Abstract
The cytokine interleukin-6 (IL-6) plays key roles in the immune and inflammatory responses, acute-phase reaction and hematopoiesis. Such biological actions of IL-6 are characterised by both the activation and the inhibition of gene transcription. Unfortunately, in contrast to gene activation, the mechanism by which IL-6 suppresses transcription remains largely unclear. We have, therefore, investigated this aspect using the Xenopus laevis CCAAT/enhancer binding protein-α (C/EBPα) gene promoter as a model. We show by transient transfection assays of various promoter–luciferase DNA constructs into hepatoma cells that a C/EBP recognition sequence in the proximal promoter region is essential for the IL-6-mediated repression. Electrophoretic mobility shift assays showed that C/EBPα was the major protein that bound to this site and, consistent with its expression pattern, the binding was reduced when the cells were exposed to IL-6. Co-transfection assays revealed for the first time that the ability of C/EBPα, but not C/EBPβ or Sp1, to transactivate the promoter was decreased dramatically when the cells were incubated with IL-6. These studies, therefore, identify a novel mechanism for IL-6-mediated repression of gene transcription that involves a reduction in C/EBPα-mediated activation.
INTRODUCTION
Interleukin-6 (IL-6) is a pleiotropic cytokine that mediates a variety of functions in different tissues/cell types, including growth and differentiation of haematopoietic cells, proliferation of hepatocytes, mesangial cells and keratinocytes, regulation of acute-phase protein synthesis in the liver and osteoclast development (1–4). The cytokine can also act in a negative manner by inhibiting the proliferation or differentiation of some cell types and acting protectively in certain diseases by counteracting the manifestation of specific inflammatory responses (1–6). The levels of IL-6 are elevated in several pathophysiological conditions (e.g. acute-phase response, certain neurological conditions and pathogenic infections, multiple myelomas) and the cytokine has been implicated to play a key role in the pathogenesis of these diseases (1–3).
The actions of IL-6 are characterised by the transcriptional activation and suppression of numerous genes (1–10). The signal transduction pathways and the transcription factors that are involved in the IL-6-mediated induction of the expression of such genes have been deciphered in detail in the last few years (1–3). Thus, the cytokine activates gene transcription predominantly through the JAK–STAT (Janus kinase–signal transducer and activator of transcription) pathway (1–3). The activation of STATs in such cases is transient and several mechanisms have been identified for their inactivation, including dephosphorylation, proteasome degradation and feedback inhibition through the induction of suppressor-of-cytokine signalling (SOCS) proteins (1–3). Activation of such SOCS proteins has also been implicated in the inhibition of IL-6 signalling by certain extracellular mediators (11–13).
Despite the recent advances in our understanding of the mechanisms involved in the IL-6-mediated activation of gene transcription, very little is known about the intracellular signalling pathways and the transcription factors through which this cytokine directly suppresses gene expression. Further studies on this aspect are required in light of the potential importance of such regulation. For example, IL-6 inhibits the transcription of several genes during the acute phase response in the liver (4,7,8). Apart from gene-specific biological actions, such regulation is necessary to allow for an increased synthesis of positive acute-phase proteins without changing the overall rate of protein synthesis in the liver (4,7,8). In addition, the transcription of key genes that promote growth is decreased during the IL-6-mediated inhibition of cellular proliferation or activation of differentiation (1–3). Furthermore, the anti-inflammatory action of IL-6 in certain conditions involves the down-regulation of pro-inflammatory cytokine expression (5,6). It is, therefore, essential that further studies are carried out on the mechanisms by which IL-6 inhibits the expression of genes, particularly those that code for key transcription factors. CCAAT/enhancer binding protein-α (C/EBPα) falls in this category.
C/EBPα belongs to a family of transcription factors that all contain a highly conserved basic-leucine zipper (bZIP) domain at the C-terminus, which is required for DNA binding and subunit dimerisation (see 14 for a recent review). In contrast, the N-termini of these proteins, which carry the regulatory and trans-activation domains, are quite divergent (14). Six distinct C/EBP isoforms have been identified to date (C/EBPα to ζ) with the majority of these able to recognise similar DNA sequences, at least in vitro, activate gene transcription in vivo and form heterodimers in intrafamilial combinations (14). Additionally, in the case of C/EBPα and C/EBPβ, polypeptides of different sizes and trans-activating capabilities can be produced either by alternative use of translational initiation codons or by limited proteolysis (14). For example, two polypeptides of 42 (p42) and 30 kDa (p30) can be produced from the C/EBPα mRNA, with the latter having a lower trans-activation potential than the former (14). In the case of C/EBPβ, at least four isoforms can be produced: full-length (FL) C/EBPβ (38 kDa), 35 kDa LAP (liver-enriched transcriptional activating protein), 20 kDa LIP (liver-enriched transcriptional inhibitory protein) and a smaller 16 kDa isoform. Because the low molecular weight isoform, LIP, lacks most of the trans-activation domain but contains an intact DNA binding and dimerisation domain, it has been proposed to function as a dominant negative regulator of FL or LAP forms of C/EBPβ (14,15).
The function of C/EBPα has recently been investigated in detail using a number of approaches and has revealed key roles for the factor in the control of growth, differentiation and metabolism along with the responses mediated by cytokines, particularly in hepatocytes, adipocytes and myelomonocytic cells (14). On the other hand, studies aimed at understanding the mechanisms that are involved in the regulation of C/EBPα gene transcription have been limited to the action of thyroid hormone, inhibition by the proto-oncogene c-myc and autoregulation (16–23). Thus, the action of thyroid hormone is mediated via a responsive element present at position –602 to –589 of the rat C/EBPα promoter (16). The c-myc protein inhibits C/EBPα gene transcription through interaction with the core promoter region, although a discrepancy exists on the precise sequences that are required for the response (17,18). In relation to autoregulation, the mouse and the rat genes were first shown to be auto-activated through a C/EBP recognition sequence present in the proximal promoter region (19–21). In contrast, the promoter region of human C/EBPα lacks any C/EBP recognition sequence and autoregulation was shown to be mediated indirectly via a C/EBPα-mediated stimulation of upstream stimulatory factor (USF) binding to the proximal promoter region (22). More recently, we have characterised the Xenopus laevis C/EBPα (xC/EBPα) gene promoter and shown that, similar to the mouse and the rat counterpart, it is subject to direct autoregulation (23).
In relation to the inhibition of C/EBPα gene transcription, it was shown recently that the levels of the LIP form of C/EBPβ are elevated in the liver of mice following injection of lipopolysaccharide (LPS), which causes the release of numerous cytokines from a number of cell types, and after partial hepatectomy (24). Electrophoretic mobility shift assays (EMSAs) showed that this was accompanied by increased binding of the isoform to the C/EBP consensus site in the mouse C/EBPα promoter (24). Because a reduction of C/EBPα mRNA levels occurs in both these biological situations, it was proposed that this increased binding of the repressor LIP may represent a potential mechanism of action (24). Unfortunately, no data were provided in this study on the functional importance of the C/EBP site by demonstrating, for example, that mutations in this sequence abolish the response and/or that the site can confer the response to a heterologous promoter. This point is particularly important given that Baer and Johnson (25) have recently shown that truncated C/EBPβ isoforms can be generated by in vitro proteolysis during the isolation of tissue/cellular extracts, thereby suggesting that any findings on gene regulation based on LIP have to be interpreted with caution and need to be confirmed in vivo by functional analysis. It is, therefore, important that further studies are carried out on understanding the mechanisms involved in the inhibition of C/EBPα gene expression.
