Abstract
Background
Previous studies have shown that the ADIPOR1, ADORA1, BTG2 and CD46 genes differ significantly between long-term survivors of breast cancer and deceased patients, both in levels of gene expression and DNA copy numbers. The aim of this study was to characterize the expression of the corresponding proteins in breast carcinoma and to determine their correlation with clinical outcome.
Methods
Protein expression was evaluated using immunohistochemistry in an independent breast cancer cohort of 144 samples represented on tissue microarrays. Fisher's exact test was used to analyze the differences in protein expression between dead and alive patients. We used Cox-regression multivariate analysis to assess whether the new markers predict the survival status of the patients better than the currently used markers.
Results
BTG2 expression was demonstrated in a significantly lower proportion of samples from dead patients compared to alive patients, both in overall expression (P = 0.026) and cell membrane specific expression (P = 0.013), whereas neither ADIPOR1, ADORA1 nor CD46 showed differential expression in the two survival groups. Furthermore, a multivariate analysis showed that a model containing BTG2 expression in combination with HER2 and Ki67 expression along with patient age performed better than a model containing the currently used prognostic markers (tumour size, nodal status, HER2 expression, hormone receptor status, histological grade, and patient age). Interestingly, BTG2 has previously been described as a tumour suppressor gene involved in cell cycle arrest and p53 signalling.
Conclusions
We conclude that high-level BTG2 protein expression correlates with prolonged survival in patients with breast carcinoma.
Background
Breast cancer is the most common malignancy among women, and accounted for approximately 1.15 million new cases and 411,000 deaths worldwide in 2002 [1]. During the last decade, the survival rate for breast cancer patients has increased dramatically due to earlier detection and new treatment protocols [2]. Presently, various clinical and pathological markers including axillary lymph node status, hormone receptor status, histological grade, tumour size, patient age, HER2 expression and vascular invasion are used to predict breast cancer prognosis and provide accurate treatment [3]. However, these markers are insufficient and approximately 20 to 30% of breast cancer patients will die from the disease within five years of diagnosis [4]. It is, therefore, of great importance to identify novel molecular markers to further refine prognosis and response to treatment. Gene expression analysis has been used to develop gene expression signatures that predict clinical outcome in breast cancer patients [5-9]. Previously, we analysed breast tumours from lymph node-negative patients using gene expression microarray and array-CGH to identify genes with altered levels of expression and aberrant chromosomal regions revealing prognostic values [7,10]. By integrating the expression and array-CGH results, 27 genes were identified which differed significantly (P < 0.05) in both gene expression and DNA copy numbers between deceased patients and 10-year survivors [10]. Based on their involvement in breast cancer and the availability of commercial antibodies, the ADIPOR1, ADORA1, BTG2 and CD46 genes were selected among the 27 previously identified genes to further investigate the association of protein expression levels to overall patient survival. In the present investigation, protein expression was analysed by immunohistochemistry on tissue microarrays in an independent cohort of breast cancer patients, and correlated to 5-year survival.
Methods
Patients and tissue microarray construction
The breast cancer samples were obtained from 144 patients undergoing surgical resection at Malmö University Hospital, Malmö, Sweden, between 2001 and 2002. One patient lacked five years follow-up time resulting in the exclusion of this sample from the 5-year survival analysis, although not from the multivariate analysis. The 5-year survival analysis was performed based on overall survival, including 111 samples from alive and 32 samples from dead patients. Further clinical information is compiled in Table 1. Tissue microarrays (TMAs) containing duplicate 1.00 mm cores from each tumour were constructed as previously described [11]. The utilization of the tumour material for research purposes was approved by regional ethical committees in Lund, Sweden.
Table 1.
