Summary
We have characterized a rodent-specific gene family designated α-takusan (meaning “many” in Japanese). We initially identified a member of the family whose expression is upregulated in mice lacking the NMDAR subunit NR3A. We then isolated cDNAs encoding 46 α-takusan variants from mouse brains. Most variants share a ~130-aa long sequence, which contains the previously identified DUF622 (domain of unknown function 622) and is predicted to form coiled-coil structures. Single-cell PCR analyses indicate one neuron can express multiple α-takusan variants, and particular variants may predominate in certain cell types. Forced expression in cultured hippocampal neurons of two variants, α1 or α2, which bind either directly or indirectly to PSD-95, leads to an increase in PSD-95 clustering, dendritic-spine density, GluR1 surface expression, and AMPAR activity. Conversely, treating cultured neurons with RNAi targeting α-takusan variants resulted in the opposite phenotype. Hence, α-takusan represents a novel gene family that regulates synaptic activity.
Introduction
NMDA-type glutamate receptors (NMDARs) are thought to act as integrators of coincident synaptic signals (Malenka and Nicoll, 1999), while their hyperactivation causes neurodegeneration (Cull-Candy et al., 2001; Dingledine et al., 1999; Lipton and Rosenberg, 1994). The conventional NMDAR requires two distinct subunits, NR1 and NR2 subunits, to form functional channels (Dingledine et al., 1999; Hollmann and Heinemann, 1994). We and others have identified a third family of NMDAR subunits, designated NR3 (Ciabarra et al., 1995; Das et al., 1998; Sucher et al., 1995). In heterologous expression systems, inclusion of NR3A decreases the amplitude, Ca2+ permeability, and Mg2+ sensitivity of NR1/NR2 channels (Chatterton et al., 2002; Ciabarra et al., 1995; Das et al., 1998; Perez-Otano et al., 2001; Sasaki et al., 2002; Sucher et al., 1995). Consistent with these findings, the amplitude of NMDA currents in NR3A knock-out (KO) neurons is larger than that of wild-type (WT) neurons (Das et al., 1998; Sasaki et al., 2002). Hence, NR3A is considered to act as an inhibitory subunit of NMDAR. Concomitantly, NR3A might control trafficking of NMDARs (Perez-Otano et al., 2006).
NMDARs are clustered in the postsynaptic density (PSD) at excitatory synapses (Nourry et al., 2003; Sheng, 2001; Sheng and Sala, 2001). This is likely mediated by physical association between C-terminal ends of NR2 and PDZ domains of postsynaptic scaffolding proteins such as PSD-95 (Kornau et al., 1995; Niethammer et al., 1996). PDZ domains are modular protein domains of ~90 amino-acids that are used for protein-protein interactions, and each PDZ domain binds to C-terminal peptides in a sequence-specific manner (Kim et al., 1995; Kornau et al., 1995; Mori et al., 1998; Niethammer et al., 1996; Songyang et al., 1997; Steigerwald et al., 2000). For example, a class I PDZ domain prefers the C-terminal tail -S/T-X-V/L/I as its binding partner. In addition to NR2 subunits, PSD-95 binds to various other proteins, such as stargazin (Schnell et al., 2002), and organizes postsynaptic supramolecular complexes (Husi et al., 2000; Kim and Sheng, 2004). Interestingly, forced expression of PSD-95 in cultured hippocampal neurons enhances postsynaptic clustering of AMPA, but not NMDA, receptors (El-Husseini et al., 2000) and the function of PSD-95 is further regulated by its palmitoylation (El-Husseini Ael et al., 2002). These and other studies have identified molecules that regulate trafficking and localization of AMPA receptor subunits (reviewed in Barry and Ziff, 2002; Malinow and Malenka, 2002; Nicoll et al., 2006; Song and Huganir, 2002).
Since neurons in NR3A KO mice manifest increased NMDA-induced currents, we reasoned that these mice might allow us to identify components of signal transduction pathways downstream to NMDAR hyperactivation. In turn, these genes may represent candidate molecules that are involved in manifestation of the phenotypes observed in NR3A KO mice, including increased dendritic spines (Das et al., 1998). Specifically, we screened for genes whose expression was altered in NR3A KO brains compared to WT brains. We identified such a gene and found that it belonged to a novel, very large gene family whose members shared a domain of ~130 amino acids (aa). A portion of this domain had previously been termed DUF622 (domain of unknown function 622), which is 85 aa in length and predicted to form a coiled-coil structure. One example of a protein that contains DUF622 is the human tumor suppressor gene Dlg (discs large) 5 that also contains PDZ and guanylate-kinase domains (Stoll et al., 2004). However, the majority of proteins with DUF622 are 150-250 aa long and contain no other known domains. Because of the size of this family of genes, we have named it takusan, a Japanese word that means “many.”
In this paper, we describe our initial characterization of α-takusan, a subclass of the takusan family. We show that more than 40 members of α-takusan are expressed in the mouse brain. Single-cell PCR analyses indicate that individual neurons can express multiple α-takusan variants and that different neurons may express different sets of α-takusan. In transfected COS-7 cells, certain α-takusan variants bind to PSD-95, while other variants do not, presumably due to their structural diversity at the C-termini. We then force expressed fusion proteins of enhanced green fluorescent protein (EGFP) and α1- (PSD-95 binding) or α2- (PSD-95 non binding) variants of takusan in cultured hippocampal neurons. Examining the localization of takusan family members fused to EGFP, we found that EGFP-α1 signals were often co-localized with PSD-95, while EGFP-α2 signals were co-localized with or adjacent to PSD-95 signals. In co-immunoprecipitation experiments, α1 bound directly to PSD-95, while α2 bound indirectly via complex formation with α1. Forced expression of EGFP-α1 or -α2 enhanced clustering of PSD-95, dendritic-spine density, surface expression of GluR1, and AMPA-receptor activity, whereas RNAi for takusan variants abrogated these effects. Interestingly, forced expression of EGFP-α1, but not -α2, altered desensitization of NMDA-induced currents.
Results
Identification of α1-takusan as a gene upregulated in NR3A KO mice
We searched for genes whose expression levels are altered in NR3A KO. To this end, we extracted RNAs from the whole brains of NR3A KO and littermate WT mice in a 129/SVJ background on postnatal day 15 (P15), synthesized cDNAs, and then used them to probe mouse cDNA microarrays (NIA mouse 15k, Ontario Cancer Institute). From these experiments, we identified a gene whose expression level is higher in NR3A KO brains than in WT brains by more than 2-fold (Figure 1A). We identified the gene that corresponds to the spots on the array as Riken cDNA clone 2610042 (Carninci and Hayashizaki, 1999; Kawai et al., 2001). This cDNA contains a 150 aa open reading frame, and its gene function was previously unknown. We named this gene the α1 form of takusan or α1-takusan, because it belongs to a large gene family. We verified the microarray data by quantitative (Q)-PCR in which the level of α1-takusan mRNA was normalized to that of a reference gene, glyceraldehyde-3-phosphate dehydrogenase (GADPH). We found that α1 takusan mRNA is expressed 2.6 times higher in NR3A-KO mouse brains than in WT brains at P15 (Figure 1B).
Figure 1. α1-Takusan is Upregulated in NR3A KO Mice.
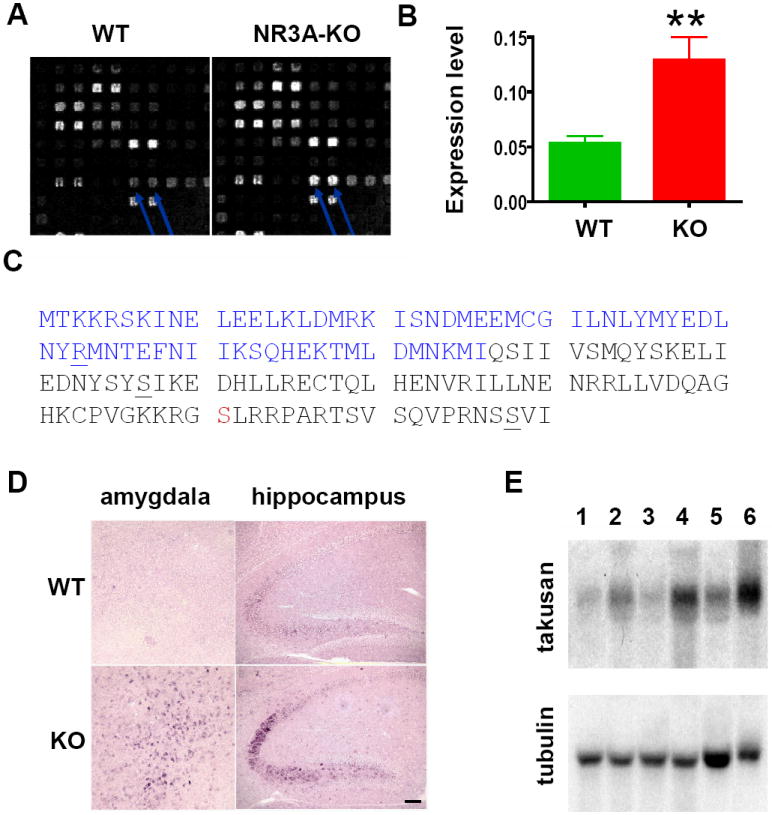
(A) Microarray probed with cDNAs prepared from WT and NR3A-KO brains. Arrows indicate duplicate spots corresponding to α1-takusan. (B) Q-PCR confirmed the results of the microarray experiments. The expression level of α-takusan mRNA in NR3A KO brain is 2.6 times higher than that in WT brain. (C) Deduced amino-acid sequence of α1-takusan. The residues in blue (1-66) show homology with DUF622. The residues at the C-terminal end form a motif that is predicted to bind to class I PDZ domains. The underlined letters (R, S, and S) indicate the end of exons 4, 5, and 6. The serine in red is a candidate for phosphorylation by protein kinases A and C. (D) In situ hybridization was performed using a pan α-takusan probe on WT and NR3A KO brain sections at P15. α-takusan mRNA is more abundant in KO than in WT in regions such as the amygdala and hippocampus. Scale bar = 100 μm. (E) Northern blot of α-takusan mRNAs in WT and NR3A KO mice in three areas of the brain. RNAs from the cerebral cortex (lane 1-WT and 2-KO), hippocampus (3-WT and 4-KO), and cerebellum (5-WT and 6-KO) were hybridized with probes for α1-takusan or control tubulin. The signals from the three areas of the KO mice are 2-3 times higher than those of WT mice.