In the present study, we show that the xC/EBPα promoter activity was also inhibited by IL-6, thereby indicating that the cytokine responsiveness was conserved between mammalian and amphibian promoters. In addition, we show that a C/EBP recognition sequence in the proximal promoter region was absolutely essential for the response and that the site interacted with C/EBPα but not other members of the family. Furthermore, we demonstrate that the ability of C/EBPα, but not C/EBPβ or Sp1 (specificity protein 1), to transactivate the promoter was inhibited by IL-6. These studies, therefore, provide novel insights into the molecular mechanisms by which IL-6 inhibits gene transcription.
MATERIALS AND METHODS
Reagents
The human hepatoma Hep3B cell line was from the European Collection of Animal Cell Cultures whereas recombinant IL-6 was from Peprotech. All the cell culture reagents and plasticware were purchased from Greiner, Helena Biosciences or Gibco/BRL. Antiserum against Sp1 and the different C/EBP isoforms was mainly from Santacruz. The Sp1 expression plasmid in the vector pEVR2 was obtained from Prof. Guntram Suske.
Cell culture and transient transfection assays
The Hep3B cell line was grown in DMEM with Stabilix™ (special l-glutamine stabiliser) supplemented with 10% (v/v) heat-inactivated fetal calf serum (1 h, 56°C) (HI-FCS), penicillin (100 U/ml) and streptomycin (100 µg/ml). The cultures were maintained at 37°C in a humid incubator with a 5% (v/v) CO2 atmosphere. DNA transfections were carried out by the calcium phosphate precipitation method (23,26) and utilised 2 µg promoter–luciferase DNA construct, 2–4 µg of expression construct (co-transfection assays only) and 0.5 µg of cytomegalovirus (CMV)-β-galactosidase plasmid to provide an internal control for transfection efficiency. After 6 h, the cells were washed with phosphate-buffered saline and left for 36 h in fresh culture medium alone or in the presence of 1000 U/ml IL-6. The luciferase and the β-galactosidase activity in cell extracts was then determined using commercially available kits (Promega). The luciferase activity was normalised to the β-galactosidase value and each transfection was repeated at least three times.
Preparation of manipulated C/EBPα promoter–reporter DNA constructs
The preparation of the –1131/+41, –321/+41 and –121/+41 xC/EBPα promoter deletion constructs in the pGL2-Basic vector and the pC/EBPx4 and pSp1x4 constructs in the pGL2-Promoter vector has been described previously (23). For the preparation of the –204/+41 xC/EBPα promoter construct, the corresponding region was amplified from the –321/+41 DNA construct by PCR using the high fidelity pwo DNA polymerase and the following two primers: 5′-TATCGGTACCTAACA CGCAC-3′ and 5′-CTGCAAGCTTCCTAGCTGTC-3′ (23). These primers were designed to allow directional cloning of the amplification product into the KpnI and the HindIII site of the pGL2-Basic vector (23). The DNA construct containing mutation in the C/EBP site in the –321/+41 context [5′-TCTGGAAAC-3′ (23) to 5′-TCTGGATCC-3′] was prepared using the Gene Editor™ in vitro site-directed mutagenesis system (Promega) and the following primer: 5′-GACAGC GGAGTGGATCCAGACAATGTAACGCCGG-3′. The sequence of all the constructs was verified before use in transfection experiments.
EMSAs
Nuclear and whole cell extracts were prepared essentially as described by Ramji et al. (27) and Timchenko et al. (22), respectively. The protease inhibitors (10 µg/ml aprotinin, 0.5 mM PMSF, 2 mM benzamidine, 10 µg/ml leupeptin, 1 mg/ml pepstatin A) and DTT (0.5 mM) were added to all the buffers before use. The concentration of proteins in the nuclear extracts was determined using the BCA protein assay kit as described by the manufacturer (Pierce).
For EMSA, 20 µg of whole cell extracts or 1–4 µg of nuclear extracts were incubated in a 20 µl total reaction volume containing 34 mM potassium chloride, 5 mM mag nesium chloride, 0.1 mM DTT and 3 µg poly(dI-dC). After 10 min on ice, 32P-labelled probes (50 000–100 000 c.p.m.) were added and the incubation continued for 20 min at room temperature. Following the addition of 7 µl of 20% (w/v) Ficoll solution to each sample, the DNA–protein complexes and the free probe were separated by electrophoresis on 6% (w/v) polyacrylamide gels in 0.5× TBE buffer. The gels were dried under vacuum and exposed to X-ray film. For antibody supershift assays, samples of the extracts were incubated for 30 min on ice prior to the addition of the radiolabelled probe. The sequences of the oligonucleotides used are shown below. These were radiolabelled by ‘fill-in’ reactions using [α-32P]dCTP and Klenow DNA polymerase.
xC/EBP: 5′-CAGTGTTTCCAGAC-3′ and 5′-TTGGTCT GGAAACA-3′
mC/EBP: 5′-AGTCAGTGGGCGTTGCGCCACGATCT-3′ and 5′-TCAGTAGATCGTGGCGCAACGCCCACT-3′
xSp1: 5′-TAGCAGGGGCGTGGC-3′ and 5′-GTGGCCA CGCCCCT-3′
Sp1: 5′-TAGATTCGATCGGGGCGGGGCGAG-3′ and 5′-GCCCTCGCCCCGCCCCGATCGAAT-3′
CRP: 5′-GGGGCATAGTGGCGCAAACTCCCTTACTG-3′ and 5′-GGGGCAGTAAGGGAGTTTGCGCCACTATG-3′
β-casein: 5′-GCAGAATTTCTTGGGAAAGAAAA-3′ and 5′-GCTATTTTCTTTCCCAAGAAATT-3′
xNF-1: 5′-GCCTTGGCATTA-3′ and 5′-GCTAATGC CAAG-3′
Reverse transcription–polymerase chain reaction (RT–PCR)
Total cellular RNA was prepared using the RNeasy™ total RNA isolation kit (Qiagen) according to the instructions from the manufacturer. Each isolated RNA sample (1 µg) was then subjected to RT–PCR essentially as described before (28,29), except that the total number of cycles and the annealing temperature varied: 19 cycles and 63°C for C/EBPα and 16 cycles and 60°C for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). These conditions were in the exponential phase of amplification and, therefore, provided a direct correlation between the amount of products and RNA template abundance in the samples. The sequences of the primers were 5′-ACTTGGTGCGTCTAAGATGAGG-3′ and 5′-AGAGACC TCCACCTTCATGTAG-3′ for C/EBPα and 5′-CCCTTC ATTGACCTCAACTACATGG-3′ and 5′-AGTCTTCTGGG TGGCAGTGATGG-3′ for GAPDH (28). The PCR products were size-fractionated on a 2% (w/v) agarose gel, photographed using a Syngene gel documentation system (GRI), converted to an uncompressed TIFF file format, and quantified using the Quantiscan computer package (Biosoft).
Western blot analysis
Samples (10–40 µg of whole cell extracts or 10 µg of nuclear extracts) were size-fractionated under reducing conditions using 10–15% polyacrylamide gels containing SDS, and then transferred to Immobilon-P PVDF membranes (Millipore) by blotting (30). The blotted membranes were incubated first for 1 h at room temperature in blocking solution [1× Tris buffered saline (TBS) containing 5% (w/v) non-fat milk powder and 0.1% (v/v) Tween-20] in order to reduce any non-specific interaction of the antiserum with the membrane. Following washing for 15 min in 1× TBS containing 0.1% (v/v) Tween-20, the membrane was incubated with the primary antibody for 1 h in 1× TBS containing 5% (w/v) non-fat milk powder and 0.1% (v/v) Tween-20. After washes as above, the membrane was incubated with secondary horseradish peroxidase (HRP)-conjugated anti-rabbit antibody for 30 min at room temperature in 1× TBS containing 5% (w/v) non-fat milk powder and 0.1% (v/v) Tween-20. After washing once more as above, the membranes were developed using an enhanced chemiluminescence detection kit (Amersham) and XAR sensitive film (Kodak). The sizes of the proteins were determined by comparison with Rainbow molecular weight markers (Amersham) that had been subjected to electrophoresis on the same gel as the test samples.
RESULTS
IL-6 reduces the steady-state levels of C/EBPα mRNA and polypeptides in Hep3B cells
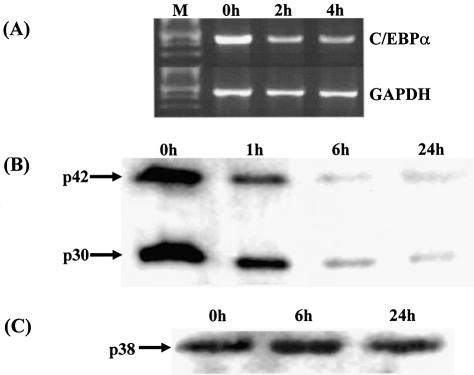
The human hepatoma Hep3B cell line is an extensively used model system for investigating the mechanisms that are involved in the IL-6-mediated regulation of gene transcription, with demonstrated similarities to what is seen in vivo (4,31). We have previously investigated the effect of IL-6 on C/EBPβ and C/EBPδ expression in Hep3B cells (27). However, the action of IL-6 on the expression of C/EBPα in these cells has not yet been determined. This was, therefore, investigated by both semiquantitative RT–PCR and western blot analysis using antisera against C/EBPα, which detects both the p42 and the p30 isoforms. As shown in Figure 1A, addition of IL-6 to the cells resulted in an immediate and marked decrease in C/EBPα mRNA expression. In addition, there was a time-dependent decrease in the steady-state levels of both the p42 and p30 polypeptides (Fig. 1B). Such a decrease was specific to C/EBPα and not seen when western blot analysis was carried out with representative samples from the same experiment and probed with an antiserum against C/EBPβ, which predominantly recognises the p38 polypeptide (27) (Fig. 1C). Thus, the previously noted IL-6-mediated reduction of C/EBPα expression in the liver (32,33) was also seen in Hep3B cells.
Figure 1.
The steady-state levels of C/EBPα mRNA and polypeptides are reduced in Hep3B cells following exposure to IL-6. (A) Hep3B cells were stimulated with IL-6 for the indicated period of time and total RNA was isolated and subjected to RT–PCR using primers for C/EBPα and GAPDH. The amplification products were size-fractionated by agarose gel electrophoresis along with molecular size markers (M). (B) and (C) Western blot analysis was carried out using whole cell extracts from Hep3B cells exposed to IL-6 for the indicated time and probed with antiserum against either C/EBPα, which recognises both the p42 and p30 isoforms (B) or C/EBPβ, which detects predominantly the p38 form (C).
The activity of the Xenopus laevis C/EBPα gene promoter in hepatocytes is decreased by IL-6
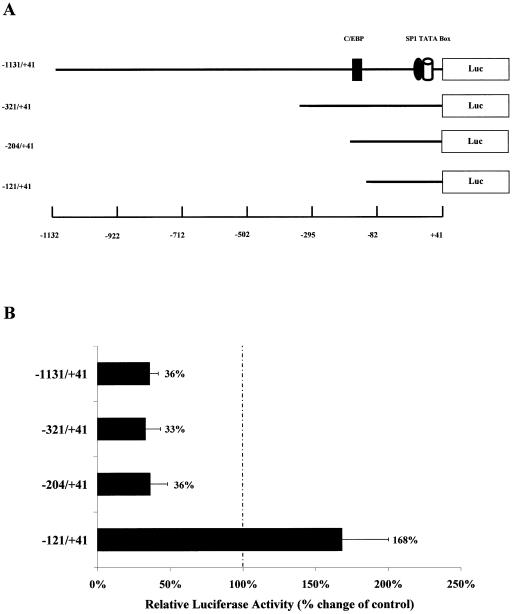
Previous studies have shown that the expression of C/EBPα is decreased in the liver and a number of other tissues during the acute phase response, which is mainly mediated via IL-6, and that this effect occurs at the transcriptional level (14,34,35). In addition, IL-6 has been shown to inhibit C/EBPα gene expression in vivo and in vitro (14,32,33 and Fig. 1). However, the action of this cytokine on the activity of the corresponding promoter has, as yet, not been determined. We have previously characterised the X.laevis C/EBPα (xC/EBPα) gene promoter in detail (23). We, therefore, first evaluated its regulation by IL-6 using a luciferase-based construct containing the largest promoter fragment (–1131/+41) (Fig. 2A) via transient transfection assays in Hep3B cells (23). These cells have been used extensively for the analysis of promoter elements that are involved in both the constitutive and regulated expression of genes in hepatocytes from different species, including the C/EBP family (4,31). Indeed, we have shown previously that the activity of the C/EBPβ and C/EBPδ gene promoters in these cells is increased following exposure to IL-6 (36,37). In contrast to these findings, the activity of the –1131/+41 xC/EBPα promoter–luciferase DNA construct was found to be reduced by ∼64% when Hep3B cells were incubated with IL-6 (Fig. 2B). This profile was, therefore, consistent with that seen at the level of gene expression and demonstrates that the –1131/+41 region of the xC/EBPα gene promoter contains sufficient information for the IL-6 response.
Figure 2.
Identification of the minimal IL-6SEs in the xC/EBPα gene promoter. (A) Schematic representation of the xC/EBPα promoter–luciferase DNA constructs. The relative position of the putative TATA box and binding sites for the transcription factors C/EBP and Sp1 are shown (see text for details). (B) Transient transfection assays with the indicated xC/EBPα promoter–luciferase DNA constructs were carried out as described in Materials and Methods. The transfected cells were then either left untreated or exposed to IL-6 for 36 h. The luciferase activity was normalised to the β-galactosidase activity. For each DNA construct, the normalised luciferase activity in unstimulated cells has been arbitrarily assigned as 100%, with that from IL-6-treated cells being represented as a percentage of this value (indicated to the right of each histogram). Each value is the average of four independent experiments.
As part of our other studies, we have produced a number of DNA constructs containing specific deletions or mutations of the xC/EBPα gene promoter (23). We, therefore, decided to use these DNA constructs to rapidly delineate the minimal regulatory sequence(s) required for the action of IL-6. Thus, three further DNA constructs, containing 5′ truncations of the xC/EBPα gene promoter in the pGL2-Basic vector (–321/+41, –204/+41 and –121/+41) (Fig. 2A), were transfected into the Hep3B cell line and the reporter gene activity in cells that were either left untreated or exposed to IL-6 was determined. As shown in Figure 2B, an IL-6-dependent reduction in reporter gene activity of 64–67% was obtained with DNA constructs containing the –321/+41 and the –204/+41 region. In contrast, a further deletion to –121 resulted in a loss of the IL-6-mediated suppression in promoter activity (Fig. 2B). These results, therefore, indicate that the minimal IL-6 suppressive elements (IL-6SEs) reside between the –204 and the –121 region of the promoter.
The C/EBP site in the promoter is responsible for the IL-6 response
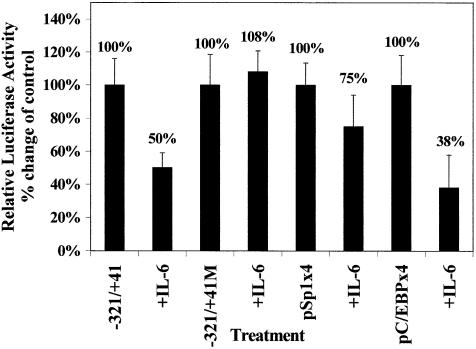
The –204 to the –121 region of the xC/EBPα promoter contains a binding site for the C/EBPs, and we have previously shown that this site is essential for auto-activation (23). As autoregulation is a critical control mechanism in the activation of the C/EBPs, including C/EBPα (14), we hypothesised that the C/EBP site in the xC/EBPα gene promoter may also play a crucial role in the action of IL-6. If this were the case then mutation of the C/EBP site should prevent the IL-6-mediated reduction in reporter gene activity and multiple copies of this site, but not for any other transcription factors present downstream of position –121, should be able to confer the IL-6 response to a heterologous promoter. We thus mutated the C/EBP site in the –321/+41 context and carried out transient transfection assays in Hep3B cells. As expected from the requirement of the C/EBP site for autoregulation (23), the mutation reduced the basal activity (i.e. in the absence of IL-6) by ∼77% (data not shown). In addition, the IL-6-mediated decrease in the activity seen with the –321/+41 promoter region was abolished (Fig. 3A). In order to further confirm the importance of the C/EBP recognition sequence, DNA constructs containing four copies of the C/EBP site (–171/–163) and the Sp1 site (control: –43/–34) linked to the minimal SV40 promoter (designated as pC/EBPx4 and pSp1x4, respectively) (23) were transfected into Hep3B cells and the reporter gene activity in untreated cells and those exposed to IL-6 was determined. The values obtained using the pGL2-Promoter vector were subtracted from those with the pC/EBPx4 and pSp1x4 constructs. As shown in Figure 3, exposure of the cells to IL-6 resulted in a dramatic decrease in the reporter gene activity of the pC/EBPx4 construct by ∼62% (Fig. 2B). In contrast, only a marginal reduction in reporter gene activity was seen with the pSp1x4 construct (Fig. 3). These experiments, therefore, show that the C/EBP site in the xC/EBPα gene promoter plays a crucial role in the IL-6 response.
Figure 3.
Analysis of the action of IL-6 on the activity of DNA constructs containing mutations of the C/EBP site or multimers of the C/EBP and Sp1 sites from the xC/EBPα promoter. Transient transfection assays were carried out in Hep3B cells using the intact –321/+41 DNA construct (–321/+41) along with that containing mutations in the C/EBP recognition sequence (–321/+41M), and the constructs pC/EBPx4 and pSp1x4. The relative luciferase activity in the absence or presence of IL-6 was determined as described in Materials and Methods. The normalised luciferase activity in unstimulated cells has arbitrarily been assigned as 100% with that from IL-6-treated cells (+IL-6) shown with respect to this value (indicated above each histogram). Each value is the average of four independent experiments.
The binding of proteins to the C/EBP recognition sequence is affected by IL-6
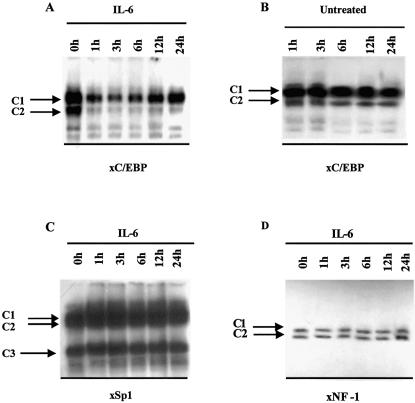
Having established that the C/EBP recognition sequence in the xC/EBPα gene promoter is crucial for the IL-6 response, we next investigated the interaction of DNA binding proteins with this site by EMSA. Oligonucleotides corresponding to the Sp1 and NF-1 sites in the xC/EBPα gene promoter were also included for comparative purposes. The radiolabelled oligonucleotides were incubated with whole cell extracts from Hep3B cells that were either left untreated or stimulated with IL-6 for various timed periods. With the C/EBP binding site oligonucleotide, two major DNA–protein complexes were seen reproducibly in independent experiments (designated as C1 and C2 in Fig. 4A). The signal intensity from both complexes was high when extracts from untreated Hep3B cells were used and decreased dramatically when those from cells stimulated with IL-6 for 1 h were employed. The signal from complex C2 then remained about equal throughout the 24 h incubation period with IL-6. On the other hand, the signal from complex C1 remained constant until the 6 h point and then increased marginally. A similar profile was seen when nuclear extracts were used instead of whole cell extracts (data not shown). Such a decrease in DNA binding was specific to IL-6 since it was not seen when extracts from untreated Hep3B cells at the various time points were used for EMSA (Fig. 4B). In addition, the IL-6 action was specific to the C/EBP site and not seen with an Sp1 or NF-1 binding site oligonucleotide (Fig. 4C and D, respectively).
Figure 4.
Analysis of the binding of factors to the C/EBP, Sp1 and NF-1 sites in the xC/EBPα promoter. EMSAs were carried out using whole cell extracts from cells that were exposed to IL-6 for different timed intervals (A, C and D) or left untreated for the same time points (B) and radiolabelled oligonucleotides corresponding to the C/EBP, Sp1 or NF-1 binding sites from the xC/EBPα promoter. The various DNA–protein complexes are shown by labelled arrows. The free probe has migrated off the gel. The data are representative of two independent experiments.
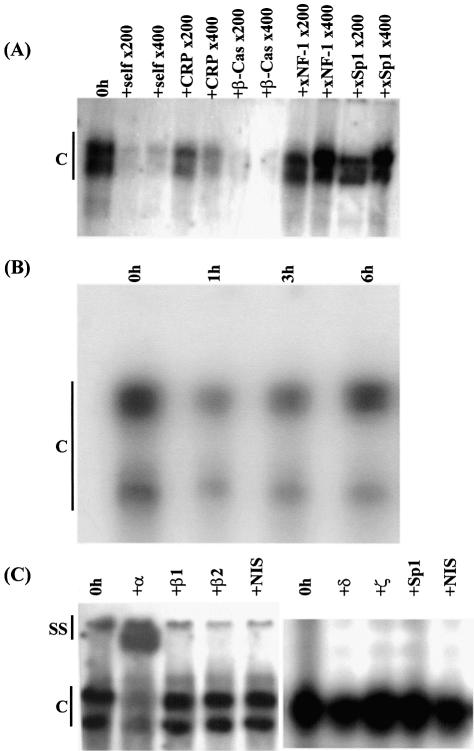
The specificity of the DNA–protein interactions seen in all the EMSA experiments was confirmed by competition assays. First, the binding of proteins to the C/EBP site with extracts from untreated cells was competed out by an excess of the corresponding sequence (self) or the C/EBP recognition sequence from the promoter of the C-reactive protein (CRP) or the β-casein genes but not by an NF-1 or Sp1 binding site oligonucleotide (Fig. 5A). Only a partial inhibition was obtained with the C/EBP recognition sequence from the CRP gene promoter because of its lower affinity for the C/EBPs compared to the site from the β-casein gene promoter (29). Secondly, the interaction of proteins with the Sp1 binding site oligonucleotide could be competed out with an excess of the corresponding sequence and a consensus Sp1 binding site oligonucleotide, but not by an oligonucleotide containing the C/EBP recognition sequence from the β-casein gene promoter or the NF-1 binding site (data not shown). Finally, the binding to the NF-1 binding site oligonucleotide could be competed out by the corresponding sequence but not by that containing the Sp1 recognition sequence (data not shown).
Figure 5.
Analysis of the binding of the C/EBPs to the recognition sequence in the C/EBPα gene promoter. (A) Competition EMSA to verify the specificity of the binding of the C/EBPs to recognition sequence in the xC/EBPα gene promoter. The analysis was carried out using whole cell extracts from untreated Hep3B cells (0 h) and radiolabelled oligonucleotides corresponding to the C/EBP binding site from the xC/EBPα gene promoter. Competitor oligonucleotides were added at 200- and 400-fold molar excess, as indicated, and included the corresponding sequence (self), the C/EBP recognition sequence from the promoters of CRP and β-casein (β-Cas), and the NF-1 and Sp1 sites from the xC/EBPα promoter (xNF-1 and xSp1, respectively). (B) Time-course profile of protein binding to the C/EBP recognition sequence from the mouse C/EBPα gene promoter. EMSAs were carried out using whole cell extracts from Hep3B cells that were treated with IL-6 for the indicated time. (C) Antibody supershift experiments using the C/EBP binding site oligonucleotide. EMSAs were carried out using the C/EBP binding site oligonucleotide from the xC/EBPα gene promoter and whole cell extracts from untreated cells (0 h) in the absence or presence of antisera against C/EBP-α (+α), -β (two different preparations; β1 and β2), -δ (+δ), -ζ (+ζ), Sp1 (+Sp1) and the non-immune serum (NIS). The two closely migrating DNA–protein complexes seen for data shown on the left side of the figure have not separated as efficiently in the case of the gel for the result shown on the right side. Vertical lines labelled C (all panels) and SS (in C only) indicate DNA–protein complexes and antibody–DNA–protein supershift complex, respectively (the free probe has migrated off the gel).
The binding of proteins to the C/EBP recognition sequence in the mouse C/EBPα gene promoter is also decreased following exposure of the cells to IL-6
The mouse C/EBPα gene promoter also contains a C/EBP recognition sequence that is essential for autoregulation (19,20). We therefore wondered whether the IL-6-dependent reduction in the binding of proteins to the C/EBP recognition sequence in the xC/EBPα gene promoter could also be seen with the site from the mouse promoter. This possibility was investigated using extracts from Hep3B cells that were either left untreated or exposed to IL-6 for representative time points. As shown in Figure 5B, two major DNA–protein complexes were obtained when extracts from untreated cells were used. The signals from both these complexes were reduced markedly when extracts from cells incubated with IL-6 for 1 h were used and then either remained constant or increased marginally.
C/EBPα is the predominant protein that interacts with the C/EBP recognition sequence
We next decided to employ supershift assays to identify the nature of the proteins that were responsible for the formation of the DNA–protein complexes with the C/EBP binding site oligonucleotide, the intensity of which decreases within 1 h of incubation of the cells with IL-6 (see Fig. 4A). For these assays, extracts were used from untreated cells and antisera against C/EBP-α, -β (two different sources), -δ and -ζ. Antiserum against C/EBPζ was included because it can act as a repressor of gene transcription mediated by the activating forms of the C/EBPs (14). In addition, the non-immune serum and antiserum against Sp1 were included as controls. As shown in Figure 5C, an inhibition of the formation of the DNA–protein complexes and the appearance of a slower migrating antibody–DNA–protein supershift complex was only obtained with antisera against C/EBPα. These results therefore indicate that the DNA–protein complexes were composed predominantly of C/EBPα, and rule out a potential involvement of the inhibitory LIP isoform of C/EBPβ or the repressor C/EBPζ in the IL-6 response.
The ability of C/EBPα to activate the transcription of its own gene is inhibited by IL-6
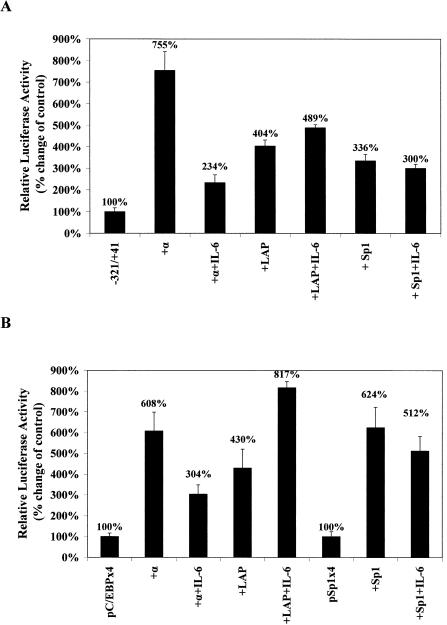
Given that the action of IL-6 was unlikely to be mediated through the inhibitory forms of C/EBP isoforms, we wondered whether the ability of C/EBPα to activate its own promoter was affected by IL-6. This was investigated by co-transfection experiments using a CMV-based pCS2+ vector expressing the Xenopus C/EBPα (xC/EBPα) gene (23) and the luciferase-based plasmid containing the –321/+41 region of the xC/EBPα gene promoter. Similar experiments with the pCS2+ vector expressing the LAP form of C/EBPβ (23) and CMV-based expression plasmid specifying for Sp1 were included for comparative purposes. As shown in Figure 6A, co-transfection of xC/EBPα produced an ∼7.5-fold increase in promoter activity. Exposure of the cells to IL-6 resulted in a dramatic reduction in this activity, which was only ∼30% of the activity obtained with the expression plasmid in the absence of the cytokine. Such a dramatic reduction of promoter activity in the presence of IL-6 was specific to C/EBPα. In the case of the LAP form of C/EBPβ, a 4-fold induction in the promoter activity was seen, which remained unchanged in the presence of IL-6. Finally, although the Sp1 expression plasmid produced a 3-fold increase in promoter activity, no significant changes were seen in the presence of IL-6.
Figure 6.
Investigation of the action of IL-6 on the activation of xC/EBPα promoter activity by different expression plasmids. Transfection experiments were carried out in Hep3B cells using the promoter construct –321/+41 (A), pC/EBPx4 or pSp1x4 (B) and either the parent pCS2+ plasmid [–321/+41 in (A) and pC/EBPx4 or pSp1x4 in (B)] or expression plasmids specifying for C/EBPα (+α), the LAP form of C/EBPβ (+LAP) or Sp1 (+Sp1). The luciferase activity was normalised to the β-galactosidase activity, and the values obtained using the pGL2-Basic or pGL2-Promoter vectors as negative control were subtracted from these. +IL-6 indicates that the values have been derived from cells that have been treated with this cytokine. The normalised luciferase activity in cells co-transfected with the pCS2+ plasmid alone has been arbitrarily assigned as 100% with the others being represented with respect to this value. Each value is the average of three independent experiments.
In order to investigate whether the IL-6-mediated reduction in the ability of C/EBPα to trans-activate could also be seen with multiple copies of the C/EBP site linked to the heterologous SV40 promoter, the experiment was repeated using the pC/EBPx4 construct. A similar experiment using the pSp1x4 construct and the Sp1 expression plasmid was also included for comparative purposes. As shown in Figure 6B, co-transfection of both C/EBP isoforms produced an increase in reporter gene activity. For C/EBPα this activity was reduced in the presence of IL-6. In contrast, the activation seen by the expression plasmid coding for the LAP form of xC/EBPβ was augmented by IL-6. Finally, no significant IL-6-dependent changes in the action of the Sp1 expression plasmid was seen when the experiment was carried out using the pSp1x4 DNA construct (Fig. 6B).
DISCUSSION
The expression of the C/EBPα gene has been shown to be inhibited during several pathophysiological conditions, including the acute-phase response, liver regeneration, diabetes mellitus and hepatocellular carcinomas (14,21,32–35,38–40). These diseases are characterised by perturbations in the local and circulating levels of several cytokines (41) including IL-6, a major inhibitor of C/EBPα gene transcription (14,32,33,35). However, no studies have yet been carried out that have investigated the mechanisms by which IL-6 suppresses the expression of this gene. This statement is also generally applicable to the majority of genes whose expression is inhibited by this cytokine. We have identified here a novel mechanism for the action of this cytokine on C/EBPα gene transcription that involves an inhibition of autoregulation via a reduction in the ability of the factor to activate the proximal promoter region.
As far as the suppression of gene transcription by the C/EBPs is concerned, two isoforms have been found to play a prominent role: the LIP form of C/EBPβ and C/EBPζ (14). For example, during adipocyte differentiation, C/EBPζ has been shown to interact with, and thereby repress, the action of other C/EBP isoforms by heterodimerisation, thereby delaying the activation of C/EBPα during adipogenesis until the mitotic clonal expansion is initiated (42). In addition, during the LPS-induced acute-phase response in the liver and following partial hepatectomy, the expression of LIP has been found to be induced and this is associated with its increased binding to the C/EBP consensus site found within the mouse C/EBPα promoter in vitro (24). It has been postulated, therefore, that this might in turn be responsible for the inhibition of C/EBPα promoter activity (24). In contrast to this study, EMSA using extracts from IL-6-treated Hep3B cells during periods when the expression of the C/EBPα gene was decreasing (Fig. 1) showed that there was a cytokine-dependent reduction in the binding of factors to the site and that C/EBPα was the major isoform that was engaged in DNA–protein interactions (Figs 4 and 5). A major limitation of the previous study, which implicated LIP in the suppression of mouse C/EBPα gene transcription during the LPS-induced acute phase response and partial hepatectomy, was that it relied mainly on DNA–protein interaction studies, and thereby lacked functional data on the importance of the C/EBP recognition sequences in the responses (24). Thus, studies were not carried out to investigate whether mutations of the C/EBP site curtail the responses and/or if multimers of the site confer the responses to a heterologous promoter. This point is particularly important because it has been shown that the production of LIP can be an artefact of the conditions of cell culture or preparation of extract (25). It has been suggested, therefore, that the presence of LIP in cell extracts must be interpreted with caution and the in vivo relevance of these isoforms should be re-evaluated (25). An additional limitation of models based on LIP is that most systems where increased expression of LIP has been noted (e.g. LPS-induced acute-phase response) are also characterised by the activation and/or induced expression of FL C/EBPβ, and this has been implicated in the transcriptional activation of numerous genes (14,32,33,35). It is difficult, therefore, to explain why LIP does not affect the expression of these genes.
Transcription factors can be regulated at multiple levels, including changes in expression, nucleo-cytoplasmic transport, DNA binding activity and ability to trans-activate (43). Although C/EBP isoforms can also be regulated at multiple levels (14), regulation of their ability to trans-activate promoters has so far been shown for only C/EBPβ (44–47). For example, the ability of C/EBPβ to trans-activate has been shown to be stimulated by agents that activate calcium-calmodulin-dependent protein kinases, protein kinase C and MAP kinases via phosphorylation of Ser276, Ser105 and The235, respectively (44–46). More importantly, IL-6 has been shown to induce the activation potential of C/EBPβ probably via activation of MAP kinases (14,46,47). Consistent with this finding, the ability of C/EBPβ to activate the pC/EBPx4 construct in this study was also found to increase in response to IL-6 (Fig. 6B). However, we have also been able to show for the first time that the ability of C/EBPα to trans-activate specific promoters is also regulated by IL-6, and this is responsible for the reduction of its own transcription by the cytokine. The precise mechanism by which this is achieved remains to be determined but could be triggered by phosphorylation of the protein, as for C/EBPβ (44–46), limited availability of co-activators shared by factors whose expression is induced by IL-6 (e.g. C/EBPδ, JunB) (14,48) or cytokine-mediated interactions with specific co-repressors.
IL-6 is known to affect the action of three members of the C/EBP family: increasing the expression of C/EBP-β and -δ (14,27,32–35), inducing the trans-activation potential of C/EBPβ (14,46,47) and reducing the expression of C/EBPα and its ability to trans-activate specific promoters (14,32,33 and this study). An important question that emerges is how IL-6 differentially regulates the expression of genes whose promoter regions contain C/EBP recognition sequences. There are several potential ways in which such specificity could be achieved. For example, although no differences have yet been noted in the binding site preference of these three C/EBP isoforms in vitro (14), it is possible that selectivity may occur in vivo, and there may be an additional modulation of this along with the affinity of binding by specific regulatory proteins. It is also possible that the action of the C/EBPs on gene promoters containing the cognate recognition sequence is modulated by factors that bind to other sites. This point is particularly important for genes that are activated by IL-6 since the promoter regions of virtually all such genes contain binding sites for STAT-3, which is primarily responsible for the induction of transcription (2). As the activation of STAT-3 by IL-6 is transient, it is likely that C/EBP-β and -δ may be involved in ensuring prolonged activation (14).
In conclusion, we have identified a novel mechanism for the IL-6-dependent inhibition of cellular gene transcription, which involves a cytokine-mediated reduction in the ability of the protein to activate the proximal promoter region. Future studies will investigate the mechanisms by which such an action of IL-6 is achieved.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. Guntram Suske for the Sp1 expression plasmid and the Biotechnology and Biological Sciences Research Council (BBSRC) for financial support.
REFERENCES
- 1.Akira S. (1997) IL-6-regulated transcription factors. Int. J. Biochem. Cell Biol., 29, 1401–1418. [DOI] [PubMed] [Google Scholar]
- 2.Heinrich P.C., Behrmann,I., Muller-Newen,G., Schaper,F. and Graeve,L. (1998) Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J., 334, 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirano T. (1998) Signal transduction through cytokine receptors. Int. Rev. Immunol., 16, 249–284. [DOI] [PubMed] [Google Scholar]
- 4.Ramji D.P., Cortese,R. and Ciliberto,G. (1993) Regulation of C-reactive protein, haptoglobin and hemopexin gene expression. In Mackiewicz,A., Kushner,M.D. and Baumann,H. (eds), Acute Phase Proteins: Molecular Biology, Biochemistry and Clinical Applications. CRC Press, Boca Raton, FL, pp. 162–242. [Google Scholar]
- 5.Jones S.A., Horiuchi,S., Topley,N., Yamamoto,N. and Fuller,G.M. (2001) The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB J., 15, 43–58. [DOI] [PubMed] [Google Scholar]
- 6.Xing Z., Gauldie,J., Cox,G., Baumann,H., Jordana,M., Lei,X.F. and Archong,M.K. (1998) IL-6 is an antiinflammatory cytokine required for controlling local or systemic acute inflammatory responses. J. Clin. Invest., 101, 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aldred A.R. and Schreibler,G. (1996) The negative acute phase proteins. In Mackiewicz,A., Kushner,M.D. and Baumann,H. (eds), Acute Phase Proteins. CRC Press, Boca Raton, FL, pp. 21–40. [Google Scholar]
- 8.Citarella F., Felici,A., Brouwer,M., Wagstaff,J., Fantoni,A. and Hack,C.E. (1997) Interleukin-6 downregulates factor XII production by human hepatoma cell line (HepG2). Blood, 90, 1501–1507. [PubMed] [Google Scholar]
- 9.Ferry A.E., Baliga,S.B., Monteiro,C. and Pace,B.S. (1997) Globin gene silencing in primary erythroid cultures. An inhibitory role for interleukin-6. J. Biol. Chem., 272, 20030–20037. [DOI] [PubMed] [Google Scholar]
- 10.Liao H.S., Matsumoto,A., Itakura,H., Doi,T., Honda,M., Kodama,T. and Geng,Y.J. (1999) Transcriptional inhibition by interleukin-6 of the class A macrophage scavenger receptor in macrophages derived from peripheral monocytes and the THP-1 monocytic cell line. Arterioscler. Thromb. Vasc. Biol., 19, 1872–1880. [DOI] [PubMed] [Google Scholar]
- 11.Terstegen L., Maassen,B.G., Radtke,S., Behrmann,I., Schaper,F., Heinrich,P.C., Graeve,L. and Gatsios,P. (2000) Differential inhibition of IL-6-type cytokine-induced STAT activation by PMA. FEBS Lett., 478, 100–104. [DOI] [PubMed] [Google Scholar]
- 12.Sengupta T.K., Schmitt,E.M. and Ivashkiv,L.B. (1996) Inhibition of cytokines and JAK-STAT activation by distinct signaling pathways. Proc. Natl Acad. Sci. USA, 93, 9499–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bode J.G., Nimmesgern,A., Schmitz,J., Schaper,F., Schmitt,M., Frisch,W., Haussinger,D., Heinrich,P.C. and Graeve,L. (1999) LPS and TNF-α induce SOCS3 mRNA and inhibit IL-6-induced activation of STAT3 in macrophages. FEBS Lett., 463, 365–370. [DOI] [PubMed] [Google Scholar]
- 14.Ramji D.P. and Foka,P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J., 365, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Descombes P. and Schibler,U. (1993) A liver-enriched protein, LAP and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell, 67, 569–579. [DOI] [PubMed] [Google Scholar]
- 16.Menendez-Hurtado A., Santos,A. and Perez-Castillo,A. (2000) Characterization of the promoter region of the rat CCAAT/enhancer-binding protein-α gene and regulation by thyroid hormone in rat immortalized brown adipocytes. Endocrinology, 141, 4164–4170. [DOI] [PubMed] [Google Scholar]
- 17.Antonson P., Pray,M.G., Jacobsson,A. and Xanthopoulos,K.G. (1995) Myc inhibits CCAAT/enhancer-binding protein-α gene expression in HIB-1B hibernoma cells through interactions with the core promoter regions. Eur. J. Biochem., 232, 397–403. [DOI] [PubMed] [Google Scholar]
- 18.Li L.H., Nerlov,C., Prendergast,G., MacGregor,D. and Ziff,E.B. (1994) c-myc represses transcription in vivo by a novel mechanism dependent on the initiator element and myc box II. EMBO J., 13, 4070–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christy R.J., Haestner,K.H., Geiman,D.E. and Lane,M.D. (1991) CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc. Natl Acad. Sci. USA, 88, 2593–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legraverend C., Antonson,P., Flodby,P. and Xanthopoulos,K.G. (1993) High level activity of the mouse CCAAT/enhancer binding protein (C/EBPα) gene promoter involves autoregulation and several ubiquitous transcription factors. Nucleic Acids Res., 21, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rana B., Xie,Y., Mischoulon,D., Bucher,N.L.R. and Farmer,S.R. (1995) The DNA binding activity of C/EBP transcription factor is regulated in the G1 phase of the hepatocyte cell cycle. J. Biol. Chem., 270, 18123–18132. [DOI] [PubMed] [Google Scholar]
- 22.Timchenko N., Wilson,D.R., Taylor,L.R., Abdelsayed,S., Wilde,M., Sawadogo,M. and Darlington,G.J. (1995) Autoregulation of the human C/EBPα gene by stimulation of upstream stimulating factor binding. Mol. Cell. Biol., 15, 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tura-Kockar F., Foka,P., Hughes,T.R., Kousteni,S. and Ramji,D.P. (2001) Analysis of the Xenopus laevis CCAAT-enhancer binding protein α gene promoter demonstrates species-specific differences in the mechanisms for both auto-activation and regulation by Sp1. Nucleic Acids Res., 29, 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welm A.L., Mackay,S.L., Timchenko,L.T., Darlington,G.J. and Timchenko,N.A. (2000) Translational induction of liver-enriched transcriptional inhibitory protein during acute phase response leads to repression of CCAAT/enhancer binding protein-α mRNA. J. Biol. Chem., 275, 27406–27413. [DOI] [PubMed] [Google Scholar]
- 25.Baer M. and Johnson,P.F. (2000) Generation of truncated C/EBPβ isoforms by in vitro proteolysis. J. Biol. Chem., 275, 26582–26590. [DOI] [PubMed] [Google Scholar]
- 26.Graham F.L. and van der Eb,A.J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology, 52, 456–467. [DOI] [PubMed] [Google Scholar]
- 27.Ramji D.P., Vitelli,A., Tronche,F., Cortese,R. and Ciliberto,G. (1993) The two C/EBP isoforms, IL-6DBP/NF-IL6 and C/EBPδ/NF-IL6β, are induced by IL-6 to promote acute-phase gene transcription via different mechanisms. Nucleic Acids Res., 21, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mead J.R., Hughes,T.R., Irvine,S.A., Singh,N.N. and Ramji,D.P. (2003) Interferon-γ stimulates the expression of the inducible cAMP early repressor in macrophages through the activation of casein kinase 2. J. Biol. Chem., 278, 17741–17751. [DOI] [PubMed] [Google Scholar]
- 29.Sabatakos G., Davies,G.E., Grosse,M., Cryer,A. and Ramji,D.P. (1998) Expression of the genes encoding CCAAT-enhancer binding protein isoforms in the mouse mammary gland during lactation and involution. Biochem. J., 334, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes T.R., Tengku-Muhammad,T.S., Irvine,S.A. and Ramji,D.P. (2002) A novel role of Sp1 and Sp3 in the interferon-γ-mediated suppression of macrophage lipoprotein lipase gene transcription. J. Biol. Chem., 277, 11097–11106. [DOI] [PubMed] [Google Scholar]
- 31.Ciliberto G., Colantuoni,V., DeFrancesco,R., DeSimone,V., Monaci,P., Nicosia,A., Ramji,D.P., Toniatti,C. and Cortese,R. (1993) Transcriptional control of gene expression in hepatic cells. In Karin,M. (ed.), Gene Expression: General and Cell-Type-Specific. Birkhauser, Boston, MA, pp. 162–242. [Google Scholar]
- 32.Isshiki H., Akira,S., Sugita,T., Nishio,Y., Hashimoto,S., Pawlowski,T., Suematsu,S. and Kishimoto,T. (1991) Reciprocal expression of NF-IL6 and C/EBP in hepatocytes: possible involvement of NF-IL6 in acute phase protein gene expression. New Biol., 3, 63–70. [PubMed] [Google Scholar]
- 33.Akira S. and Kishimoto,T. (1997) NF-IL6 and NF-κB in cytokine gene regulation. Adv. Immunol., 65, 1–46. [PubMed] [Google Scholar]
- 34.Alam T., An,M.R. and Papaconstantinou,J. (1992) Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem., 267, 5021–5024. [PubMed] [Google Scholar]
- 35.Poli V. (1998) The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem., 273, 29279–29282. [DOI] [PubMed] [Google Scholar]
- 36.Foka P., Kousteni,S. and Ramji,D.P. (2001 Molecular characterization of the Xenopus CCAAT-enhancer binding protein β gene promoter. Biochem. Biophys. Res. Commun., 285, 430–436. [DOI] [PubMed] [Google Scholar]
- 37.Davies G.E., Sabatakos,G., Cryer,A. and Ramji,D.P. (2000) The ovine CCAAT-enhancer binding protein δ gene: cloning, characterization and species-specific autoregulation. Biochem. Biophys. Res. Commun., 271, 346–352. [DOI] [PubMed] [Google Scholar]
- 38.Zador I.Z., Hsieh,C.C. and Papaconstantinou,J. (1998) Renal CCAAT/enhancer-binding proteins in experimental diabetes mellitus. Nephron, 79, 313–316. [DOI] [PubMed] [Google Scholar]
- 39.Xu L., Hui,L., Wang,S., Gong,J., Jin,Y., Wang,Y., Ji,Y., Wu,X., Han,Z. and Hu,G. (2001) Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res., 61, 3176–3181. [PubMed] [Google Scholar]
- 40.Flodby P., Liao,D.Z., Blanck,A., Xanthopoulos,K.G. and Hallstrom,I.P. (1995) Expression of the liver-enriched transcription factors C/EBPα, C/EBPβ, HNF-1 and HNF-4 in preneoplastic nodules and hepatocellular carcinoma in rat liver. Mol. Carcinog., 12, 103–109. [DOI] [PubMed] [Google Scholar]
- 41.Mire-Sluis A. and Thorpe,R. (eds) (1998) Cytokines. Academic Press, New York, NY. [Google Scholar]
- 42.Tang Q.Q. and Lane,M.D. (2000) Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer binding protein β during adipogenesis. Proc. Natl Acad. Sci. USA, 97, 12446–12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calkhoven C.F. and Geert,A.B. (1996) Multiple steps in the regulation of transcription-factor level and activity. Biochem. J., 317, 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wegner M., Cao,Z. and Rosenfeld,M.G. (1992) Calcium-regulated phosphorylation within the leucine zipper of C/EBPβ. Science, 256, 370–373. [DOI] [PubMed] [Google Scholar]
- 45.Trautwein C., Caelles,C., van der Geer,P., Hunter,T., Karin,M. and Chojkier,M. (1993) Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature, 364, 544–547. [DOI] [PubMed] [Google Scholar]
- 46.Nakajima T., Kinoshita,S., Sasagawa,T., Sasaki,K., Naruto,M., Kishimoto,T. and Akira,S. (1993) Phosphorylation at threonine-235 bcy a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc. Natl Acad. Sci. USA, 90, 2207–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poli V., Mancini,F.P. and Cortese,R. (1990) IL-6DBP, a nuclear protein involved in interleukin-6 signal transduction defines a new family of leucine zipper proteins related to C/EBP. Cell, 63, 643–653. [DOI] [PubMed] [Google Scholar]
- 48.Nakajima K., Kusafuka,T., Takeda,T., Fujitani,Y., Nakae,K. and Hirano,T. (1993) Identification of a novel interleukin-6 response element containing an Ets-binding site and a CRE-like site in the junB promoter. Mol. Cell. Biol., 13, 3027–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]