Clinicopathological features of the 144 breast tumour specimens included in this study
| deceased patients | 5 year survivors | lack 5 years follow-up | Total | |
|---|---|---|---|---|
| Median age at diagnosis | 77 | 63 | 75 | 65 |
| Recurrence free for 5 years | ||||
| yes | 0 | 102 | 0 | 102 |
| no | 16 | 7 | 1 | 24 |
| missing | 16 | 2 | 0 | 18 |
| Total | 32 | 111 | 1 | 144 |
| Type | ||||
| ductal | 26 | 77 | 1 | 104 |
| lobular | 4 | 23 | 0 | 27 |
| tubular | 1 | 6 | 0 | 7 |
| medullary | 1 | 2 | 0 | 3 |
| missing | 0 | 3 | 0 | 3 |
| Total | 32 | 111 | 1 | 144 |
| Size | ||||
| median (mm) | 27 | 19 | 27 | 20 |
| 20 mm and below | 11 | 62 | 0 | 73 |
| above 20 mm | 21 | 49 | 1 | 71 |
| Total | 32 | 111 | 1 | 144 |
| Nodal status | ||||
| positive | 17 | 38 | 1 | 56 |
| negative | 10 | 63 | 0 | 73 |
| missing | 5 | 10 | 0 | 15 |
| Total | 32 | 111 | 1 | 144 |
| Estrogen receptor status | ||||
| positive | 23 | 101 | 1 | 125 |
| negative | 9 | 10 | 0 | 19 |
| Total | 32 | 111 | 1 | 144 |
| Progesterone receptor status | ||||
| positive | 15 | 85 | 0 | 100 |
| negative | 17 | 26 | 1 | 44 |
| Total | 32 | 111 | 1 | 144 |
| Her2 status | ||||
| positive | 7 | 6 | 0 | 13 |
| negative | 22 | 101 | 0 | 123 |
| missing | 3 | 4 | 1 | 8 |
| Total | 32 | 111 | 1 | 144 |
Immunohistochemistry (IHC)
The expression of ADIPOR1, ADORA1, BTG2 and CD46 proteins was investigated using IHC. Prior to hybridisation to the tissue microarrays, antibodies corresponding to the selected genes were optimised on paraffin-embedded sections of breast tumours. After deparaffinisation in Xylene, the tissue microarrays were autoclaved for at least one hour in buffer S1699 or S2367 (Dako Norden A/S, Denmark) or Borgs Decloaker pH9 buffer solution (Biocare Medical, CA, USA) (Table 2). The immunohistochemical staining was performed in an automated immunostainer (TechMate 228 500 Plus; Dako Norden A/S, Denmark). The TMA sections were incubated with the different antibodies at a dilution of 1:300 for ADIPOR1 (Phoenix Pharmaceuticals, Inc, CA, USA), 1:500 for ADORA1 (Genway Biotech, Inc, CA USA), 1:1000 for BTG2 (Genway Biotech, Inc, CA, USA), and 1:40 for CD46 (BD Biosciences, New Jersey, USA); (Table 2). The antibodies were visualised with the EnVision (K5007, Dako Norden A/S, Denmark) or LSAB (K5001, Dako Norden A/S, Denmark) visualization system according to the manufacturer's instructions (Table 2). EnVision uses a secondary antibody against both rabbit and mouse that is directly labelled with HRP (horseradish peroxidase) reacting with DAB, whereas LSAB uses a secondary antibody against rabbit and mouse, labelled with biotin, then streptavidin-HRP is added and the staining is done with DAB. The TMA sections were then washed in water, dehydrated in an alcohol gradient followed by Xylen treatment, and mounted.
Table 2.
Technical data of the laboratory procedure for each specific antibody
| Antibody | Antibody dilution | Manufacturer | Catalogue number | Type of antibody | Time of incubation (RT) | Pre-treatment buffer | visualization system |
|---|---|---|---|---|---|---|---|
| AdipoR1 | 1:300 | Phoenix Pharmaceuticals | H-001-44 | Rabbit polyclonal | 30 min | Borgs decloaker | LSAB |
| Adora1 | 1:500 | Genway Biotech | 18-461-10001 | Rabbit polyclonal | 30 min | Borgs decloaker | LSAB |
| BTG2 | 1:1000 | Genway Biotech | 18-003-42396 | Rabbit polyclonal | 30 min | S2367 | Envision |
| CD46 | 1:40 | BD Biosciences | 555948 | Mouse monoclonal | 30 min | S1699 | Envision |
Evaluation of IHC
The immunostained tissue microarray sections were analysed by a pathologist (AK). CD46 protein is a cell membrane protein, while ADIPOR1 and ADORA1 are cytoplasm proteins. The subcellular location of BTG2 has varied in previous publications, and in this study, the cytoplasm and cell membrane were stained. The cell membrane staining intensities were graded as no expression (0), low expression (+), moderate expression (++), and high expression (+++). The expression of the cytoplasm proteins was also graded from 0-3 (ranging from no expression to high expression). The proportion of tumour cells expressing membrane and/or cytoplasm protein was determined. In the evaluation of ADIPOR1, ADORA1 and CD46, any level of sample staining was considered positive. For the BTG2 protein, moderate to strong staining of at least 50% of the cells was required for the sample to be scored as positive.
Statistical analysis
The difference in expression of the four proteins between tumours from alive and dead patients was tested using two-tailed Fisher's exact test. Kaplan-Meier survival curves were produced using the SPSS version 16 software to demonstrate the difference in overall survival between overall BTG2-positive and overall BTG2-negative samples, as well as BTG2 membrane-positive and BTG2 membrane-negative samples. Significant differences between the curves were compared using the Breslow-Wilcoxon test [12]. Additionally, a multivariate analysis (Cox-regression) was performed to evaluate the clinical significance of using current prognostic markers (tumour size, nodal status, HER2 expression, hormone receptor status, histological grade, and patient age) versus using a model containing the proposed markers HER2 expression, patient age, and increasingly used marker Ki67 in combination with BTG2 expression. To cope with the reduced validity of the scientific inference due to misrepresentation we used Multiple Imputation by Chained Equations. The effect of the markers on survival probability was modelled by Proportional Hazards Model. Variable selection was based on a combination bootstrap and information theory approach [13]. As an external validating measure we used time-dependent AUC and Concordance index (C-index) [14]. The time-dependent AUC characterises the temporal changes in predictive accuracy. Concordance index offers an easy way to interpret global accuracy measure that varies between 0.5 and 1. A concordance index of 1 means that with 100% precision we can rank the patient's survival time given the recorded marker information. If the concordance index converges to 0.5 the ranking of survival times becomes more and more driven by chance, and becomes completely random at 0.5. To quantify the impact of a single marker on the predictive accuracy we removed one marker at the time from the final model and refitted a Proportional Hazards Model and re-estimated the C-index. An estimation of the correlation between expression of the different proteins were performed using Pearson correlation.
Results
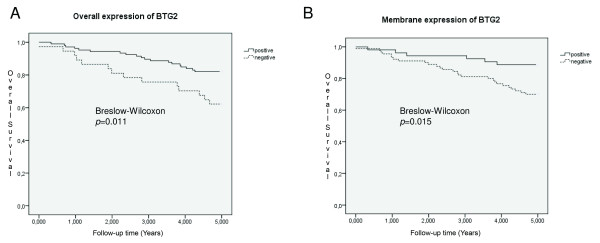
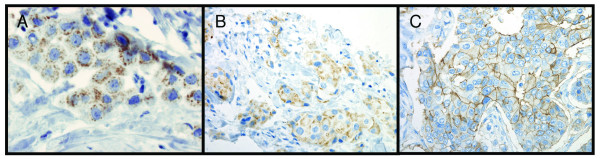
Of the 144 specimens present in the tissue microarray, 136-141 were interpretable for protein expression (Table 3). Excluded samples had few tumour cells, large tissue loss or affluence of necrotic tissue. BTG2 was expressed in both the cytoplasm and the cell membrane (Figure 1). In one sample, BTG2 showed expression in the cytoplasm and the membrane in one area and was expressed in the membrane exclusively in another area (Figure 2). The proportion of BTG2 protein expression was higher in tumours from 5-year overall survivors than among the tumours from deceased patients. The overall expression differed significantly between alive and dead patients (P = 0.026), although there was a stronger association with membrane specific expression (P = 0.013) (Table 3). Kaplan-Meier curves visualize the difference in overall survival between patients with tumours positive versus negative for overall and cell membrane specific BTG2 expression in Figure 3. Moreover, the difference between the curves was significant using the Breslow-Wilcoxon test [12] for both overall BTG2 (P = 0.011) and cell membrane expression (P = 0.015). Cytoplasm or cell membrane expression of BTG2 was observed in 78% of the samples and cell membrane specific expression in 39% of the samples. None of the remaining three analysed proteins (ADORA1, ADORA1 and CD46) showed a statistically significant difference in expression between alive and dead patients in this study (Table 3). The ADIPOR1 protein was expressed in the cytoplasm in 18% of the samples. The majority of the positive samples showed primarily granular staining (Figure 4a). Approximately 24% of the samples were positive for ADORA1 staining, also displaying primarily granular staining in the cytoplasm (Figure 4b). Fourteen percent of the samples expressed CD46 in the cell membrane (Figure 4c).
Table 3.
Difference in protein expression between tumours from 5-year overall survivors and tumours from deceased patients
| Dead patients | Alive patients | |||||
|---|---|---|---|---|---|---|
| Protein | Protein expression positive (%) | Protein expression negative (%) | Protein expression positive (%) | Protein expression negative (%) | not available (No of samples) | P-value dead vs. alive patients |
| AdipoR1 | 26 | 74 | 17 | 83 | 3 | 0.29 |
| Adora1 | 30 | 70 | 23 | 77 | 8 | 0.47 |
| BTG2 | 61 | 39 | 82 | 18 | 6 | 0.026* |
| -membrane only | 19 | 81 | 44 | 56 | 6 | 0.013* |
| -cytoplasm only | 52 | 48 | 68 | 32 | 6 | 0.14 |
| CD46 | 16 | 84 | 14 | 86 | 4 | 0.77 |
P-values were calculated using a two-tailed Fisher's exact test. The samples designated as not available had few tumour cells, large tissue loss or affluence of necrotic tissue.
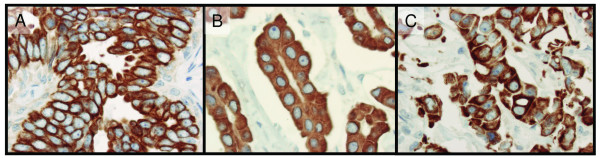
Figure 1.
Sub-cellular location of the BTG2 protein in this study. Brown colour represents BTG2 staining. BTG2 protein immunohistochemistry staining using tissue microarrays show exclusive cell membrane specific expression (A), exclusive cytoplasm expression (B), and both cell membrane and cytoplasm expression of BTG2 in the same sample (C).
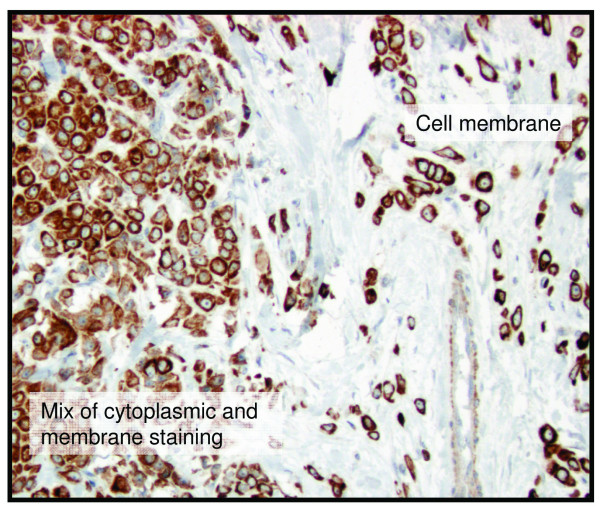
Figure 2.
Various sub-cellular location of the BTG2 protein in even within one sample. One TMA sample showing cytoplasm and cell membrane expression of BTG2 in a part of the sample and exclusively membranous expression in another part of the sample. Brown colour indicates BTG2 staining.
Figure 3.
Kaplan-Meier survival curves illustrating the effect of BTG2 expression. The Kaplan-Meier curves show the difference in survival between patients with tumours that revealed any BTG2 expression and patients whose tumours did not (A), as well as the difference in survival between patients with tumours that revealed cell membrane specific BTG2 expression and patients whose tumours did not (B). Solid lines represent patients whose tumours expressed BTG2 and dashed lines represent patients whose tumours did not. The p-values for the difference between the curves were calculated using a generalized Wilcoxon test.
Figure 4.
Sub-cellular location for samples staining positive for ADIPOR1, ADORA1 and CD46. Brown colour represents staining of the specific protein. Immunohistochemistry staining performed for these proteins on tissue microarrays show staining of ADIPOR1 (A) and ADORA1 (B) as granular staining in the cytoplasm, and expression of CD46 concentrated to the cell membrane (C).
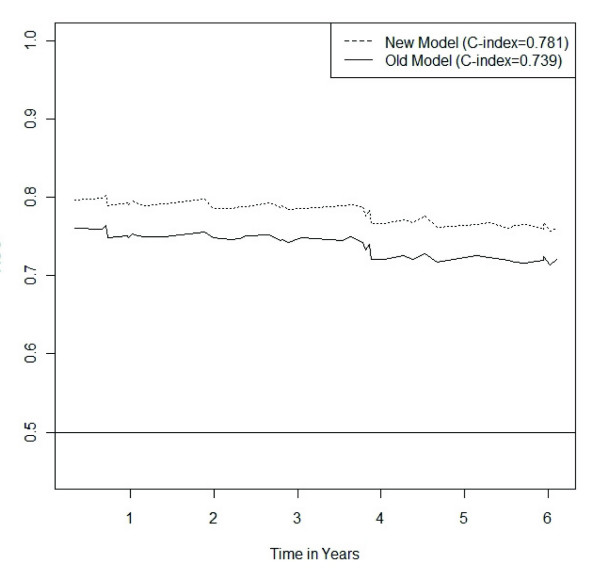
The multivariate analysis showed that the model containing BTG2 expression had better predictive power than the model built on current classical pathological markers (Table 4). The BTG2 model revealed a C-value of 0.781 compared to the slightly lower C-value of 0.772 for the model of currently used markers. If only the markers displaying statistical significance are used from the model of current prognostic markers (HER2 expression and patient age), a C-value of 0.739 was achieved. Figure 5 shows the classification accuracy of a Cox-regression model based on the current markers compared to the classification accuracy of a new model based on the new markers considered in the present study. Over the whole time-span considered the new markers offer superior classification accuracy. For both the new markers and the old ones the classification accuracy shows a slight decreasing trend with time. The strongest correlation of protein expression were between ADIPOR1 and ADORA1 (k = 0.749) and between BTG2 overall expression and BTG2 cytoplasm expression (k = 0.723) (Figure 6).
Table 4.
The effect on the survival status of currently used markers compared to BTG2 expression
| A | Odds Ratio | 95%CI | P-value | C-index |
|---|---|---|---|---|
| Age | 1.058 | 1.031; 1.087 | < 0.0001* | 0.711 |
| BTG2 both | 0.338 | 0.336; 0.339 | < 0.0001* | 0.768 |
| BTG2 cytoplasm | 0.699 | 0.697; 0.700 | < 0.0001* | 0.768 |
| BTG2 membrane | 0.980 | 0.977; 0.983 | < 0.0001* | 0.768 |
| HER2 | 3.331 | 3.248; 3.416 | < 0.0001* | 0.768 |
| Ki67 | 2.441 | 1.202; 4.956 | 0.013* | 0.758 |
| B | Odds Ratio | 95%CI | P-value | C-index |
| Age | 1.058 | 1.032; 1.085 | < 0.0001* | 0.696 |
| HER2 | 2.668 | 2.640; 2.696 | < 0.0001* | 0.761 |
| Histological grade 2 | 0.954 | 0.294; 3.094 | 0.938 | 0.770 |
| Histological grade 3 | 1.463 | 0.432; 4.949 | 0.539 | 0.770 |
| Hormone receptor status | 0.567 | 0.248; 1.296 | 0.179 | 0.767 |
| Nodal Status | 1.673 | 0.823; 3.398 | 0.154 | 0.768 |
| Tumour size | 1.569 | 0.739; 3.330 | 0.240 | 0.765 |
A Cox-regression multivariate analysis was performed to test the effect on survival status of the markers used in current clinical praxis and the changes in the model predictive power induced by the removal of a single marker. The predictive power of the full new model as measured by the C-index is 0.781 (A). The C-index for the old model is 0.772 (B), and the model with only the statistically significant variables in the old model has a predictive power of 0.739.
Figure 5.
Variation of the predictive power depending on survival time. The new Cox-regression model containing BTG2 expression, HER2 expression, patient age and Ki67 expression performed better, i.e. revealed higher prediction accuracy, than a Cox-regression model containing the currently used prognostic markers that gained statistical significance (HER2 expression, and patient age). This difference in predictive power was stable, independent of survival time.
Figure 6.
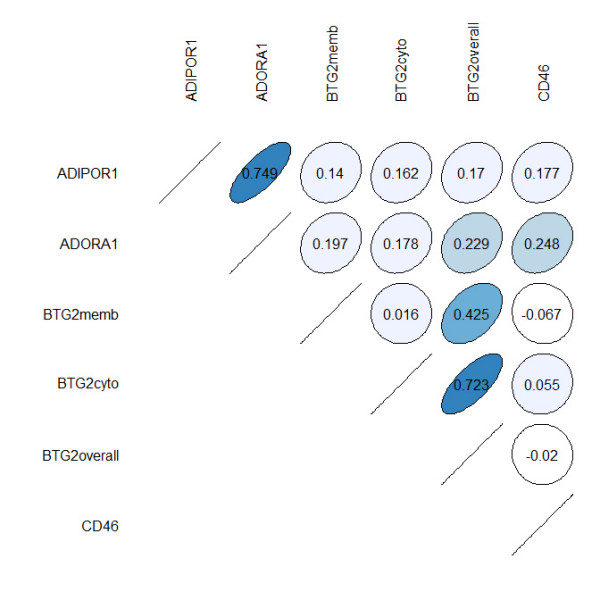
Correlation plot for the expression of the analyzed proteins. The strongest correlations were found between ADIPOR1 and ADORA1, as well as BTG2 overall expression and BTG2 cytoplasm expression. The course of the circle indicates whether the correlation is positive or negative.
Discussion
In the present investigation, the expression of four proteins (ADIPOR1, ADORA1, BTG2 and CD46) was studied by IHC using tissue microarrays. The aim was to evaluate the association between protein expression of these four genes and 5-year overall survival in breast cancer patients. Protein expression of BTG2 was found to be significantly more prevalent in tumours from alive patients compared to tumours from dead patients. Furthermore, a multivariate analysis showed that a model containing BTG2 expression in combination with HER2 expression, patient age and Ki67 expression performed better, i.e. revealed a higher prediction accuracy, than a model containing the currently used prognostic markers (tumour size, nodal status, HER2 expression, hormone receptor status, histological grade, and patient age). In this model, BTG2 expression as well as HER2 expression and patient age were highly significant (P < 0.0001), whereas Ki67 expression displayed a lower significance (P = 0.013). These results further strengthen that BTG2 expression could be a useful complement to the currently used markers, and in addition, suggest Ki67 expression as a useful marker of breast cancer survival. This is the first report of a large quantitative analysis demonstrating that BTG2 expression is associated with breast cancer patient survival.
A portion of the samples demonstrated cell membrane specific expression only, several showed only cytoplasm expression, and many of the samples showed expression in both the cell membrane and the cytoplasm (Figure 1). One sample showed distinct expression of BTG2 in the cytoplasm and cell membrane in one area and in another area BTG2 was exclusively expressed in the cell membrane (Figure 2). In previous studies of BTG2, the sub-cellular location of the protein was diverse. The protein has been reported to be located in the cytoplasm [15-17] and in the nucleus [18]. Immunostained lung, kidney and small intestine tissue display cell membrane expression of BTG2 in one report [19]. In the present investigation, we observed cytoplasm and cell membrane expression, but no nuclear expression. The various sub-cellular locations of BTG2 could indicate that the protein is expressed in different cellular compartments during altering conditions, such as cell cycle phase, differentially expressed in diverse tissue types, or due to varying specificity of different antibodies.
The BTG2 gene is located at the 1q32 chromosomal region, which was gained in a significantly higher proportion of 10-year survivors than in deceased lymph node-negative breast cancer patients in a previous study [10]. The gene belongs to the structurally homogeneous BTG family of which five genes have been identified in human, BTG1, BTG2, BTG3, Tob and Tob2. The BTG2 protein is highly conserved and shares 94% homology with the murine equivalent [20]. BTG2 is a tumour suppressor gene [21-23] which is directly regulated by p53 and involved in p53-mediated response to DNA damage [24]. According to the literature, BTG2 is involved in cell cycle arrest in the transition from G1 to S phase [25,26]. In addition, BTG2 can regulate G2 to M cell cycle arrest independent of p53 [27,28]. BTG2 is known to mediate chemotherapy induced apoptosis in cancer cells [29-31] and a study by Lim et al. indicates that BTG2 enhances cancer cell death by accumulation of H2O2 [32].
Down-regulation of BTG2 has been observed in several cancer types such as prostate cancer, breast cancer and gliomas [17,18,33]. In this study 78% of the breast cancer samples showed moderate to high expression of BTG2 in the majority of tumour cells. Nevertheless, BTG2 was significantly down-regulated in tumours from dead patients compared to tumours from alive patients, both in overall expression and cell membrane specific expression. This finding suggests that high total BTG2 or specific cell membrane expression may contribute to a prolonged survival. A previous study analysed BTG2 protein expression and correlated decreased nucleus expression to a more aggressive phenotype of breast cancer, although they did not detect a significant difference in survival [34]. This discrepancy could be due to the use of different BTG2 antibodies since Kawakubo et al. detected staining predominantly in the nucleus. However, our finding supports the theory that down-regulation of BTG2 contributes to a more malignant behaviour of the cells. BTG2 is therefore a promising prognostic marker in breast cancer.
The proteins ADIPOR1, ADORA1 and CD46 did not demonstrate differences in expression between tumours from alive and dead patients. High mRNA levels of ADIPOR1 have earlier been associated with lower risk of breast cancer [35]. Previously, Mirza et al. speculated that increased expression of ADORA1 may contribute to tumour cell growth and decreased apoptosis in breast tumour cells [36]. The intensity of CD46 expression has been negatively correlated with histological grade and type, tumour size, and tumour recurrence but not to overall survival [37], which is supported by the results from this study. These proteins may still be involved in breast cancer progression, although no significant difference was seen in expression between alive and dead patients in the current investigation. The strongest correlations in protein expression were between ADIPOR1 and ADORA1, as well as BTG2 overall expression and BTG2 cytoplasm expression. The BTG2 correlation was expected since BTG2 overall expression is a combination of BTG2 cytoplasm expression and BTG2 membrane specific expression. The correlation between ADIPOR1 and ADORA1 is however difficult to explain since they to our knowledge do not interact, although both were expressed in similar proportions of the samples in this study (Table 3), and they are located at 1q32, within 300 000 base pairs from each other. This correlation might be interesting for further investigation.
Conclusions
We conclude that high BTG2 expression levels correlate with prolonged breast cancer survival. Furthermore, BTG2 protein expression may be used as a prognosticator for breast cancer as well as a possible molecular target in breast cancer treatment. Further studies in independent tumour sets are needed to validate and establish BTG2 protein expression as a prognostic marker.
List of abbreviations
AUC: Area Under the Receiver Operating Characteristic Curve; C-index: Concordance index; CGH: Compatative Genome Hybridisation; DNA: Deoxyribonucleic Acid; IHC: Immunohistochemistry; TMA: Tissue Microarray.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KL, EK and AD performed the immunohistochemistry. AK evaluated the immunostained tissue microarrays. EK performed the 5-year survival analysis. SN performed the multivariate analysis. DB performed the molecular analyses (such as ER, PR, HER2) and KJ provided the tissue microarrays as well as the clinical information. EK, UD, AD, TP, PK, KJ and KH interpreted the results and wrote the paper. All authors read and approved the final manuscript. KH was responsible for supervision as well as providing the funding.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Elin Möllerström, Email: elin.mollerstrom@neuro.gu.se.
Anikó Kovács, Email: aniko.kovacs@vgregion.se.
Kristina Lövgren, Email: kristina.lovgren@med.lu.se.
Szilard Nemes, Email: nemes.szilard@oc.gu.se.
Ulla Delle, Email: ulla.delle@oncology.gu.se.
Anna Danielsson, Email: anna.danielsson@oncology.gu.se.
Toshima Parris, Email: toshima.parris@oncology.gu.se.
Donal J Brennan, Email: donal.brennan@ucd.ie.
Karin Jirström, Email: Karin.Jirstrom@med.lu.se.
Per Karlsson, Email: per.karlsson@oncology.gu.se.
Khalil Helou, Email: khalil.helou@oncology.gu.se.
Acknowledgements
This work was performed at the Department of Oncology, Göteborg University, Sweden and was supported by The King Gustav V Jubilee Clinic Cancer Research Foundation.
We gratefully acknowledge the help of Ulric Pedersen in preparing some of the illustrations.
References
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD, Feuer EJ. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Progress and promise: highlights of the international expert consensus on the primary therapy of early breast cancer 2007. Ann Oncol. 2007;18:1133–1144. doi: 10.1093/annonc/mdm271. [DOI] [PubMed] [Google Scholar]
- Coleman MP, Quaresma M, Berrino F, Lutz JM, De Angelis R, Capocaccia R, Baili P, Rachet B, Gatta G, Hakulinen T. Cancer survival in five continents: a worldwide population-based study (CONCORD) Lancet Oncol. 2008;9:730–756. doi: 10.1016/S1470-2045(08)70179-7. [DOI] [PubMed] [Google Scholar]
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Karlsson E, Delle U, Danielsson A, Olsson B, Abel F, Karlsson P, Helou K. Gene expression variation to predict 10-year survival in lymph-node-negative breast cancer. BMC Cancer. 2008;8:254. doi: 10.1186/1471-2407-8-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naderi A, Teschendorff AE, Barbosa-Morais NL, Pinder SE, Green AR, Powe DG, Robertson JF, Aparicio S, Ellis IO, Brenton JD, Caldas C. A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene. 2006. [DOI] [PubMed]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Mollerstrom E, Delle U, Danielsson A, Parris T, Olsson B, Karlsson P, Helou K. High-resolution genomic profiling to predict 10-year overall survival in node-negative breast cancer. Cancer Genet Cytogenet. 2010;198:79–89. doi: 10.1016/j.cancergencyto.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Ryden L, Landberg G, Stal O, Nordenskjold B, Ferno M, Bendahl PO. HER2 status in hormone receptor positive premenopausal primary breast cancer adds prognostic, but not tamoxifen treatment predictive, information. Breast Cancer Res Treat. 2008;109:351–357. doi: 10.1007/s10549-007-9660-2. [DOI] [PubMed] [Google Scholar]
- Gehan EA. A Generalized Wilcoxon Test for Comparing Arbitrarily Singly-Censored Samples. Biometrika. 1965;52:203–223. [PubMed] [Google Scholar]
- Heymans MW, van Buuren S, Knol DL, van Mechelen W, de Vet HC. Variable selection under multiple imputation using the bootstrap in a prognostic study. BMC Med Res Methodol. 2007;7:33. doi: 10.1186/1471-2288-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Guardavaccaro D, Starace G, Tirone F. Overexpression of the nerve growth factor-inducible PC3 immediate early gene is associated with growth inhibition. Cell Growth Differ. 1996;7:1327–1336. [PubMed] [Google Scholar]
- Varnum BC, Reddy ST, Koski RA, Herschman HR. Synthesis, degradation, and subcellular localization of proteins encoded by the primary response genes TIS7/PC4 and TIS21/PC3. J Cell Physiol. 1994;158:205–213. doi: 10.1002/jcp.1041580125. [DOI] [PubMed] [Google Scholar]
- Ficazzola MA, Fraiman M, Gitlin J, Woo K, Melamed J, Rubin MA, Walden PD. Antiproliferative B cell translocation gene 2 protein is down-regulated post-transcriptionally as an early event in prostate carcinogenesis. Carcinogenesis. 2001;22:1271–1279. doi: 10.1093/carcin/22.8.1271. [DOI] [PubMed] [Google Scholar]
- Kawakubo H, Carey JL, Brachtel E, Gupta V, Green JE, Walden PD, Maheswaran S. Expression of the NF-kappaB-responsive gene BTG2 is aberrantly regulated in breast cancer. Oncogene. 2004;23:8310–8319. doi: 10.1038/sj.onc.1208008. [DOI] [PubMed] [Google Scholar]
- Melamed J, Kernizan S, Walden PD. Expression of B-cell translocation gene 2 protein in normal human tissues. Tissue Cell. 2002;34:28–32. doi: 10.1054/tice.2001.0220. [DOI] [PubMed] [Google Scholar]
- Duriez C, Moyret-Lalle C, Falette N, El-Ghissassi F, Puisieux A. BTG2, its family and its tutor. Bull Cancer. 2004;91:E242–253. [PubMed] [Google Scholar]
- Boiko AD, Porteous S, Razorenova OV, Krivokrysenko VI, Williams BR, Gudkov AV. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006;20:236–252. doi: 10.1101/gad.1372606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim IK, Lee MS, Lee SH, Kim NK, Jou I, Seo JS, Park SC. Differential expression of TIS21 and TIS1 genes in the various organs of Balb/c mice, thymic carcinoma tissues and human cancer cell lines. J Cancer Res Clin Oncol. 1995;121:279–284. doi: 10.1007/BF01209594. [DOI] [PubMed] [Google Scholar]
- Elmore LW, Di X, Dumur C, Holt SE, Gewirtz DA. Evasion of a single-step, chemotherapy-induced senescence in breast cancer cells: implications for treatment response. Clin Cancer Res. 2005;11:2637–2643. doi: 10.1158/1078-0432.CCR-04-1462. [DOI] [PubMed] [Google Scholar]
- Cortes U, Moyret-Lalle C, Falette N, Duriez C, Ghissassi FE, Barnas C, Morel AP, Hainaut P, Magaud JP, Puisieux A. BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol Carcinog. 2000;27:57–64. doi: 10.1002/(SICI)1098-2744(200002)27:2<57::AID-MC1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D, Corrente G, Covone F, Micheli L, D'Agnano I, Starace G, Caruso M, Tirone F. Arrest of G(1)-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol Cell Biol. 2000;20:1797–1815. doi: 10.1128/MCB.20.5.1797-1815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim IK, Lee MS, Ryu MS, Park TJ, Fujiki H, Eguchi H, Paik WK. Induction of growth inhibition of 293 cells by downregulation of the cyclin E and cyclin-dependent kinase 4 proteins due to overexpression of TIS21. Mol Carcinog. 1998;23:25–35. doi: 10.1002/(SICI)1098-2744(199809)23:1<25::AID-MC4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Ryu MS, Lee MS, Hong JW, Hahn TR, Moon E, Lim IK. TIS21/BTG2/PC3 is expressed through PKC-delta pathway and inhibits binding of cyclin B1-Cdc2 and its activity, independent of p53 expression. Exp Cell Res. 2004;299:159–170. doi: 10.1016/j.yexcr.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Hong JW, Ryu MS, Lim IK. Phosphorylation of serine 147 of tis21/BTG2/pc3 by p-Erk1/2 induces Pin-1 binding in cytoplasm and cell death. J Biol Chem. 2005;280:21256–21263. doi: 10.1074/jbc.M500318200. [DOI] [PubMed] [Google Scholar]
- Tirone F. The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol. 2001;187:155–165. doi: 10.1002/jcp.1062. [DOI] [PubMed] [Google Scholar]
- Chen JG, Yang CP, Cammer M, Horwitz SB. Gene expression and mitotic exit induced by microtubule-stabilizing drugs. Cancer Res. 2003;63:7891–7899. [PubMed] [Google Scholar]
- Islaih M, Halstead BW, Kadura IA, Li B, Reid-Hubbard JL, Flick L, Altizer JL, Thom Deahl J, Monteith DK, Newton RK, Watson DE. Relationships between genomic, cell cycle, and mutagenic responses of TK6 cells exposed to DNA damaging chemicals. Mutat Res. 2005;578:100–116. doi: 10.1016/j.mrfmmm.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Lim YB, Park TJ, Lim IK. B cell translocation gene 2 enhances susceptibility of HeLa cells to doxorubicin-induced oxidative damage. J Biol Chem. 2008;283:33110–33118. doi: 10.1074/jbc.M804255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolari F, Appolloni I, Tutucci E, Caviglia S, Terrile M, Corte G, Malatesta P. Tumor progression and oncogene addiction in a PDGF-B-induced model of gliomagenesis. Neoplasia. 2008;10:1373–1382. doi: 10.1593/neo.08814. following 1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakubo H, Brachtel E, Hayashida T, Yeo G, Kish J, Muzikansky A, Walden PD, Maheswaran S. Loss of B-cell translocation gene-2 in estrogen receptor-positive breast carcinoma is associated with tumor grade and overexpression of cyclin d1 protein. Cancer Res. 2006;66:7075–7082. doi: 10.1158/0008-5472.CAN-06-0379. [DOI] [PubMed] [Google Scholar]
- Kaklamani VG, Sadim M, Hsi A, Offit K, Oddoux C, Ostrer H, Ahsan H, Pasche B, Mantzoros C. Variants of the adiponectin and adiponectin receptor 1 genes and breast cancer risk. Cancer Res. 2008;68:3178–3184. doi: 10.1158/0008-5472.CAN-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza A, Basso A, Black S, Malkowski M, Kwee L, Pachter JA, Lachowicz JE, Wang Y, Liu S. RNA interference targeting of A1 receptor-overexpressing breast carcinoma cells leads to diminished rates of cell proliferation and induction of apoptosis. Cancer Biol Ther. 2005;4:1355–1360. doi: 10.4161/cbt.4.12.2196. [DOI] [PubMed] [Google Scholar]
- Madjd Z, Durrant LG, Pinder SE, Ellis IO, Ronan J, Lewis S, Rushmere NK, Spendlove I. Do poor-prognosis breast tumours express membrane cofactor proteins (CD46)? Cancer Immunol Immunother. 2005;54:149–156. doi: 10.1007/s00262-004-0590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]