Figure 1C shows the deduced amino-acid sequence of α1-takusan that we isolated from WT brain of C57BL/6J. The coding sequence of α1-takusan was assigned to an open reading frame (ORF) near the 5’ end of the cDNA, based on the codon usage index and its surrounding sequences. This assignment is likely correct, because this ORF would produce a protein that contains a portion of the domain that is conserved among a large number of proteins. This domain, previously designated DUF622 at Pfam (www.sanger.ac.uk/Software/Pfam), is ~85 aa long and overlaps with the N-terminal half (residues 1-76) of α-takusan. Garnier-Robson and Chou-Fasman algorithms predict that α1 takusan forms an α-helical structure through much of the coding region. Furthermore, the COILS program (www.ch.embnet.org/software/COILS_form.html) predicts two separate coiled-coil domains (residues 1-37 and 84-123) will be formed in this protein. The Scansite 2.0 program (Obenauer et al., 2003) identified a phosphorylation site for both protein kinase A (PKA) and protein kinase C (PKC) at Serine 131, as well as a candidate motif for PDZ binding at the C-terminal of α1-takusan. We also determined exon boundaries within the coding region of α1-takusan gene by comparing cDNA and genomic sequences using a search program BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat). The coding region of this gene is encoded by four separate exons (exons 4-7), which are preceded by three exons encoding 5’-UTR.
Expression patterns of α-takusan mRNA in WT and NR3A KO mice
Using a part of the α1-takusan cDNA as a probe, we performed in situ hybridization on NR3A KO and WT at P15 (Figure 1D). The probe spans 363 nucleotide (nt) of the α1-takusan coding region and 279 nt of the 3’UTR sequences. This probe contains the entire exons of 5 and 6, and would thus likely hybridize with other α-takusan variants (see below). We found that α-takusan mRNA is expressed in WT brains in the hippocampus, cerebral cortex, amygdala and cerebellum. In contrast, the level of α-takusan expression is significantly increased in NR3A KO mice in brain areas including the cerebral cortex layer V, hippocampal CA2-3 region, amygdala, and cerebellar Purkinje cells. These areas normally express NR3A in WT mice (Wong et al., 2002). These data suggest that, at least initially, the upregulation of α-takusan mRNA by a genetic deletion of NR3A occurs in a cell-autonomous manner; namely, α-takusan is upregulated in KO mice in the cells in which NR3A expression has been abolished.
We next examined α-takusan expression by Northern analyses. At P13, the level of α-takusan expression is too low to be detectable in WT and KO mice (data not shown). At P25, when normalized to tubulin expression, KO mice expressed higher levels of α-takusan than WT mice by 2-3 fold in the areas of the brain we examined (Figure 1E). Again, the probe used in this study would likely hybridize to other α-takusan variants. We currently do not know which form(s) of α-takusan is upregulated in KO mice.
The studies described above focused on the upregulation of α-takusan in NR3A KO mice. Throughout these studies, we noticed WT brains express α-takusan mRNAs, albeit their expression levels are lower than those in KO brains. To gain insight into the functions of α-takusan, we examined tissue distribution of α-takusan mRNA in WT mice. As shown in Supplemental Figure 1A, we found that α-takusan mRNA is expressed in two adult tissues, the brain and testis, but not in heart, lung, liver, spleen, skeletal muscles, or kidney. We next performed in situ hybridization on male 6-week old 129/SVJ WT brains. As shown in Supplemental Figure 1B, adult WT mouse expresses α-takusan in various areas of the brain. These areas include glomerular and mitral cells of the olfactory bulb, the pyramidal layer and dentate gyrus of the hippocampus, scattered cells in the amygdala, cerebral cortex and facial nucleus, and Purkinje neurons of the cerebellum.
α-takusan is an unusually large gene family
Prior to this study, there had been multiple cDNA entries whose DNA sequences were highly homologous to α1-takusan in GenBank. Many of these cDNAs were isolated from the testis and whole embryos by the Riken cDNA project. By comparing these genes, we found that these genes were most conserved within the area encoded by exon 5 of α1-takusan (residues 43 – 87, see Figure 1C). When we used exon 5 of α1-takusan as a query in BLASTN, we found 115 hits in the mouse genome. Most hits correspond to genome sequences that are predicted to encode proteins with >70% identity to exon 5 of α1-takusan. We consider these genes to be candidate members of the α-takusan gene family (see below on how we define this family).
We next attempted to determine which members of α-takusan were expressed in the brain. To this end, four PCR primer pairs were designed from conserved sequences of 5’- and 3’- UTRs of α-takusan. RT-PCR was performed from RNAs extracted from a forebrain of a WT C57BL/6 (male, 6-week old) mouse. We chose this strain, because it was used for the genome sequencing project. We identified 31 distinct members of α-takusan expressed in a single brain (see Figure 2A and Supplemental Figure 2 for their deduced amino-acid sequences). Additional α-takusan variants may also be expressed in the brain (see below).
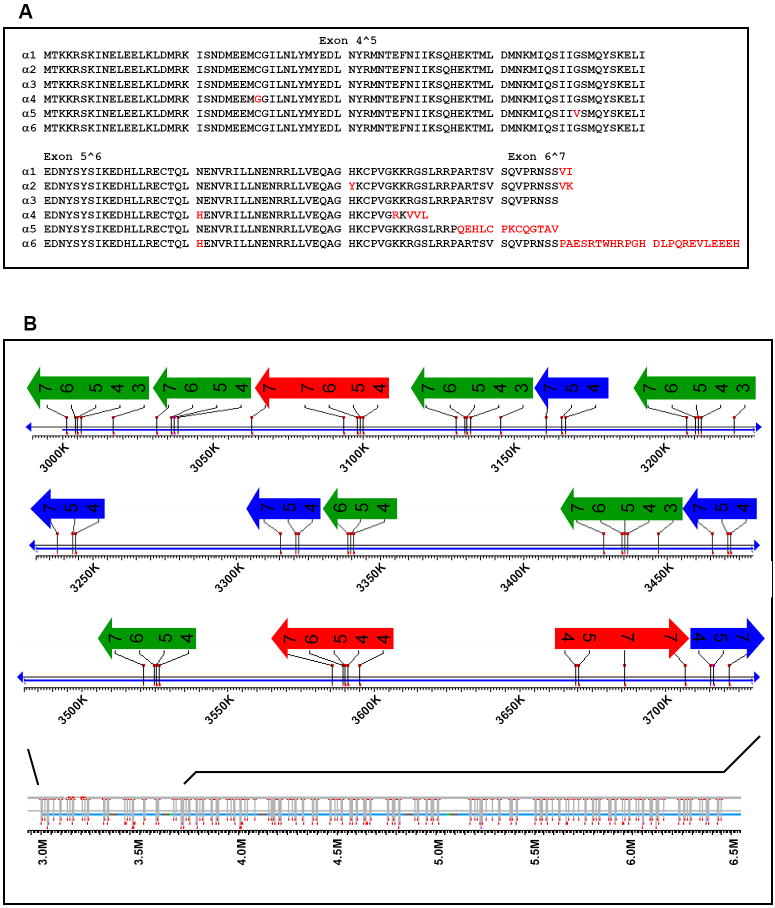
Figure 2. Deduced Amino-Acid Sequences and Gene Organization of Selected α-Takusan Variants.
(A) Deduced amino-acid sequences of α1-α6 takusan cDNAs isolated from WT C57BL/6 mouse brain. These six variants were used for the experiments presented in Figure 4C. Twenty-five other variants (α7 - α31) were also isolated from the same brain, and their deduced amino-acid sequences are presented in Supplemental Figure 2. Typical exon boundaries are marked by ˆ. Non-consensus amino acids are indicated in red. (B) Location of exons encoding α-takusan protein sequences on a segment of chromosome 14. Exons 3-7 (the potential protein coding region) of α-takusan cDNA sequences were compared to the mouse genome using megaBLAST. The diagram at the bottom shows the positions of these hits on a 3.5 Mb stretch using the Entrez Genomes MapViewer. An enlarged map of the 720-kb area that contains 15 apparent α-takusan loci is presented in three lines. See text for explanation of arrows in different colors.
The alignment shown in Figure 2A and Supplemental Figure 2 reveals highly conserved amino-acid sequences among α-takusan variants we isolated. We compared cDNA and genomic sequences, and found that variations observed in cDNAs were also present in the mouse genome sequences. Therefore, it is unlikely these variations are products of RNA editing or mutations introduced during PCR. From these alignments, we made the following observations. (1) At the N-terminal, the presence (or absence) of a termination codon at the 5’-UTR creates variations in the starting ATG position. Due to this and other reasons, α-takusan variants that we isolated differ in size ranging from 46 to 237 aa long (see also Supplemental Figure 4A). (2) The C-termini vary in multiple forms. Interestingly, some α-takusan variants encode C-terminal sequences, such as –SSVI and –GTAV, that are predicted to bind a class I PDZ domain, according to Scansite predictions (Songyang et al., 1997). Other variants contain C-terminal sequences that are predicted not to bind PDZ domains. These include variants that end with –SSVK and –EEEH. (3) Multiple amino-acid changes are present within the takusan domain, although many of these changes are conservative and may not alter protein properties. (4) Some variants such as α22 appear to lack exon 6 (see below).
Apparent loci for α-takusan are located on chromosomes 12, 14 and 17. In particular, a cluster of approximately 75 α-takusan loci is found within a 16-Mb region that corresponds to 14A1 and 14A2 on chromosome 14. Figure 2B shows the organization of exons for the potential coding region of α-takusan in this area. The bottom map shows the location of all hits with exons 3-7 of α1-takusan in a 3.5-Mb region in the 16-Mb area. The upper diagram shows exon-intron organization of 15 apparent α-takusan loci, each of which encodes a different variant within a ~720 Kb stretch. As shown in the figure, α-takusan genes are located in both strands of the chromosome. The exon organization in these loci sheds light on the origin of the diversity observed among various α-takusan cDNAs. The first class of loci, indicated by green arrows in the figure, consists of consecutive exons (3-7, 4-7 or 4-6), and each locus appears to encode a typical “full-length” variant that is 148 – 205 aa long (see Supplemental Figure 2). The second class of loci, indicated by blue arrows, lack exon 6, and thus these loci might directly encode shorter variants such as α22 (additional examples are presented in Supplemental Figure 4A). This observation is interesting because these shorter variants could potentially be produced from the first class of loci by alternative splicing. Alternatively, most of the shorter variants might be transcribed from loci that lack exon 6. The third class of loci, indicated by red arrows, contain tandem repeats of exon 4 or 7 within a locus. These loci could possibly produce two variants whose diversity is restricted to the N- or C-terminus by alternative splicing. Examples of two variants that might be produced by this mechanism are α14 and α18. It is not possible, however, to definitively determine the locus from which these two cDNAs are transcribed because there are multiple loci whose sequences are very similar to each other and to the two cDNAs. Finally, it is also possible, at least in theory, that alternative splicing occurs across different loci. For example, an exon of one locus might be spliced into an exon of another locus. In summary, an examination of the organization of the α-takusan genes in the mouse genome leads to the notion that diversity among variants is likely created by two distinct mechanisms: gene multiplication during evolution and alterative splicing.
Moreover, α-takusan appears to be only one branch of a larger gene family. To examine this possibility, we used α7-takusan, which contains an elongated N-terminal sequence, as a query to search for similar sequences in the nr database at NCBI by BLASTP. We found 446 similar proteins and extracted 157 mouse sequences, the majority of which were annotated as hypothetical proteins. We then selected 51 sequences after removing highly similar proteins (identity ≥95%), added α7-takusan, and built an alignment using ClustalW (Supplemental Figure 3A) to construct a sequence similarity dendrogram (Supplemental Figure 3B).
The dendrogram shows that α7-takusan belongs to one (presented in green in Supplemental Figure 3B) of three distinct branches of the gene family. Based on this observation, we define genes that belong to this branch as members of the α-takusan gene family. The average sequence identity within this branch is 84.3%, while the minimum sequence identity within this group is 69.4%. We refer to the members of the other branches as non-α forms of takusan. The exact size of the entire takusan gene family in the mouse genome remains to be determined. The majority of takusan genes, both α- and non-α- family members, encode proteins of 150-220 aa. These proteins share a conserved domain of ~130 aa long, which contains the previously identified DUF622 within its boundaries. Strikingly, among the species for which genomic information is available, the takusan gene family is present only in mice and rats. As described in Introduction, the human tumor suppressor protein dlg 5 contains DUF622.
Different neurons express different sets of α-takusan variants
Experiments described above show that mouse brain expresses a large number of α-takusan variants. How does the CNS exploit this molecular diversity? We therefore asked (1) whether a single neuron expresses more than one α-takusan, and (2) whether different neurons express different sets of α-takusan variants. To address these questions, we examined cDNAs expressed in individual neurons cultured from the hippocampus and cerebral cortex of C57BL/6 mice dissected at E18 or P0 by single-cell PCR techniques (Sucher and Deitcher, 1995).
We collected cytoplasmic samples from 22 hippocampal and 22 cortical neurons and subjected them to RT-PCR, which yielded PCR products of the appropriate size from 11 hippocampal and 9 cortical cells. We then cloned these products and determined the sequences of 5 clones per cell. We identified a total of 22 distinct variants expressed in these neurons (see Supplemental Figure 4 for deduced amino-acid sequences). Seven of these sequences were identical to those isolated from the forebrain of the adult mouse, while the other 15 were distinct. Therefore, between the whole-brain and single-cell PCR analyses, we have isolated 46 distinct α-takusan variants from a single mouse strain (C57BL/6J).
In Table 1, we tabulate how each of the 22 α-takusan variants is expressed in the 20 neurons we examined in detail. Each variant is noted with its size, type based on exon composition of the coding region (see Supplemental Figure 4B), and four amino-acid residues at the C-termini. As shown below, the C-terminal residues of each takusan variant determine the ability to bind PDZ domains. The variants that are predicted to bind PDZ domains are presented in red, while those that are predicted not to bind to PDZ domains are in black in Table 1. Overall, we made the following observations. (1) Many neurons express more than one α-takusan variant. (2) The repertoire of α-takusan variants is different from neuron to neuron. Since identical PCR conditions were used for all experiments, this likely reflects the difference in expression patterns of α-takusan variants among these cells. Interestingly, there may be a preference in the expression of certain types of α-takusan variants by particular cell types. For example, type I α-takusan variants that end with XSVK were isolated in only 1 out of 11 hippocampal neurons, but in 5 out of 9 cortical neurons (p<0.05, Fisher’s exact test). Conversely, α-takusan variants that are predicted to bind PDZ domains were detected in 6 out of 11 hippocampal neurons (25 of 55 clones), while these variants were detected in only 2 out of 9 cortical neurons (8 of 45 clones; p<0.01 for clones, Fisher’s exact test).
Table 1.
Distribution of α-Takusan Variants in Hippocampal and Cortical Neurons.
| takusan | size (aa) |
type | C-terminal residues |
hippocampal cell | cortical cell | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||||
| α1 | 150 | I | XSVI | 2 | 5 | ||||||||||||||||||
| α32 | 150 | I | XSVI | 1 | |||||||||||||||||||
| α19 | 150 | I | XSVI | 4 | 1 | ||||||||||||||||||
| α33 | 150 | I | XSVI | 2 | |||||||||||||||||||
| α5 | 148 | I | GTAV | 1 | |||||||||||||||||||
| α25 | 148 | I | GTAV | 2 | 3 | 3 | |||||||||||||||||
| α38 | 237 | II | XSVI | 5 | |||||||||||||||||||
| α9 | 205 | II | XSVI | 1 | 1 | ||||||||||||||||||
| α39 | 205 | II | XSVI | 1 | |||||||||||||||||||
| α40 | 89 | III | XSVI | 1 | |||||||||||||||||||
| α34 | 150 | I | XSVK | 2 | |||||||||||||||||||
| α35 | 150 | I | XSVK | 3 | 2 | 2 | 1 | 5 | 2 | ||||||||||||||
| α36 | 150 | I | XSVK | 1 | |||||||||||||||||||
| α23 | 172 | I | EEEH | 3 | 5 | ||||||||||||||||||
| α37 | 172 | I | EEEH | 5 | 5 | 1 | |||||||||||||||||
| α24 | 156 | I | RTWH | 2 | 2 | 5 | 5 | ||||||||||||||||
| α41 | 72 | III | XSVK | 1 | |||||||||||||||||||
| α42 | 72 | III | XSVK | 1 | |||||||||||||||||||
| α43 | 72 | III | XSVK | 1 | |||||||||||||||||||
| α44 | 72 | III | XSVK | 3 | 3 | ||||||||||||||||||
| α45 | 46 | III | XSVK | 2 | 1 | 1 | 2 | ||||||||||||||||
| α46 | 107 | III | XSVK | 1 | |||||||||||||||||||
The numbers inside squares represent the numbers of clones isolated.
α-Takusan proteins are primarily expressed in neurons
We generated antibodies against α-takusan variants using a peptide TKKRSKINELEELKLDMRK whose sequence was taken from the 2-20 position of α1-takusan. The protein sequences at this position are divergent between α-takusan and non-α-takusan variants (see Supplemental Figure 3A); thus, the antibody we generated should be specific to α-takusan. The sequence is strongly conserved among all α-takusan variants that we isolated by PCR, thus the antibody likely recognizes these α-takusan variants. However, there are α-takusan variants whose sequence similarity to α1-takusan in this area drops down to approximately 50%. We do not know whether our antibody recognizes these α-takusan variants. Figure 3A shows an immunoblot of the PSD fraction from the forebrain of an adult C57BL6 WT mouse. The blot shows a major band at ~ 17 kD which was competed out when the antibody was preincubated with antigenic peptides (data not shown). Since the calculated MW of the deduced α1-takusan protein is 17.7 kD, this band is likely to represent α-takusan variants with a protein size near 150 aa. It thus appears that type I is the predominant form of α-takusan variants in the forebrain. The blot also shows two other minor bands, estimated at 24 and 30 kD, which were also competed out with peptide preincubation. These bands may represent minor species of α-takusan variants that are present in the forebrain. For example, the calculated MW of the deduced α7-takusan protein is 24.4 kD and several α-takusan variants shown in Supplemental Figure 3A could encode ~30 kD proteins. It appears that the intensity of α-takusan bands is increased in the NR3A KO (data not shown).
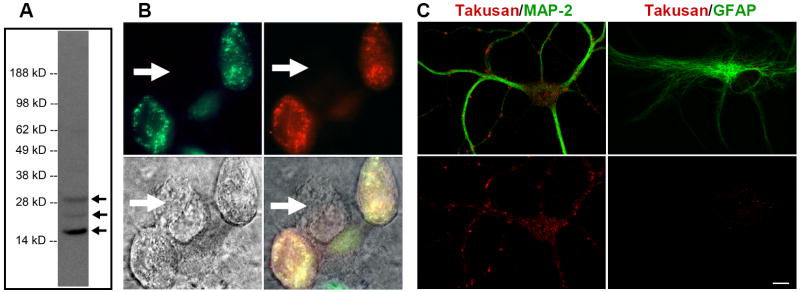
Figure 3. Characterization of Endogenous α-Takusan Proteins.
(A) Immunoblot of the PSD fraction prepared from adult C57BL/6 WT mouse brain using α-takusan antibody. The antibody detected a major band at ~17 kD and minor bands at ~24 and ~30 kD in this preparation. (B) Immunocytochemistry with α-takusan antibody of HEK293 cells expressing exogenous EGFP-α1. Images represent EGFP-α1 fluorescence (green, top left), anti-α-takusan staining (red, top right), Nomarsky (bottom left), and overlay (bottom right). Staining pattern of α-takusan approximates that of EGFP; additionally, a cell not expressing EGFP-α1 does not stain for α-takusan (indicated by the arrow). (C) Selective expression of α-takusan in MAP-2 positive neurons. Mixed neuronal/glial cultures of hippocampal cells were co-stained for α-takusan/MAP-2 or α-takusan/GFAP. The two images at the top represent an overlay of α-takusan (red) and either MAP-2 (green; left) or GFAP (green; right). The images on the bottom represent α-takusan staining only. The α-takusan antibody detected strong signals in MAP-2 positive (neuronal) cells, but only weak signals in GFAP-positive (astrocytic) cells. In the MAP-2 positive cells, α-takusan localized to both dendrites and cell bodies. Scale bar = 10 μm.
We next tested whether our antibody is useful for immunocytochemistry. For this purpose, HEK293 cells were transfected with cDNAs encoding a fusion protein in which EGFP was fused at the N-terminus of α1-takusan (referred to as EGFP-α1). The antibody detected EGFP-α1, as evidenced by co-localization of green fluorescence and immunosignal (Figure 3B). No anti-takusan signals were detected in the cells expressing EGFP alone (data not shown). We then performed immunocytochemistry on mixed neuronal/glial cell cultures prepared from hippocampi isolated from E18 C57BL/6 mice. Figure 3C shows cells co-stained with either α-takusan/MAP-2 or α-takusan/GFAP. α-takusan signals are present in neurites and cell bodies of MAP-2 positive cells, while only weak signals are seen in GFAP positive cells. These experiments suggest that α-takusan is primarily expressed in neurons rather than astrocytes.
α-Takusan dimerizes (or oligomerizes) in transfected HEK-293 cells
The conserved sequence among α-takusan proteins consist of a predicted coiled-coil domain, which often mediates protein-protein interactions as well as dimerization or oligomerization (Cohen and Parry, 1990). In order to test if α-takusan dimerizes, we performed co-immunoprecipitation (IP) experiments cells transfected with two fusion proteins of α1-takusan. We chose α1 among the α-takusan variants because we frequently observed this clone in many experiments on mouse brain. In addition, α1-takusan belongs to the type I form of α-takusan variants, which appear to predominate in the mouse forebrain (Figure 3A). The two fusion proteins used in this study were EGFP-α1 and myc-α1, the latter containing a myc-tag fused at the N-terminus of α1-takusan. We co-transfected HEK-293 cells with EGFP-α1 and myc-α1. Lysates prepared from transfected cells (inputs) were directly probed with anti-EGFP and anti-myc antibodies to verify expression of the fusion proteins (Figure 4A; two top panels). We then performed co-IP experiments using anti-EGFP and anti-myc antibodies in either order. Thus, we precipitated lysates with anti-myc and blotted with anti-EGFP (Figure 4A, third panel from the top), or precipitated with anti-EGFP and blotted with anti-myc (Figure 4A, bottom panel). The bands in the third lane of the bottom two panels show that EGFP-α1 had associated with α1-myc in transfected HEK-293 cells. These data are consistent with the notion that the coiled-coil domains of α-takusan proteins mediate self-dimerization or oligomerization of these proteins.
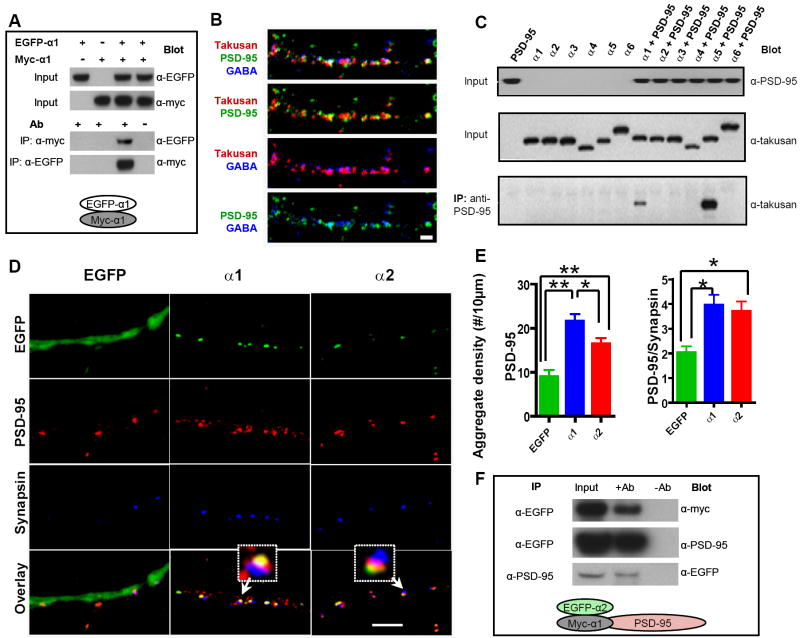
Figure 4. α1-Takusan Dimerizes and α-Takusan Variants with Different C-terminals Manifest Differential Ability to Associate with PSD-95.
(A) Self-dimerization of α1-takusan. HEK293 cells were transfected with EGFP-α1 alone, myc-α1 alone, or the combination of both, as indicated at the top of the panel. The top two panels are immunoblots of cell extracts (inputs) probed with anti-EGFP and anti-myc antibodies, respectively. The bottom two panels show co-IP experiments in which antibodies used for IP and blotting are shown. In the right most lane of these two panels, mock IPs were performed (antibodies were absent during the IP procedure). The experimental scheme is shown at the bottom. (B) Immunocytochemistry of cultured hippocampal neurons decorated by antibodies against PSD-95 (green), GABAA receptor (blue), and α-takusan (red). α-takusan proteins appear as clusters or aggregates in dendrites. Some α-takusan signals overlap with PSD-95, while others do not. α-takusan signals generally do not overlap with GABAA receptors. Scale bar = 2 μm. (C) Co-IP experiments from COS-7 cells expressing α-takusan variants and PSD-95. COS-7 cells were transfected with PSD-95 (lane 1), one of six takusan variants (α1 - α6) fused with EGFP at the N-termini (lanes 2-7), or the combination of PSD-95 and one takusan variant (lanes 8-13). In the top two panels, lysate inputs were probed with anti-PSD-95 or anti-α-takusan antibody to verify their expression. In the bottom panel, the lysates were immunoprecipitated with anti-PSD-95 antibody; precipitates were then probed with anti-takusan antibody. The data show that α1- and α5-takusan co-immunoprecipitated with PSD-95, while other variants tested did not. (D) Immunocytochemistry of cultured hippocampal neurons expressing EGFP, EGFP-α1, or EGFP-α2, stained with antibodies against PSD-95 (red) or synapsin (blue). These images show that both EGFP-α1 and EGFP-α2 appear to be localized at postsynaptic sites. In addition, forced expression of EGFP-α1 or -α2 increases the degree of PSD-95 clustering in these neurons compared to control cells expressing EGFP alone. Scale bar = 5 μm. (E) Quantification of PSD-95 aggregates in neurons expressing EGFP (n = 8), EGFP-α1 (n = 8), or EGFP-α2 (n = 9). The number of total PSD-95 aggregates in dendrites (left) and PSD-95 aggregates juxtaposed to synapsin (right) were counted, and their density calculated per 10 μm dendritic length (mean density ± SEM; *p < 0.05, **p < 0.01, one-way ANOVA). (F) Association of α2-takusan with PSD-95 in the presence of α1-takusan. Immunoblots of lysates and IPs prepared from cells co-transfected with plasmids containing myc-α1, EGFP-α2, and PSD-95. In each immunoblot, the left lanes are loaded with lysates (inputs), the middle lanes co-IP experiments, and the right lanes mock IPs. The antibodies used for IP and blotting are shown next to each panel. Top panel shows that myc-α1 associates with EGFP-α2. The two other panels show the association of EGFP-α2 and PSD-95. Since C shows that α2-takusan does not directly bind to PSD-95, this association is probably indirect via α1-takusan (shown schematically at bottom of figure).
Select α-takusan variants associate with PSD-95
We next examined the subcellular localization of endogenous α-takusan proteins by performing immunocytochemistry on cultured hippocampal neurons using our α-takusan antibody. Figure 4B shows the pattern of α-takusan immunostaining and its relative relationship with PSD-95 and α1-GABAA receptor signals. In dendrites, α-takusan was localized in small clusters. The α-takusan signal overlapped in part with PSD-95, but exhibited little overlap with GABAA receptors. Taken together with additional data shown below, this finding is consistent with the notion that α-takusan is primarily localized at excitatory synapses.
The partial but not exclusive overlap between α-takusan and PSD-95 signals observed in Figure 4B may be related to the fact that some α-takusan variants contain C-terminal sequences that are predicted to bind to class I PDZ domains, while other variants do not. We therefore tested the ability of six α-takusan proteins (α1 – α6) to bind PSD-95 in co-immunoprecipitation experiments. Figure 4C shows that α1 and α5-takusan associated with PSD-95 when these proteins were co-expressed in COS-7 cells. In contrast, α2, α3, α4 and α6 did not bind PSD-95 in COS-7 cells. These results match the predictions of the Scansite program. The data suggest that disparate α-takusan variants possess different functional properties.
Additionally, if the physical association between α1-takusan and PSD-95 is physiologically relevant; the proteins might be expected to co-localize in neurons. To test this idea, we constructed another fusion protein EGFP-α2 in which EGFP is linked to the N-terminus of α2-takusan, and tested it along with EGFP-α1. We chose these two variants for two reasons: (1) α1 and α2 are two of the most prevalent species that we isolated in our cloning efforts. (2) the amino-acid sequences of α1 and α2 are identical except for two residues (H/Y at residue 121 and I/K at residue 150 for α1/α2, respectively). Therefore, any difference we observe between two variants can be attributed to these two residues. We then transduced these fusion proteins into cultured hippocampal neurons with a Semliki Forest Virus (SFV) vector. Figure 4D shows the results of immunocytochemical experiments on neurons expressing EGFP-α1 or -α2. The green fluorescence patterns show the localization of the fusion proteins in cells co-stained with antibodies against PSD-95 or the pre-synaptic protein synapsin. Both EGFP-α1 and -α2 form puncta within dendrites (Figure 4D, green). These proteins thus appeared to localize close to PSD-95. In contrast, there was little, if any, overlap between EGFP-α1/α2 and synapsin. Upon closer examination (Figure 4D, Overlay), there was a difference between the localization of EGFP-α1 and α2 with respect to PSD-95. Namely, EGFP-α1 and PSD-95 often overlapped, indicating their co-localization within dendrites. In contrast, some EGFP-α2 fluorescence overlapped with PSD-95, while other fluorescent signals did not. The distinct localizations of these two α-takusan variants may be due to the difference in their ability to associate with PSD-95. Furthermore, the patterns of signals generated by the α-takusan antibody (Figure 4B) can be explained as a mixture of localization patterns of the two types of α-takusan variants, those that directly bind PSD-95 (such as α1) and those that do not (such as α2).
Interestingly, overexpressing α-takusan in cultured neurons might mimic conditions of NR3A-deficient neurons to some extent. We thus examined whether forced expression of α1- or α2- takusan could induce physiological effects in the transduced neurons. When neurons expressing EGFP-α1 or EGFP-α2 were stained with PSD-95 antibodies, we observed an increase in the number of PSD-95 clusters compared to neurons expressing the control protein EGFP. We quantified the number of PSD-95 clusters in neurons expressing EGFP, EGFP-α1 or EGFP-α2, and then calculated their density per 10 μm dendritic length. As shown in Figure 4E (left), we found a significant increase in the number of PSD-95 clusters after forced expression of EGFP-α1 (21.62 ± 1.56; n = 8) or EGFP-α2 (16.49 ± 1.28; n = 9) compared to that in control neurons (8.97 ± 1.47; n = 8). EGFP-α1 manifested a greater effect than EGFP-α2. In addition, we quantified the number of PSD-95 clusters that were juxtaposed to synapsin (Figure 4E, right). This value measures the degree of PSD-95 clustered at presumed synaptic sites. We observed a significant increase in the number of PSD-95/synapsin cluster after forced expression of EGFP-α1 (3.97 ± 0.40) or EGFP-α2 (3.71 ± 0.39) compared to control neurons (2.04 ± 0.25). These results suggest that forced expression of EGFP-α1 or EGFP-α2 promotes synaptic clustering of PSD-95.
The aforementioned results reveal only subtle differences between the subcellular localization and synaptic effects of EGFP-α1 and EGFP-α2. The actions α1-takusan may be accounted for at least in part via association with PSD-95. However, since α1-takusan can both interact directly with PSD-95 and also dimerize (or oligomerize) with itself, we wondered whether α2-takusan might associate with α1-takusan, thereby indirectly interacting with PSD-95. To test this idea, HEK-293 cells were transfected with myc-α1, EGFP-α2, and PSD-95 cDNAs. Cell lysates were prepared and three different co-IP experiments were performed. First, lysates were precipitated with anti-EGFP and blotted with anti-myc (Figure 4F, top panel). This experiment showed that myc-α1 and EGFP-α2 bind to each other in cell lysates. Next, lysates were either precipitated with anti-EGFP and blotted with anti-PSD-95 (middle panel) or precipitated with anti-PSD-95 and blotted with anti-EGFP (Figure 4F, bottom panel). These experiments show that EGFP-α2 and PSD-95 associate each other in cell lysates. Since we have shown that EGFP-α2 does not directly bind to PSD-95 (Figure 4C), EGFP-α2 likely associates with PSD-95 via its interaction with myc-α1 (Figure 4F, scheme).
Forced expression of α1- or α2-takusan increases dendritic-spine density and surface expression of GluR1 protein
Forced expression of PSD-95 in cultured neurons has been shown to promote maturation of dendritic spines and synaptic clustering of the AMPAR subunit GluR1, but not NR1 (El-Husseini et al., 2000). We thus hypothesized that the increase in the number of PSD-95 clusters observed in neurons expressing EGFP-α1 or EGFP-α2 might lead to similar consequences. To test this hypothesis, we transfected hippocampal neurons cultured for ≤7 days in vitro (DIV) with a lentiviral vector encoding EGFP in order to visualize dendritic structures. Cells were then superinfected with SFV encoding mCherry, mCherry-α1, or mCherry-α2 at 21 DIV. mCherry-α1 and mCherry-α2 are fusion proteins between mCherry and α1-takusan or α2-takusan, respectively. Twenty-four hours later, cells were fixed, the number of dendritic spines counted, and their density calculated. As shown in Figures 5A and B, forced expression of mCherry-α1 or mCherry-α2 resulted in a significant increase in the density of dendritic spines compared to control neurons. There was no significant difference between the effects of mCherry-α1 and mCherry-α2.
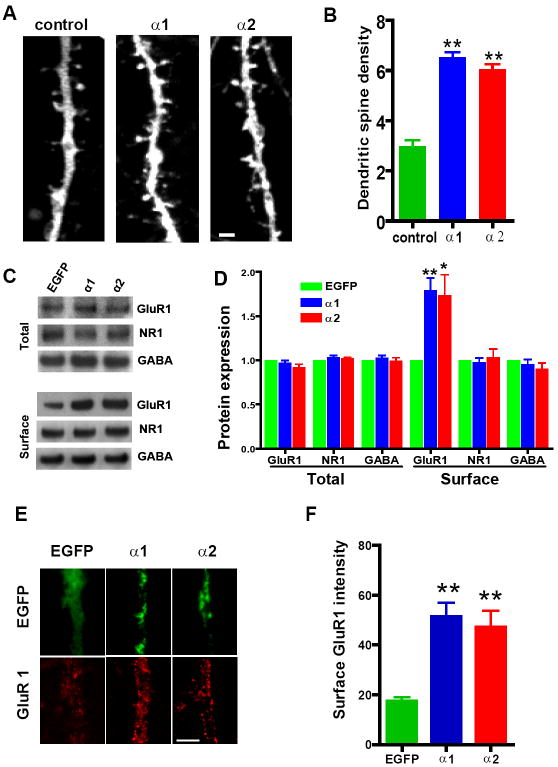
Figure 5. Forced Expression of α1- or α2-Takusan in Cultured Hippocampal Neurons Increases Dendritic-Spine Density and Surface GluR1 Labeling.
(A) Dendritic spines of cultured hippocampal neurons expressing mCherry alone (left), mCherry-α1 (center), or mCherry-α2 (right). Dendritic structures were visualized by EGFP expressed from a lentivirus. The size and number of dendritic spines increased in neurons expressing mCherry-α1 or -α2 compared to cells expressing mCherry alone. Scale bar = 5 μm. (B) Quantification of dendritic-spine densities in hippocampal cells expressing control mCherry alone (n = 7), mCherry-α1 (n = 7), or mCherry-α2 (n = 7). The density of dendritic spines (the number of spines per 10 μm) is significantly higher in cells expressing mCherry-α1- or mCherry–α2 than in control (**p < 0.01, one-way ANOVA). (C) Immunoblots of total and biotin-labeled (surface) proteins in hippocampal neurons infected with EGFP alone, EGFP-α1 or EGFP-α2. Proteins were subjected to immunoblotting using antibodies against GluR1, NR1, or α1-subunit of GABAA. (D) Quantification of total or surface GluR1, NR1, or GABAA-α1 proteins. Intensities of immunoreactive bands were quantified relative to the control bands in 4 different experiments. Surface expression of GluR1, but not NR1 or GABAA receptor subunits, significantly increased after forced expression of EGFP-α1 or EGFP-α2 (*p < 0.05, **p < 0.01, one-way ANOVA). (E) Immunocytochemistry of GluR1 surface labeling (red, bottom panel) on neurons expressing EGFP, EGFP-α1 or EGFP-α2. Scale bar = 5 μm. (F) Quantification of surface GluR1 labeling on dendrites. Mean intensity and SEM of GluR1 surface staining measured on dendritic membrane (n = 40 per sample). Forced expression of EGFP-α1 or -α2 led to an increase in GluR1 surface labeling (**p < 0.01, one-way ANOVA). Scale bar = 5 μm.
We next examined whether forced expression of EGFP-α1 or -α2 might enhance surface expression of GluR1. Twenty-four hours after transduction, we prepared total or biotin-labeled (surface) lysates from infected cells. As shown in Figure 5C top, forced expression of EGFP-α1 and EGFP-α2 showed no effect on the level of expression of GluR1, NR1 or the α1-subunit of the GABAA receptor in total cell lysates. In contrast, forced expression of EGFP-α1 or EGFP-α2 increased the level of expression of GluR1, but not that of NR1 or α1-GABAA, in biotin-labeled fractions of lysates (Figure 5C, bottom). We then quantified the intensity of bands in multiple immunoblots and determined that forced expression of either EGFP-α1 or -α2 increased surface expression of GluR1 by 1.7-1.8 fold (Figure 5D).
We next performed immunohistochemistry for surface GluR1 on neurons expressing EGFP, EGFP-α1 or EGFP-α2 (Figure 5E). We quantified the intensity of GluR1 signal on the surface of dendrites (Figure 5F) and cell bodies (data not shown). In both cases, GluR1 signal was significantly higher in neurons expressing EGFP-α1 (51.23 ± 5.71, n = 40) or EGFP-α2 (47.10 ± 6.64, n = 40) than in control neurons (17.38 ± 1.71, n = 40; p < 0.01). The effect of EGFP-α1 on GluR1 surface staining was not significantly different from that of EGFP-α2. In summary, the experiments presented in Figure 5 show that forced expression of α1- or α2- takusan in hippocampal neurons increased dendritic spine maturation and surface expression of GluR1.
Forced expression of α1- and α2- takusan differentially modifies AMPA- and NMDA-receptor properties
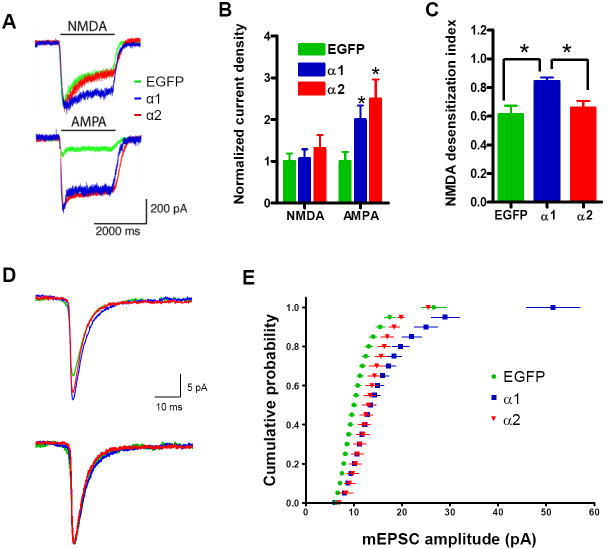
We next tested whether increased surface expression of GluR1 in neurons expressing EGFP-α1 or -α2 is accompanied by altered electrophysiological properties of these cells. To this end, we performed whole-cell patch recordings from these neurons. Figure 6 shows representative traces and a summary of the results of these electrophysiological studies. As shown in Figures 6A and 6B, AMPA-induced currents were enhanced after forced expression of EGFP-α1 (2.00 ± 0.34, n = 9) or EGFP-α2 (2.50 ± 0.46, n = 8) compared to those of control neurons (normalized to 1 ± 0.22, n = 7; p < 0.001). As an additional control, we recorded from non-transfected cells and EGFP-transfected cells under the same conditions and did not observe a significant difference (data not shown). These effects of EGFP-α1 and EGFP-α2 on AMPA-induced currents are consistent with the observation that forced expression of these molecules increased surface expression of GluR1 (Figure 5).
Figure 6. Forced Expression of α1- or α2-Takusan Differentially Modifies AMPA- and NMDA-Receptor Activity in Cultured Hippocampal Neurons.
(A) Representative traces of NMDA- and AMPA-induced whole-cell currents in neurons infected with SFV encoding EGFP (green), EGFP-α1 (blue), or EGFP-α2 (red). (B) Normalized density of NMDA- and AMPA-induced currents from neurons expressing EGFP (n = 7), EGFP-α1 (n = 9), or EGFP-α2 (n = 9 for NMDA; n = 8 for AMPA). The current density of neurons expressing EGFP was arbitrarily set at 1. (C) Desensitization index (the ratio of the steady state to the peak current amplitudes) of NMDA-induced currents in neurons expressing EGFP (n = 7), EGFP-α1 (n = 9), or EGFP-α2 (n = 9). *p < 0.05 (D) Representative traces of mEPSCs recorded from cultured hippocampal neurons after infection with SFV encoding EGFP, EGFP-α1 or -α2 (top). To compare the shape of the currents, the same traces are presented with the peaks normalized to the same point (bottom). No significant difference was found in the shape of the currents. (E) Cumulative amplitude histograms are plotted from neurons infected with EGFP (n = 12 cells; 200 events/cell), EGFP-α1 (n = 11 cells; 200 events/cell), or EGFP-α2 (n = 10 cells; 200 events/cell). mEPSC amplitude increased in EGFP-α1 and EGFP-α2 compared to EGFP-infected neurons (p < 0.001 for each by Kolmogorov–Smirnov test). The difference between the effects of α1- and α2-takusan was not significant. Values are mean ± SEM.
In contrast, steady-state NMDA current amplitude (Figures 6A and 6B) was not significantly different among neurons expressing EGFP (normalized to 1 ± 0.19, n = 7), EGFP-α1 (1.06 ± 0.22, n = 9), or EGFP-α2 (1.31 ± 0.31, n = 9). However, forced expression of EGFP-α1, but not EGFP-α2, resulted in a change in the desensitization kinetics of NMDA-induced currents (Figures 6A and 6C). Specifically, NMDA-induced currents in neurons expressing EGFP-α1 (index 0.84 ± 0.19, n = 9) exhibited less apparent desensitization than control neurons (index 0.61 ± 0.23, n = 7) and neurons expressing EGFP-α2 (index 0.66 ± 0.22, n = 9). Based on forced expression studies, α-takusan variants appear to modulate excitatory neurotransmission by two distinct mechanisms. Namely, they may enhance trafficking of AMPAR subunits to the surface via a direct or indirect interaction with PDZ-containing proteins, whereas they may modify NMDAR properties through a direct interaction with PDZ-containing proteins (i.e., mediated by α1- but not α 2-takusans).
Finally, forced expression of EGFP-α1 or EGFP-α2 in cultured hippocampal neurons increased the amplitude of AMPAR-mediated miniature excitatory postsynaptic currents (mEPSCs) (Figures 6D and 6E). Although the amplitude of these currents was enhanced by forced expression of EGFP-α1 or EGFP-α2 (Figure 6D top), the shape of currents was not significantly altered (Figure 6D bottom). The difference between the effects of α1 and α2 on the amplitude was not significant (Figure 6E). These results suggest that both α1- and α2-takusan increase the number of AMPAR subunits at functional synapses.
Transfection of hippocampal neurons with siRNAs that target α-takusan variants reduces surface expression of GluR1
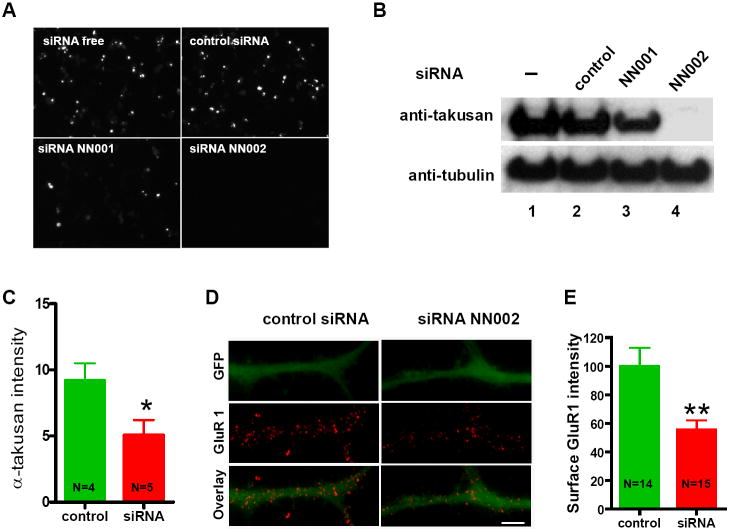
We next tried to interfere with the activity of endogenous α-takusan molecules using RNAi methods. We designed siRNAs that would knock down as many α-takusan variant mRNAs as possible in the following manner. We analyzed the coding sequence of α1-takusan using siRNA designing programs and identified two 21-nt long sequences, NN001 and NN002 (see Supplemental Experimental Procedures for their sequences), each of which was completely conserved among all of the α-takusan variant cDNAs that we isolated. As expected, the mouse genome carries multiple sites that are homologous to these sequences. Specifically, we found 35 and 54 sites that showed 100% homology with the NN001 and NN002 sequences, respectively. We examined the sequences surrounding all of these sites, and determined that they all encoded α-takusan variants. Furthermore, 16 out of 18 α-takusan variant cDNAs that are presented in Supplemental Figure 3B manifest a perfect match with NN002, while the other two have a single-base mismatch. These observations indicate that NN001 and NN002 siRNAs, particularly the latter, can potentially knock down most, if not all, of the endogenous α-takusan variants.
We co-transfected HEK293 cells with NN001, NN002 or a negative control siRNA (siCONTROL™ non-targeting pool, Dharmacon), together with an expression vector carrying EGFP-α1. Figures 7A and 7B show fluorescence microscopy and Western blotting using anti-takusan antibody performed 24 hours after the transfection. While NN001 reduced the level of newly synthesized EGFP-α1 by ~50% compared to control, NN002 eliminated EGFP-α1 expression altogether (within the limits of protein detection).
Figure 7. α-Takusan siRNA Knocks Down Exogenous Expression of α1-Takusan in HEK293T Cells and Reduces Surface Expression of GluR1 in Cultured Neurons.
(A and B) HEK293 cells were transfected with EGFP-α1 alone or EGFP-α1 together with control siRNA (siCONTROL), siRNA NN001, or siRNA NN002. Twenty-four hours after transfection, cells were examined under fluorescence microscopy (A) or subjected to immunoblotting (B) using anti-takusan or anti-tubulin antibodies. The results show that the level of exogenously expressed α1-takusan was significantly reduced by both siRNAs but not control. NN002 was more effective than NN001 in the ability to knock down α1-takusan expression. (C to E) Effects of NN002 transfection into cultured hippocampal neurons at 12 DIV. The EGFP plasmid (pEGFP-N2) was co-transfected in these cells to visualize cell bodies and dendrites. (C) Twenty-four hours after siRNA transfection, immunocytochemistry using anti-α-takusan was performed and signal intensity quantified. Transfection of NN002 reduced α-takusan signals by ~50% compared to transfection of siCONTROL. (D) Surface staining of GluR1 on dendrites treated with siCONTROL or NN002. Scale bar = 5 μm (E) Quantification of the intensity of surface GluR1 signal localized over dendrites filled with EGFP. Transfection of NN002 into cultured neurons reduced the surface expression of GluR1 by ~50% in the dendrites of these cells compared to control siRNA transfection.
We next examined the effects of NN002 on cultured hippocampal neurons. We transfected neurons with siRNAs together with an EGFP expression vector (pEGFP-N2, Clontech). We used the latter vector in order to visualize transfected neurons and their dendrites. Initially, we assessed the effects of NN002 on the expression levels of α-takusan proteins. As shown in Figure 7C, transfection of NN002 in cultured hippocampal neurons reduced the intensity of α-takusan immune signals by ~50% compared to that in neurons treated with siCONTROL, indicating that NN002 effectively knocked down endogenous takusan. The level of knock down by NN002 in cultured neurons was not as efficient as observed in HEK293 cells. This is likely due to the fact that there are pre-existing α-takusan proteins in cultured neurons prior to siRNA treatment, while HEK293 cells do not express endogenous α-takusan proteins. Alternatively, α-takusan variants may exist in neurons that are not entirely knocked down by the NN002 siRNA.
Having established that NN002 significantly reduces the level of α-takusan proteins in cultured hippocampal neurons, we next performed surface staining for GluR1 on these neurons (Figure 7D) and quantified the intensity of GluR1 signal localized over dendritic processes filled with EGFP (Figure 7E). The results show that transfection of cultured neurons with NN002 reduced the surface expression of GluR1 by ~50% in the dendrites. Since the NN002 sequence bore no sequence homology with GluR1 mRNAs, the reduction in GluR1 surface expression in these cells was likely caused by the knock down of α-takusan proteins. The loss of function observed in these experiments suggests that endogenous α-takusan proteins facilitate surface expression of AMPAR subunits such as GluR1.
Transfection of hippocampal neurons with shRNAs that target α-takusan variants reduces GluR1 surface expression, PSD-95 synaptic clusters, and dendritic filopodia-like protrusions
We next utilized shRNA technology via infection with lentiviral vectors because this method has the advantage of achieving significantly higher transduction efficiencies in cultured neurons compared to transfection methods available for siRNA. The shRNA sequence derived from the NN002 siRNA sequence was inserted downstream to the pH1 promoter in a lentiviral vector that also contained a cDNA encoding EGFP driven by the human phosphoglycerate kinase promoter (hPGK). This approach allowed us to identify infected cells by their EGFP expression.
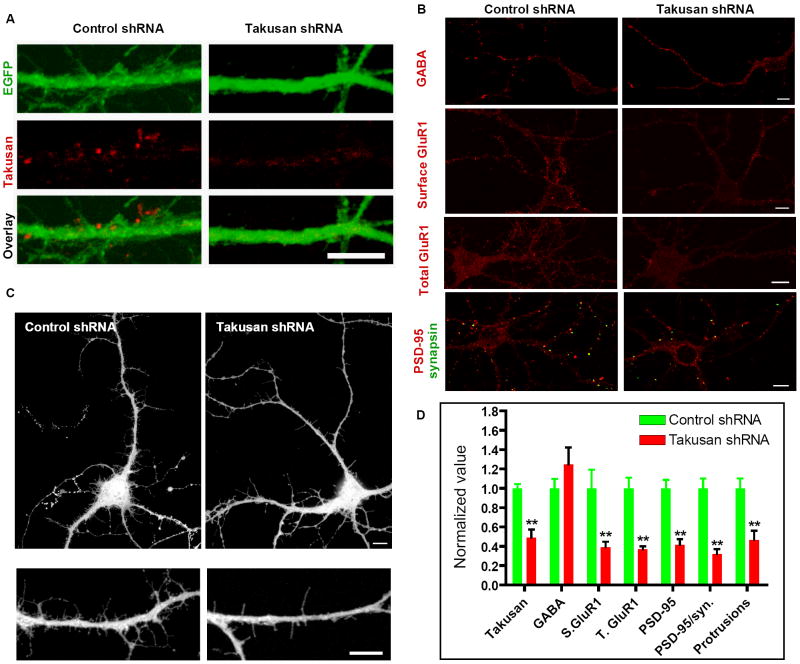
We infected cultured hippocampal neurons with lentiviral vectors encoding scrambled non-targeting control shRNA or α-takusan shRNA, and stained them with anti-α-takusan antibody (Figure 8A). Quantification of the signals showed that α-takusan expression was significantly reduced in neurons expressing α-takusan shRNA (0.49 ± 0.08; n = 10; value normalized to control) compared to neurons expressing control shRNA (1 ± 0.05; n = 10; Figure 8D). This result shows that the shRNA effectively knocked down α-takusan proteins in these neurons. We next examined the effect of the α-takusan shRNA on the expression of GABAA receptors by immunocytochemistry. Figure 8B (top panel) presents α1-GABAA receptor immunostaining of hippocampal neurons expressing control shRNA or α-takusan shRNA. Quantification of the signals revealed no significant difference between cells expressing α-takusan shRNA (1.25 ± 0.18; n = 10) and cells expressing control shRNA (1 ± 0.10; n = 10). This result is consistent with the observation that α-takusan does not co-localize with GABAA receptors in neurons (Figure 4B). It is also consistent with the finding that forced expression of α-takusan did not alter GABAA receptor expression (Figures 5C and 5D).
Figure 8. α-Takusan shRNA Knocks Down Endogenous α1-Takusan and Alters Dendritic Phenotype in Cultured Neurons.
(A) Reduction of α-takusan protein in neurons infected with a lentiviral vector expressing α-takusan shRNA. Cultured hippocampal neurons were infected with lentiviral vectors encoding scrambled non-targeting-control or α-takusan shRNA. The vectors carried EGFP cDNA whose expression allowed identification of infected cells. (B) Effects of α-takusan shRNA on GABAA receptor, surface GluR1, total GluR1, and PSD-95/synapsin signals. Cultured hippocampal neurons were infected with lentiviral vectors encoding control or α-takusan shRNA. Cells were subsequently subjected to immunohistochemistry using various antibodies as indicated. The images obtained here were used for the quantitative analyses presented in D, below. (C) Effect of α-takusan shRNA on dendritic morphology. Neurons infected with lentiviral vectors encoding control or α-takusan shRNA were identified by expression of EGFP and their images captured by confocal microscopy. The gross cell morphology was not affected by α-takusan shRNA (top panel). The number of dendritic filopodia-like protrusions, however, appeared to be reduced by α-takusan shRNA (bottom panel). (D) Quantification of immunosignals, PSD-95 clustering, and dendritic morphology in neurons infected with lentivirus expressing control or α-takusan shRNA. For the first four columns, the density of immunosignal for α-takusan, GABAA, surface GluR1 (S. GluR1), or total GluR1 (T. GluR1) was quantified in cells expressing control or α-takusan shRNA. For the next two columns, the density of PSD-95 clustering and PSD-95 clustering juxtaposed to synapsin was quantified. For the last column, the density of dendritic filopodia-like protrusions was quantified. For each measurement, the values obtained for cells expressing α-takusan shRNA were normalized to the mean value obtained for cells expressing control shRNA. Expression of α-takusan shRNA did not alter GABAA receptor signal in these neurons, but reduced signal intensities for α-takusan, surface GluR1, and total GluR1. Similarly, α-takusan shRNA reduced the density of PSD-95 aggregates, PSD-95 aggregates juxtaposed to synapsin, and dendritic protrusions in these neurons. **p < 0.01; Scale bars = 10 μm.
We next examined the effects of α-takusan shRNA on events that we had observed were altered with forced expression of α-takusan cDNAs in hippocampal neurons. Specifically, using immunocytochemistry we examined surface expression of GluR1, and synaptic clustering of PSD-95 in neurons expressing α-takusan (Figure 8B). When quantified (Figure 8D), the intensity of surface GluR1 signal in neurons expressing α-takusan shRNA (0.39 ± 0.06; n = 12) was significantly lower than that in control neurons (1.0 ± 0.19; n = 12). Surprisingly, the intensity of total GluR1 signal in neurons expressing α-takusan shRNA (0.37 ± 0.03; n = 8) was reduced compared to control neurons (1.0 ± 0.11; n = 8). We then quantified PSD-95 clustering by measuring the density of PSD-95 aggregates or the density of PSD-95 aggregates juxtaposed to synapsin signals. The density of PSD-95 aggregates in neurons expressing α-takusan shRNA (0.41 ± 0.06; n = 10) was significantly lower than that in control neurons (1.0 ± 0.09; n = 10). Similarly, the density of PSD-95 aggregates juxtaposed to synapsin in neurons with α-takusan shRNA (0.32 ± 0.05; n = 10) was lower than that in control cells (1.0 ± 0.10; n = 10).
Finally, these neurons were examined for dendritic morphology (Figure 8C). For technical reasons, the neurons we used in shRNA experiments were young and did not yet possess mature dendritic spines. Nonetheless, the effect of α-takusan shRNA expression was evident when we quantified the number of dendritic filopodia-like protrusions (Figure 8D), which are known to be harbinger for spine formation (Papa and Segal, 1996). The density of these protrusions was significantly reduced in neurons expressing α-takusan shRNA (0.47 ± 0.10; n = 6) compared to control cells (1.0 ± 0.10; n = 8). The ramifications of these observations are discussed below.
Discussion
We have identified a new gene family, designated α-takusan, whose expression is upregulated in the brains of NR3A KO mice compared to WT mice. Since the absence of NR3A has been shown to increase NMDAR activity (Das et al., 1998; Sasaki et al., 2002), our findings are consistent with the notion that upregulation of α-takusan gene expression is likely caused by NMDAR hyperactivation in mice lacking NR3A. It is also possible, however, that takusan upregulation in NR3A KO mice might be caused by downregulation of the putative excitatory glycine-receptor activity composed of NR1 and NR3A subunits (Chatterton et al., 2002). Either way, the observation that upregulation of α-takusan occurs in areas of the brain where NR3A is normally expressed, suggests that these effects are likely cell autonomous.
In our microarray experiments, Q-PCR determinations, and Northern blot analyses, the probes we used do not distinguish among the various α-takusan variants. Therefore, if only a small fraction of the takusan genes are regulated by an NR3A-dependent mechanism, our results may underestimate the true impact of NR3A deletion.
Intriguingly, we have found that multiple α-takusan variants are expressed in the mouse brain. From WT adult brain and E18-cultured neurons, we have isolated cDNAs encoding 46 different α-takusan variants. It should be noted that our cloning strategy targeted narrow branches of the α-takusan family due to inherent biases imposed by the PCR primers used in our studies. Furthermore, we limited our RNA sources to tissues extracted from forebrain and from cultured hippocampal or cortical neurons. Other areas of the brain that express α-takusan mRNA such as the cerebellum might express unique and additional sets of α-takusan variants.
Despite our best efforts to minimize the occurrence of mutations during the PCR, we cannot completely exclude the possibility that at least some variations were introduced. However, several lines of evidence suggest that most of the variations observed in α–takusan sequences are authentic: (1) Variations occur only at certain positions in the sequence. (2) Variations are reproducible from one PCR reaction to another. (3) Each variation found in the PCR products is present in genome sequences deposited in GenBank.
We have made some intriguing, perhaps even peculiar, observations regarding the diversity of the α-takusan PCR products that we isolated. Cortical cell #4 (Table 1), for example, expressed three variants (α34-36) that are extremely similar to one other, presumably indicating similar protein properties. Again, these variations are likely to be authentic because they occur at positions where the same variations are present in the genome. However, the physiological role in the nervous system of such subtle diversity remains to be seen. The analysis presented in Supplemental Figure 3B shows that the α-takusan family is merely one branch of a much larger takusan gene family. Some non-α-takusan cDNAs were isolated from the whole body, testis, ovary, uterus, mammary tumors, and spinal cord of mice and rats. Eight of these genes have been reported to be members of a family of testis-specific genes (Spiess et al., 2003).
Single-cell PCR experiments from hippocampal and cortical cultures show that an individual neuron expresses more than one α-takusan variant, and different cells express disparate variants (Table 1). Furthermore, there might be a cell-type specific preference in the repertoire of these variants. A note or caution, however, is appropriate in that that the repertoire of α-takusan expression we observe in cultured neurons may not reflect the situation in vivo. This would be particularly true if electrical activity modifies the repertoire of expression. Further studies are needed to determine the expression of α-takusan variants in vivo and if such expression is cell-type dependent.
In the present study, we chose two variants, α1 which directly binds PSD-95 and α2 which does not, for further gain-of-function studies. When these proteins were forced expressed in cultured neurons as fusion proteins with EGFP, both α1 and α2 led to an increase in the number of synaptic PSD-95 clusters, density of dendritic spines, surface expression of GluR1, AMPA-induced whole-cell currents and AMPAR-mediated mEPSCs. It has been established that the abundance of AMPA receptors correlates with the size of the synapse and dimensions of the dendritic spine head (Matsuzaki et al., 2001; Nusser, 2000; Takumi et al., 1999) and overexpression of GluR2 in hippocampal neurons increases spine size and density (Passafaro et al., 2003). Together with the observation that forced expression of PSD-95 also leads to increased dendritic spine density, synaptic expression of GluR1 and AMPAR-mediated EPSCs (El-Husseini et al., 2000), the effects of takusan proteins on PSD-95 clustering, dendritic spine density and GluR1 surface expression are likely related.
An important question concerns the mechanism whereby α1- and α2-takusan induce synaptic events in transfected neurons. We found evidence that α1-takusan directly associates with PSD-95 to reorganize postsynaptic density complexes. Although EGFP-α2-takusan did not directly bind to PSD-95 in our experiments, we found that EGFP-α2 (but not EGFP alone) could form a complex with PSD-95 in the presence of α1. Thus, it is probable that forced expression of α2 leads to dimerization with endogenous α-takusan variants (such as α1 or α5), which in turn bind to PSD-95; these α-takusan dimers or oligomers may well be responsible for the observed increase in PSD-95 clustering and other downstream effects.
We also found that EGFP-α1, but not EGFP-α2, modifies desensitization of NMDA-induced currents. Thus, the effect on NMDA-evoked currents is likely mediated by direct interaction between EGFP-α1 and PDZ-containing proteins since α2 does not manifest this protein-protein interaction. Along these lines, overexpression of PSD-95 has been shown to reduce desensitization of NMDA-induced currents in immature neurons independent of NMDAR subunit composition (Li et al., 2003). Alternatively, EGFP-α1, together with PSD-95 or another PDZ-containing protein, might modify the composition of NR2-containing receptors that could alter desensitization of NMDA-induced currents.
Additionally, we have previously reported that cortical neurons of layer V in NR3A KO mice manifest increased dendritic spine density (Das et al., 1998). In the present study, we show that α-takusan mRNAs are upregulated in the same location in NR3A KO mice, and forced expression of α-takusan increases synaptic clustering of PSD-95 and large-headed spine density. Thus, α-takusan may be involved in the manifestation of this dendritic-spine phenotype in NR3A KO mice. Perhaps somewhat surprisingly, we had not observed increased AMPA currents in cortical neurons isolated from NR3A KO mice relative to WT neurons (Das et al., 1998). It is possible, however, that loss of NR3A leads to additional events other than the upregulation of α-takusan in these neurons, and thus molecular contexts of NR3A-deficient cortical neurons might differ from those of cultured hippocampal neurons transfected with α-takusan.
Finally, we performed loss-of-function experiments using siRNAs and shRNAs that can target the majority of α-takusan variants. These experiments showed that knock down of endogenous α-takusan in hippocampal neurons generally manifested the opposite effects of overexpression or gain-of-function. Namely, expression of α-takusan shRNAs in hippocampal cultured neurons led to reduction in surface expression of GluR1, synaptic clustering of PSD-95, and density of dendritic filopodia-like protrusions. Surprisingly, total GluR1 staining is also reduced by α-takusan shRNA expression. Therefore, the loss of GluR1 surface staining in these experiments might reflect down regulation of protein levels.
We have thus far identified extreme amplifications of takusan genes only in mice and rats. Our preliminary study indicates that the rat genome contains >400 loci encoding takusan variants. A detailed analysis of rat takusan genes is a subject of future study. Possibly related to our discovery of the takusan family, it was recently reported that the human genome contains a previously unknown gene family that is expressed in the brain (Popesco et al., 2006). Members of that gene family encode proteins displaying multiple copies of DUF1220, which incidentally has no structural homology to DUF622 of the takusans. Analogous to the takusan gene family in rodents, however, the DUF1220-encoding gene family is human-lineage specific. Although it is not clear whether takusan and DUF1220 proteins share functional similarities, the mechanisms by which both gene families tremendously amplified their size during evolution are likely to be similar.
In summary, α-takusan proteins are likely to be involved in the organization of postsynaptic molecules. Since disparate α-takusan variants are expressed in different types of neurons, it is possible that the binding partners and organizational attributes of the α-takusan may vary from cell to cell. Moreover, the structural diversity of α-takusan variants expressed in these neurons might contribute to the functional diversity of these cells. In the future, generating mutant mice in which the various α-takusan variants are knocked down will help identify additional cellular functions of this newly recognized gene family.
Experimental Procedures
See Supplemental Experimental Procedures for details of cDNA microarray, in situ hybridization, Northern Blots, isolation of α-takusan cDNAs, single-cell RT-PCR, immunoprecipitation, immunocytochemistry, electrophysiological recordings, siRNA and shRNA treatments.
Cell Culture
Low-density primary hippocampal and cerebrocortical cultures were prepared from newborn or E16-18 mice and maintained in a modified neural basal medium. See Supplemental Experimental Procedures for more details.
Electrophysiological Recordings
Conventional whole-cell recordings were performed to record GABA, AMPA, or NMDA induced currents and AMPA mEPSCs on cultured hippocampal neurons. See Supplemental Experimental Procedures for details.
siRNA and shRNA Treatment
Takusan siRNAs NN001 and NN002 were designed using siRNA Target Finder (Ambion). For siRNA experiments, we used chemically-synthesized dsRNAs. For shRNA experiments, NN002 or its scrambled non-targeting shRNA was expressed from a lentiviral vector. See Supplemental Experimental Procedures for details.
Supplementary Material
Acknowledgments
We would like to thank Dr. Alexey Terskikh for providing us lentiviral vectors and Dr. Ruchi Bajpai for technical advice. This work was supported in part by NIH grants P01 HD29587, R01 EY05477 (to S.A.L.) and K12AG00975 (to G.T.). B.S. was supported by a fellowship from the PhRMA foundation. S.A.L. was a Senior Scholar in Aging Research of the Ellison Medical Research Foundation.
Footnotes
Accession Numbers The accession numbers for the forty-six α–takusan genes (α1 to α46) described in this paper are EF651798 to EF651843.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barry MF, Ziff EB. Receptor trafficking and the plasticity of excitatory synapses. Curr Opin Neurobiol. 2002;12:279–286. doi: 10.1016/s0959-4388(02)00329-x. [DOI] [PubMed] [Google Scholar]
- Carninci P, Hayashizaki Y. High-efficiency full-length cDNA cloning. Methods Enzymol. 1999;303:19–44. doi: 10.1016/s0076-6879(99)03004-9. [DOI] [PubMed] [Google Scholar]
- Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, et al. Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature. 2002;415:793–798. doi: 10.1038/nature715. [DOI] [PubMed] [Google Scholar]
- Ciabarra AM, Sullivan JM, Gahn LG, Pecht G, Heinemann S, Sevarino KA. Cloning and characterization of chi-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J Neurosci. 1995;15:6498–6508. doi: 10.1523/JNEUROSCI.15-10-06498.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen C, Parry DA. Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, Dikkes P, Conner DA, Rayudu PV, Cheung W, et al. Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature. 1998;393:377–381. doi: 10.1038/30748. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- Li B, Otsu Y, Murphy TH, Raymond LA. Developmental decrease in NMDA receptor desensitization associated with shift to synapse and interaction with postsynaptic density-95. J Neurosci. 2003;23:11244–11254. doi: 10.1523/JNEUROSCI.23-35-11244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton SA, Rosenberg PA. Excitatory amino acids as a final common pathway for neurologic disorders. N Engl J Med. 1994;330:613–622. doi: 10.1056/NEJM199403033300907. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Nicoll RA. Long-term potentiation--a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Ellis-Davies GC, Nemoto T, Miyashita Y, Iino M, Kasai H. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2001;4:1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Manabe T, Watanabe M, Satoh Y, Suzuki N, Toki S, Nakamura K, Yagi T, Kushiya E, Takahashi T, et al. Role of the carboxy-terminal region of the GluR epsilon2 subunit in synaptic localization of the NMDA receptor channel. Neuron. 1998;21:571–580. doi: 10.1016/s0896-6273(00)80567-x. [DOI] [PubMed] [Google Scholar]
- Nicoll RA, Tomita S, Bredt DS. Auxiliary subunits assist AMPA-type glutamate receptors. Science. 2006;311:1253–1256. doi: 10.1126/science.1123339. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP. PDZ domain proteins: plug and play! Sci STKE. 2003;2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- Nusser Z. AMPA and NMDA receptors: similarities and differences in their synaptic distribution. Curr Opin Neurobiol. 2000;10:337–341. doi: 10.1016/s0959-4388(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa M, Segal M. Morphological plasticity in dendritic spines of cultured hippocampal neurons. Neuroscience. 1996;71:1005–1011. doi: 10.1016/0306-4522(95)00490-4. [DOI] [PubMed] [Google Scholar]
- Passafaro M, