controls: OR = 1.34 (17); sporadic cases versus controls: OR = 3.11 (5)], whereas the remaining studies found a null relationship. A meta-analysis showed that the Leu217 variant was statistically significantly associated only with familial prostate cancer (OR = 1.37) (6). Taken together, 80% of studies showed no evidence of association between Leu217 and prostate cancer risk. Some studies indicated that the joint effect of Ser217Leu and Ala541Thr was associated with a higher risk of prostate cancer than Leu217 alone (1,9,17), whereas others failed to observe this joint effect (6,10–12,15,16).
Past studies of ELAC2 and prostate cancer risk have focused on the two common missense variants, Ser217Leu and Ala541Thr, and results have been inconsistent. However, two single-nucleotide polymorphisms (SNPs) provide a relatively small amount of information for predicting prostate cancer risk. The relationship between haplotypes in ELAC2 and the risk of sporadic prostate cancer has not been explored. Therefore, in addition to these two non-synonymous SNPs in ELAC2, we selected additional tagging SNPs and hypothesized that haplotypes of ELAC2 are associated with susceptibility to prostate cancer in the Health Professionals Follow-Up Study. We also explored the relationship between ELAC2 genetic polymorphisms and aggressiveness of prostate cancer.
Materials and methods
Study population
In this nested case–control study, incident prostate cancer cases were identified from the ongoing Health Professionals Follow-Up Study with follow-up from 1986 through 2000. A total of 51 529 USA men aged 40–75 years were enrolled in 1986. At baseline, every participant completed a mailed questionnaire on demographics, lifestyle and medical history and a semiquantitative food-frequency questionnaire. Information on exposures and diseases was updated every other year, and diet information was updated every 4 years. Deaths were identified through reports by family members or the postal system upon follow-up questionnaires or a search of the National Death Index (21). This study was approved by the Institutional Review Board at the Harvard School of Public Health.
Blood samples (collected in tubes containing sodium ethylenediaminetetraacetic acid) were obtained from 18 018 of the participants between 1993 and 1995. We obtained informed consent from each subject before blood was drawn. Samples were shipped by overnight courier and centrifuged; the aliquots, including plasma, erythrocytes and buffy coat, were stored in liquid nitrogen freezers. QIAGEN QIAamp blood extraction kit (QIAGEN, Valencia, CA) was used for DNA extraction. All DNA samples were whole-genome amplified, and quality control (QC) samples had 100% genotype concordance. Among the men who provided a blood specimen, 95% responded to the year 2000 questionnaire, and the 18 who died of prostate cancer before the end of follow-up were included in the case series.
We identified 659 incident prostate cancer cases and 656 controls; all are Caucasians. Each case was matched with one control who was alive, had not been diagnosed with cancer by the date of the case's diagnosis and had a prostate-specific antigen test after the date of blood draw. The latter criterion ensured that controls had the opportunity to have an occult prostate cancer diagnosed. All controls had a prostate-specific antigen test within 2.5 years of the date of diagnosis of their matched case. Because plasma analyses were performed on the same case–control set, cases and controls were matched on year of birth (±1 year), prostate-specific antigen test prior to blood draw (yes/no), time of blood draw (midnight to before 9 A.M., 9 A.M. to before noon, noon to before 4 P.M. and 4 P.M. to before midnight), season (winter, spring, summer and fall) and year of blood draw. To elucidate the effect of sporadic prostate cancer (prostate cancer that occurs occasionally and at random intervals in a population), we also performed analyses excluding subjects with familial prostate cancer cases (two or more family members had prostate cancer cases, n = 15 cases and 10 controls).
Laboratory assays
Eight common (frequency > 5%) tagging SNPs in ELAC2 (s1, s4, s5, s7, s11, s13, AT and s17) were selected from the study of Camp et al. (22). Laboratory personnel were blinded to case–control status. All case–control matched pairs were analyzed together using the Sequenom system. Multiplex polymerase chain reactions were carried out to generate short polymerase chain reaction products (>100 bp) containing one SNP. The details of polymerase chain reaction and matrix-assisted laser desorption ionization time-of-flight mass spectrometry are available upon request. Six control DNA samples were used for optimization. The SNP s13 in Camp et al. (22) failed both Sequenom and Taqman assay designs in our population due to low QC rates. This SNP could be replaced by snp5 (Table I) because they were in the same linkage disequilibrium group according to phylogenetic network analysis from Camp et al. (22). Analyses using the Tagger program (http://www.broad.mit.edu/mpg/tagger/) also suggested that the new set of SNPs after replacement can accurately capture the information within haplotype blocks (r2 = 0.984). Finally, a total of eight SNPs were genotyped in three plexes at the Harvard Partners Center for Genetics and Genomics (Boston, MA). For each SNP, genotyping data were missing for fewer than 5% of the study participants. Sixty-eight QC samples were obtained from 18 external participants who were recruited the same way as the study population, and each of them had two to six duplicates. These QC samples were genotyped together with all other samples in this study. All QC samples passed the QC test with discordance rate = 0.
Table I.
Characteristics of ELAC2 SNPs among Caucasians
| SNP name | SNP name used in Camp et al. (22) | rs # | Nucleotide change (amino acid change) | Location | bp relative to start codon ATG | Controls | Cases | ||
| MAF (%) | HWE, P-value | MAF (%) | HWE, P-value | ||||||
| Snp1 | S1 | rs2322779 | T → C | Promoter | −12682 | 16 | 0.14 | 19 | 0.74 |
| Snp2 | S4 | Not applicable | G → A | Promoter | −6280 | 30 | 0.99 | 33 | 0.40 |
| Snp3 | S5 | rs12600940 | C → T | Promoter | −3831 | 49 | 0.35 | 46 | 0.44 |
| Snp4 | S7 | rs2051974 | G → A | Promoter | −381 | 24 | 0.81 | 24 | 0.17 |
| Snp5 | SL | rs4792311 | C → T (Ser217Leu) | Exon | 6256 | 32 | 0.88 | 32 | 0.17 |
| Snp6 | S11 | rs2302069 | A → G | Intron | 7241 | 14 | 0.45 | 13 | 0.44 |
| Snp7 | AT | rs5030739 | G → A (Ala514Thr) | Exon | 21363 | 0 | Not applicable | 0 | Not applicable |
| Snp8 | S17 | rs17552022 | A → G | Exon | 22970 | 13 | 0.36 | 14 | 0.21 |
HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency.
Ascertainment of prostate cancer
Investigators reviewed the medical and pathology records for men with prostate cancer, reported from the follow-up questionnaire or rarely death certificate, to confirm adenocarcinoma of the prostate and to document clinical presentation, stage and Gleason sum of the tumor. The cases were categorized into regionally invasive or metastatic (stage ≥ T3c, N1 or M1), organ-confined or minimal extraprostatic extension (T1b–T3a and N0M0), higher grade (Gleason sum ≥ 7) and lower grade (Gleason sum < 7). Incidental microscopic focal tumors (stage T1a) were excluded because they are generally indolent and susceptible to detection bias due to differential rates of surgery for benign prostatic hyperplasia. In addition, men with a previous cancer (except non-melanoma skin cancer) prior to the date of blood draw were excluded. Confirmed non-T1a tumors between blood draw and 31 January 2000 were included. In the blood subcohort, 92% of cases were confirmed by medical record and 5% by other corroborating information; only 3% were based on self-report (23). We included the self-reported cases in the analyses because the concordance between self-reported and medical record-confirmed cases was high (>90%) in this cohort.
Statistical analysis
The Hardy–Weinberg equilibrium test was performed for each SNP among controls. Haplotype block structure (Figure 1) was determined using Haploview (http://www.broad.mit.edu/mpg/haploview/index.php) and Locusview (http://www.broad.mit.edu/mpg/locusview/) programs. The expectation-maximization algorithm (24,25) was applied to construct haplotypes in each block using the Tagsnp program (26). We estimated haplotype frequencies and their 95% confidence interval (CI) using the progressive ligation algorithm (as implemented in SAS PROC HAPLOTYPE).
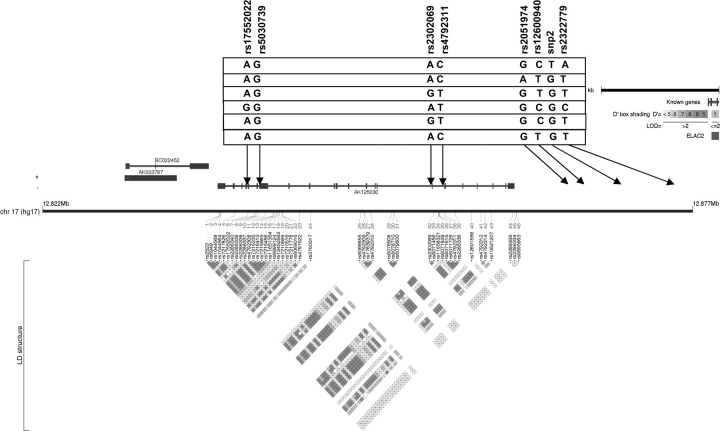
Fig. 1.
ELAC2 linkage disequilibrium plot. This plot was generated by Haploview and Locusview programs. When the approach of Gabriel et al. was used, the eight SNPs formed one block. The rs number on the top from left to right corresponds to the SNP name (e.g. snp1, snp2, etc.). The level of pairwise D′, which indicates the degree of linkage disequilibrium between two SNPs, is shown in the linkage disequilibrium structure in red. Six common haplotypes (frequency > 0.05) were identified.
Conditional logistic regression models were used to estimate ORs for disease in participants carrying either 1 or 2 versus 0 copies of the minor allele of each SNP and each multilocus haplotype. Haplotype trend regression (27) was used to test global association between ELAC2 haplotypes and prostate cancer. To assess the risk of sporadic prostate cancer, we performed the same SNP and haplotype association analyses after exclusion of subjects with more than one prostate cancer case in his family. The type I error rate was first controlled by the single multiple-degree-of-freedom test of association between ELAC2 haplotypes and prostate cancer. Given a statistically significant global test, haplotype-specific tests can provide some guidance as to which variants contribute to the statistically significant global test, although the nominal P-values we present do not control the family-wise error rate for these post hoc comparisons. Second, we performed Westfall's step-down permutation test (number of permutation tests = 100 000) for SNP and haplotype analyses to correct for multiple comparisons.
Age and family history are known risk factors for prostate cancer (28,29); previous studies found that, compared with lower body mass index (BMI) (<24.9 kg/m2), higher BMI (≥30 kg/m2) was associated with lower risk of all prostate cancer (30), as well as early-onset (<60 years old) (31) and high-grade prostate cancer (32,33), but results were not consistent across studies. Family history of prostate cancer was available in 1990, 1992 and 1996; we checked the consistency of data across these time periods and used the updated information in 1996 for analyses. We used the likelihood ratio test to evaluate how these factors modified the association between ELAC2 SNPs or haplotypes and the risk of prostate cancer by comparing a model with terms for main effects and interaction terms with the model with terms for main effects only. Because of the possible role of ELAC2 in germline proliferation and cell cycle (3), aggressiveness of prostate cancer may relate to genetic variations of ELAC2. We tested the association between ELAC2 haplotypes and aggressiveness of prostate cancer by using the definitions for tumor aggressiveness (aggressiveness: stages T3b, T4, N1, M1 or death due to prostate cancer or Gleason sum ≥7). All analyses were conducted with SAS release 9.0 (SAS Institute, Cary, NC), and all statistical tests were two sided.
Results
Eight SNPs in ELAC2 were genotyped. Except for snp7, none of the rest of the SNPs was out of Hardy–Weinberg equilibrium among controls (Table I). Because snp7 (rs5030739, Ala541Thr) was a rare allele with no heterozygote and no homozygote variants in this Caucasian population, it was dropped from the haplotype analyses. The internal blinded QC specimens did not show evidence of genotyping error.
The study population included 659 incident prostate cancer cases and 656 matched controls. Age and BMI distributions were similar for cases and controls (Table II), but family history of prostate cancer was statistically significantly different (P = 0.02). The mean age at starting smoking, lifetime average number of cigarettes/day (include non-smokers) and alcohol consumption were similar for cases and controls. The distribution of prostatitis by age group and case–control status did not show statistically significant difference between cases and controls (P = 0.09). Among cases, 79% were in tumor stages T1b–T3a, 49% had Gleason grades 5–6 and 36% had aggressive prostate cancer. Eighteen cases died of prostate cancer before 31 January 2000.
Table II.
Characteristics of Caucasians in the Health Professionals Follow-Up Study
| Variable | Cases n = 659, N (%) | Controls n = 656, N (%) |
| Age (year) (matching factor) | ||
| ≤65 | 297 (45) | 302 (46) |
| >65 | 362 (55) | 354 (54) |
| BMI (kg/m2) | ||
| ≤25 | 405 (61) | 489 (59) |
| 25–30 | 221 (34) | 221 (34) |
| >30 | 33 (5) | 46 (7) |
| Family history of prostate cancer | ||
| No | 528 (80) | 557 (85) |
| Yes | 131 (20) | 99 (15) |
| Age started smoking | 23.0 ± 5.3 | 22.9 ± 5.4 |
| Lifetime average cigarettes per day | 10.5 ± 6.3 | 10.9 ± 6.7 |
| Alcohol (g/day) | 11.4 ± 14.9 | 10.5 ± 14.7 |
| Prostate-specific antigen at diagnosis of prostate cancer (ng/ml) | 11.3 ± 21.0 (n = 479) | Not applicable |
| Stage | ||
| T1b-T3a | 517 (79) | Not applicable |
| T3b, T4, N1, M1 or death due to prostate cancer | 55 (8) | |
| Missing | 87 (13) | |
| Gleason sum | ||
| 2–4 | 44 (7) | Not applicable |
| 5–6 | 320 (49) | |
| 7 | 164 (25) | |
| 8–10 | 55 (8) | |
| Missing | 76 (12) | |
| Aggressiveness | ||
| No | 419 (64) | Not applicable |
| Yes | 240 (36) | |
| Missing | 0 | |
| Death due to prostate cancer | ||
| No | 641 (97) | Not applicable |
| Yes | 18 (3) | |
| Prostatitis | ||
| ≤60 | 35 (24) | 18 (15) |
| >60 | 111 (76) | 102 (85) |
Aggressiveness defined as stages T3b, T4, N1, M1 or death due to prostate cancer or Gleason sum ≥7.
No SNP (including the non-synonymous SNP Ser217Leu) was associated with prostate cancer risk (Table III). After dropping the rare SNP (snp7, Ala541Thr), seven SNPs spanning ELAC2 formed one block using the algorithm of Gabriel et al. (34), where blocks identified with the default settings in Haploview were merged if they had multiallelic D′ >0.8, and the cumulative frequency of common (>5% frequency) haplotypes in the merged block was above 80% (35). Six common haplotypes (frequency > 5%) were found with an accumulated frequency of 84% in controls (Table IV), and the P-value for the global test was 0.18. Men carrying one copy of the variant hap4 had a 1.39-fold increased risk of prostate cancer (95% CI = 1.05–1.85) compared with non-carriers. When subjects were restricted to sporadic prostate cancer, results changed very little (the P-value for global test became 0.03; hap4: OR = 1.41, 95% CI = 1.06–1.87; data not shown).
Table III.
SNP analysis by ELAC2 genotypes
| SNP | 0 copies | 1 copy | 2 copies | P-value* | |||
| Case/control | OR | Case/control | OR (95% CI) | Case/control | OR (95% CI) | ||
| All prostate cancer | |||||||
| Snp1 | 428/447 | 1.00 | 198/186 | 1.11 (0.87–1.41) | 21/12 | 1.82 (0.89–3.75) | 0.20 |
| Snp2 | 288/314 | 1.00 | 293/272 | 1.18 (0.94–1.49) | 64/59 | 1.19 (0.80–1.75) | 0.33 |
| Snp3 | 179/161 | 1.00 | 328/330 | 0.89 (0.69–1.16) | 133/146 | 0.82 (0.60–1.13) | 0.46 |
| Snp4 | 376/370 | 1.00 | 223/237 | 0.93 (0.73–1.17) | 44/36 | 1.21 (0.76–1.92) | 0.52 |
| Snp5 | 308/296 | 1.00 | 263/280 | 0.90 (0.71–1.13) | 72/68 | 1.01 (0.70–1.46) | 0.63 |
| Snp6 | 480/472 | 1.00 | 152/157 | 0.95 (0.73–1.22) | 9/10 | 0.89 (0.36–2.22) | 0.89 |
| Snp8 | 472/487 | 1.00 | 164/149 | 1.13 (0.88–1.46) | 9/8 | 1.16 (0.44–3.03) | 0.63 |
| Sporadic prostate cancer | |||||||
| Snp1 | 418/440 | 1.00 | 193/183 | 1.10 (0.87–1.41) | 21/12 | 1.84 (0.89–3.78) | 0.20 |
| Snp2 | 280/312 | 1.00 | 287/265 | 1.21 (0.96–1.53) | 63/58 | 1.21 (0.82–1.79) | 0.23 |
| Snp3 | 178/159 | 1.00 | 320/325 | 0.88 (0.68–1.14) | 127/143 | 0.80 (0.58–1.10) | 0.36 |
| Snp4 | 369/363 | 1.00 | 216/234 | 0.91 (0.72–1.15) | 43/36 | 1.18 (0.74–1.88) | 0.49 |
| Snp5 | 301/293 | 1.00 | 258/274 | 0.91 (0.72–1.15) | 69/67 | 1.00 (0.69–1.45) | 0.73 |
| Snp6 | 471/466 | 1.00 | 147/154 | 0.94 (0.72–1.22) | 8/9 | 0.91 (0.35–2.38) | 0.87 |
| Snp8 | 458/478 | 1.00 | 163/148 | 1.14 (0.88–1.48) | 9/8 | 1.16 (0.44–3.04) | 0.59 |
*P-value was for testing the null hypothesis: OR1 copy = OR2 copies = 1.
Table IV.
ORs between ELAC2 haplotypes and the risk of prostate cancer
| Haplotype | Prevalence among controls, % (95% CI) | 0 copies | 1 copy | 2 copies | P-value* | |||
| Case/control | OR | Case/control | OR (95% CI) | Case/control | OR (95% CI) | |||
| All prostate cancer (global test P = 0.18) | ||||||||
| Hap1: TACGCAA | 26.2 (23.8–28.6) | 330/359 | 1.00 | 277/250 | 1.21 (0.96–1.53) | 52/47 | 1.20 (0.79–1.84) | 0.24 |
| Hap2: TGTACAA | 22.8 (20.5 –25.1) | 396/386 | 1.00 | 225/239 | 0.92 (0.73–1.16) | 38/31 | 1.21 (0.73–1.99) | 0.53 |
| Hap3: TGTGTGA | 12.2 (10.4–14.0) | 520/506 | 1.00 | 133/142 | 0.90 (0.69–1.18) | 6/8 | 0.73 (0.25–2.11) | 0.65 |
| Hap4: CGCGTAG | 8.9 (7.4–10.5) | 512/542 | 1.00 | 142/109 | 1.39 (1.05–1.85) | 5/5 | 0.96 (0.26–3.51) | 0.07 |
| Hap5: TGCGCAA | 8.4 (6.9–9.9) | 556/550 | 1.00 | 101/101 | 0.98 (0.72–1.35) | 2/5 | 0.43 (0.08–2.34) | 0.59 |
| Hap6: TGTGCAA | 5.0 (3.7–6.1) | 613/596 | 1.00 | 44/55 | 0.77 (0.50–1.18) | 2/5 | 0.39 (0.08–2.04) | 0.25 |
| Others | 16.5 | |||||||
| Sporadic prostate cancer (global test P = 0.03) | ||||||||
| Hap1: TACGCAA | 26.2 (23.8–28.6) | 321/354 | 1.00 | 271/245 | 1.25 (0.98–1.58) | 52/47 | 1.23 (0.80–1.89) | 0.16 |
| Hap2: TGTACAA | 22.9 (20.6–25.2) | 388/379 | 1.00 | 219/236 | 0.90 (0.71–1.14) | 37/31 | 1.18 (0.71–1.94) | 0.50 |
| Hap3: TGTGTGA | 11.9 (10.2–13.7) | 510/500 | 1.00 | 129/139 | 0.90 (0.68–1.19) | 5/7 | 0.71 (0.22–2.25) | 0.65 |
| Hap4: CGCGTAG | 9.0 (7.4–10.5) | 498/533 | 1.00 | 141/108 | 1.41 (1.06–1.87) | 5/5 | 0.97 (0.26–3.54) | 0.06 |
| Hap5: TGCGCAA | 8.5 (7.0–10.0) | 544/540 | 1.00 | 98/101 | 0.97 (0.71–1.33) | 2/5 | 0.42 (0.08–2.33) | 0.58 |
| Others | 21.5 | |||||||
*P-value was for testing the null hypothesis: OR1 copy = OR2 copies = 1.
We looked at interactions with the following risk factors: age, BMI and family history of prostate cancer. However, none of these risk factors modify the association between sequence variants of ELAC2 and prostate cancer.
We also explored the main effects for aggressive and non-aggressive prostate cancer separately (Table V). Two SNPs showed a statistically significant association with aggressive prostate cancer (carriers versus non-carriers—snp2: OR = 1.43, 95% CI = 1.06–1.93; snp3: OR = 0.69, 95% CI = 0.50–0.95). For haplotype analysis (global test P-value = 0.11), hap1 and hap4 variant carriers had a 1.47- and 1.51-fold increased risk of aggressive prostate cancer (95% CI = 1.08–1.99 and 1.04–2.18, respectively) compared with non-carriers. Hap2 variant carriers showed 0.72-fold risk of prostate cancer (95% CI = 0.52–0.98) compared with non-carriers. Results were not statistically significant for non-aggressive prostate cancer. After correction for multiple comparisons by the permutation test, hap4 is the only haplotype that remained statistically significantly associated with sporadic prostate cancer (P for permutation test = 0.0069).
Table V.
ELAC2 SNP and haplotypes and risk of aggressive and non-aggressive prostate cancer
| Non-carriers | Carriers | P-value* | |||
| Cases/controls | OR | Cases/controls | OR (95% CI) | ||
| SNP | |||||
| Aggressive prostate cancer | |||||
| Snp1 | 158/447 | 1.00 | 78/198 | 1.10 (0.80–1.52) | 0.55 |
| Snp2 | 94/314 | 1.00 | 141/331 | 1.43 (1.06–1.93) | 0.02 |
| Snp3 | 76/161 | 1.00 | 157/476 | 0.69 (0.50–0.95) | 0.03 |
| Snp4 | 152/370 | 1.00 | 83/273 | 0.74 (0.54–1.01) | 0.05 |
| Snp5 | 112/296 | 1.00 | 122/348 | 0.92 (0.68–1.24) | 0.57 |
| Snp6 | 174/472 | 1.00 | 58/167 | 0.93 (0.66–1.32) | 0.69 |
| Snp8 | 172/487 | 1.00 | 63/157 | 1.13 (0.80–1.58) | 0.50 |
| Non-aggressive prostate cancer | |||||
| Snp1 | 270/447 | 1.00 | 141/198 | 1.18 (0.91–1.54) | 0.21 |
| Snp2 | 194/314 | 1.00 | 216/331 | 1.06 (0.83–1.36) | 0.65 |
| Snp3 | 103/161 | 1.00 | 304/476 | 1.00 (0.75–1.34) | 0.98 |
| Snp4 | 224/370 | 1.00 | 184/273 | 1.11 (0.87–1.43) | 0.40 |
| Snp5 | 196/296 | 1.00 | 213/348 | 0.92 (0.72–1.19) | 0.53 |
| Snp6 | 306/472 | 1.00 | 103/167 | 0.95 (0.72–1.26) | 0.73 |
| Snp8 | 300/487 | 1.00 | 110/157 | 1.14 (0.86–1.51) | 0.37 |
| Haplotype | |||||
| Aggressive prostate cancer (global test P = 0.11) | |||||
| Hap1: TACGCAA | 109/358 | 1.00 | 131/298 | 1.47 (1.08–1.99) | 0.01 |
| Hap2: TGTACAA | 183/542 | 1.00 | 80/269 | 0.72 (0.52–0.98) | 0.04 |
| Hap3: TGTGTGA | 160/387 | 1.00 | 51/151 | 0.91 (0.63–1.31) | 0.60 |
| Hap4: CGCGTAG | 169/383 | 1.00 | 57/114 | 1.51 (1.04–2.18) | 0.03 |
| Hap5: TGCGCAA | 197/550 | 1.00 | 43/106 | 1.14 (0.76–1.72) | 0.52 |
| Hap6: TGTGCAA | 189/505 | 1.00 | 14/60 | 0.59 (0.32–1.11) | 0.09 |
| Non-aggressive prostate cancer (global test P = 0.27) | |||||
| Hap1: TACGCAA | 221/358 | 1.00 | 198/298 | 1.08 (0.84–1.39) | 0.55 |
| Hap2: TGTACAA | 329/542 | 1.00 | 183/269 | 1.11 (0.87–1.43) | 0.40 |
| Hap3: TGTGTGA | 236/387 | 1.00 | 88/151 | 0.89 (0.65–1.20) | 0.44 |
| Hap4: CGCGTAG | 257/383 | 1.00 | 90/114 | 1.31 (0.96–1.80) | 0.09 |
| Hap5: TGCGCAA | 359/550 | 1.00 | 60/106 | 0.85 (0.59–1.22) | 0.38 |
| Hap6: TGTGCAA | 331/505 | 1.00 | 32/60 | 0.82 (0.52–1.30) | 0.40 |
Aggressive prostate cancer was defined as stages T3b, T4, N1, M1 or death due to prostate cancer or Gleason sum ≥7.
*P-value was for testing the null hypothesis: ORcarriers = ORnon-carriers = 1.
We also assessed whether prostatitis affected the association between ELAC2 genotypes and the risk of prostate cancer. Prostatitis status did not modify the association between hap1 and prostate cancer (one variant carrier of hap1 versus non-carrier: OR = 5.45, 95% CI = 1.60–18.6, P interaction = 0.12) among younger men (age ≤ 60) (data not shown). Results were also not statistically significant among older men (age > 60).
Discussion
For main effect, none of the tagging SNPs was associated with the risk of prostate cancer. In contrast, hap4 was associated with an increased risk of prostate cancer. Hap1 and hap4 were related to increased and hap2 to decreased risk of aggressive prostate cancer. This is the first demonstration that common SNPs and haplotypes in ELAC2 are associated with aggressive prostate cancer. A previous study (22) found that a very rare 8-SNP haplotype (frequency, case: 0.022, controls: 0) was associated with ‘familial early-onset’ prostate cancer. Since the outcomes selected by Camp et al. (22) (familial prostate cancer) and our study (>98% were sporadic prostate cancer) were different, our results were not comparable. The results changed very little after we restricted all cases to sporadic prostate cancer.
Two non-synonymous SNPs, Ser217Leu and Ala541Thr, corresponding to snp5 and snp7 in our study, have been widely explored previously. In our study, Ala541Thr was too rare to support statistical analyses. Ala541 of ELAC2 is located directly beside the histidine motif (1) and is important to 3′-tRNAse catalytic activity for removing a 3′ trailer from precursor tRNA (36). This may explain its association with prostate cancer in some studies (6,7,9,15). A meta-analysis (37) showed that Thr541 carriers accounted for 2% of prostate cancer in the general population. Thr541 is a very rare allele in our study population (Table I), which might be a result of genotyping error. However, Thr541 is in strong linkage disequilibrium with Leu217 (9–12,15,20), which was not associated with the risk of prostate cancer. Therefore, exclusion of Ala541Thr from haplotype analyses had a negligible effect on our results. Fewer than a quarter of previous studies found that either Ser217Leu or Ala541Thr was associated with the risk of prostate cancer. Because of the null findings in the majority of studies on these two SNPs, we included more common tagging SNPs and performed haplotype analyses to capture more genetic information than that provided by individual SNPs.
Our study does have some advantages. We restricted subjects to Caucasians, and thus population stratification is not a concern. Large sample size, high concordance rate in genotyping QC samples, few self-reported cases (3%) and high concordance between self-reported and medical record-confirmed cases (90%) increased the reliability of our findings. However, the low frequency of Thr541 in this population as compared with other Caucasian populations may be a result of sample variation across Caucasian populations. The tagging SNP approach, using a small set of representative SNPs, is a cost-effective approach for haplotype analyses.
A study in Caenorhabditis elegans showed that reduction of the gene activity of hoe-1 (the homolog of ELAC2) resulted from activation of mutation in ras and led to subsequent reduction of germline proliferation (3). In addition, the amino acid sequence of ELAC2 is similar to some proteins (1,4) with known functions, as described in the Introduction. This suggests that the sequence variants in ELAC2 may either indirectly deactivate the mutation in ras gene, which leads to inhibition of germline proliferation, or through messenger RNA modification and then the formation of subsequent carcinogenesis in the prostate. We did not observe a statistically significant association between genetic variations in ELAC2 and the risk of prostate cancer. However, ELAC2 common SNPs and haplotypes were statistically significantly related to aggressive prostate cancer. More experimental and association studies are warranted to explain the role of ELAC2 in prostate carcinogenesis.
Funding
National Institutes of Health (UO1 CA98233, CA55075).
Acknowledgments
The authors are grateful to Pati Soule and Ana-Tereza Andrade for DNA sample extraction and to the Partners High-Throughput Genotyping Center (Dr David Kwiatkowski, Alison Brown and Maura Regan) for genotyping.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BMI
body mass index
- CI
confidence interval
- ELAC2
elaC homolog 2 (Escherichia coli)
- OR
odds ratio
- QC
quality control
- SNP
single-nucleotide polymorphism
References
- 1.Tavtigian SV, et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 2.Dumont M, et al. Structure of primate and rodent orthologs of the prostate cancer susceptibility gene ELAC2. Biochim. Biophys. Acta. 2004;1679:230–247. doi: 10.1016/j.bbaexp.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Smith MM, et al. The Caenorhabditis elegans homolog of the putative prostate cancer susceptibility gene ELAC2, hoe-1, plays a role in germline proliferation. Dev. Biol. 2004;266:151–160. doi: 10.1016/j.ydbio.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 4.Dronkert ML, et al. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol. Cell. Biol. 2000;20:4553–4561. doi: 10.1128/mcb.20.13.4553-4561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi H, et al. Ser217Leu polymorphism of the HPC2/ELAC2 gene associated with prostatic cancer risk in Japanese men. Int. J. Cancer. 2003;107:224–228. doi: 10.1002/ijc.11347. [DOI] [PubMed] [Google Scholar]
- 6.Fujiwara H, et al. Association of common missense changes in ELAC2 (HPC2) with prostate cancer in a Japanese case-control series. J. Hum. Genet. 2002;47:641–648. doi: 10.1007/s100380200099. [DOI] [PubMed] [Google Scholar]
- 7.Yokomizo A, et al. HPC2/ELAC2 polymorphism associated with Japanese sporadic prostate cancer. Prostate. 2004;61:248–252. doi: 10.1002/pros.20107. [DOI] [PubMed] [Google Scholar]
- 8.Shea PR, et al. ELAC2 and prostate cancer risk in Afro-Caribbeans of Tobago. Hum. Genet. 2002;111:398–400. doi: 10.1007/s00439-002-0816-1. [DOI] [PubMed] [Google Scholar]
- 9.Rebbeck TR, et al. Association of HPC2/ELAC2 genotypes and prostate cancer. Am. J. Hum. Genet. 2000;67:1014–1019. doi: 10.1086/303096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rokman A, et al. ELAC2/HPC2 involvement in hereditary and sporadic prostate cancer. Cancer Res. 2001;61:6038–6041. [PubMed] [Google Scholar]
- 11.Wang L, et al. Role of HPC2/ELAC2 in hereditary prostate cancer. Cancer Res. 2001;61:6494–6499. [PubMed] [Google Scholar]
- 12.Xu J, et al. Evaluation of linkage and association of HPC2/ELAC2 in patients with familial or sporadic prostate cancer. Am. J. Hum. Genet. 2001;68:901–911. doi: 10.1086/319513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nupponen NN, et al. Mutational analysis of susceptibility genes RNASEL/HPC1, ELAC2/HPC2, and MSR1 in sporadic prostate cancer. Genes Chromosomes Cancer. 2004;39:119–125. doi: 10.1002/gcc.10308. [DOI] [PubMed] [Google Scholar]
- 14.Rennert H, et al. Association of susceptibility alleles in ELAC2/HPC2, RNASEL/HPC1, and MSR1 with prostate cancer severity in European American and African American men. Cancer Epidemiol. Biomarkers Prev. 2005;14:949–957. doi: 10.1158/1055-9965.EPI-04-0637. [DOI] [PubMed] [Google Scholar]
- 15.Suarez BK, et al. Polymorphisms in the prostate cancer susceptibility gene HPC2/ELAC2 in multiplex families and healthy controls. Cancer Res. 2001;61:4982–4984. [PubMed] [Google Scholar]
- 16.Meitz JC, et al. HPC2/ELAC2 polymorphisms and prostate cancer risk: analysis by age of onset of disease. Br. J. Cancer. 2002;87:905–908. doi: 10.1038/sj.bjc.6600564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanford JL, et al. Association of HPC2/ELAC2 polymorphisms with risk of prostate cancer in a population-based study. Cancer Epidemiol. Biomarkers Prev. 2003;12:876–881. [PubMed] [Google Scholar]
- 18.Severi G, et al. ELAC2/HPC2 polymorphisms, prostate-specific antigen levels, and prostate cancer. J. Natl Cancer Inst. 2003;95:818–824. doi: 10.1093/jnci/95.11.818. [DOI] [PubMed] [Google Scholar]
- 19.Adler D, et al. HPC2/ELAC2 gene variants associated with incident prostate cancer. J. Hum. Genet. 2003;48:634–638. doi: 10.1007/s10038-003-0091-6. [DOI] [PubMed] [Google Scholar]
- 20.Vesprini D, et al. HPC2 variants and screen-detected prostate cancer. Am. J. Hum. Genet. 2001;68:912–917. doi: 10.1086/319502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stampfer MJ, et al. Test of the National Death Index. Am. J. Epidemiol. 1984;119:837–839. doi: 10.1093/oxfordjournals.aje.a113804. [DOI] [PubMed] [Google Scholar]
- 22.Camp NJ, et al. Characterization of linkage disequilibrium structure, mutation history, and tagging SNPs, and their use in association analyses: ELAC2 and familial early-onset prostate cancer. Genet. Epidemiol. 2005;28:232–243. doi: 10.1002/gepi.20054. [DOI] [PubMed] [Google Scholar]
- 23.Platz EA, et al. Sex steroid hormones and the androgen receptor gene CAG repeat and subsequent risk of prostate cancer in the prostate-specific antigen era. Cancer Epidemiol. Biomarkers Prev. 2005;14:1262–1269. doi: 10.1158/1055-9965.EPI-04-0371. [DOI] [PubMed] [Google Scholar]
- 24.Clayton D. SNPHAP: a Program for Estimating Frequencies of Large Haplotypes of SNPs. Cambridge, UK: Department of Medical Genetics, Cambridge Institute for Medical Research; 2002. [Google Scholar]
- 25.Qin ZS, et al. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am. J. Hum. Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stram DO, et al. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum. Hered. 2003;55:179–190. doi: 10.1159/000073202. [DOI] [PubMed] [Google Scholar]
- 27.Zaykin DV, et al. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum. Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- 28.Kalish LA, et al. Family history and the risk of prostate cancer. Urology. 2000;56:803–806. doi: 10.1016/s0090-4295(00)00780-9. [DOI] [PubMed] [Google Scholar]
- 29.Cerhan JR, et al. Family history and prostate cancer risk in a population-based cohort of Iowa men. Cancer Epidemiol. Biomarkers Prev. 1999;8:53–60. [PubMed] [Google Scholar]
- 30.Calle EE, et al. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 31.Giovannucci E, et al. Body mass index and risk of prostate cancer in U.S. health professionals. J. Natl Cancer Inst. 2003;95:1240–1244. doi: 10.1093/jnci/djg009. [DOI] [PubMed] [Google Scholar]
- 32.Amling CL. Relationship between obesity and prostate cancer. Curr. Opin. Urol. 2005;15:167–171. doi: 10.1097/01.mou.0000165550.94663.fb. [DOI] [PubMed] [Google Scholar]
- 33.Rohrmann S, et al. Family history of prostate cancer and obesity in relation to high-grade disease and extraprostatic extension in young men with prostate cancer. Prostate. 2003;55:140–146. doi: 10.1002/pros.10211. [DOI] [PubMed] [Google Scholar]
- 34.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 35.Florez JC, et al. Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004;53:1360–1368. doi: 10.2337/diabetes.53.5.1360. [DOI] [PubMed] [Google Scholar]
- 36.Takaku H, et al. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 2003;31:2272–2278. doi: 10.1093/nar/gkg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camp NJ, et al. Meta-analysis of associations of the Ser217Leu and Ala541Thr variants in ELAC2 (HPC2) and prostate cancer. Am. J. Hum. Genet. 2002;71:1475–1478. doi: 10.1086/344516. [DOI] [PMC free article] [PubMed] [Google Scholar]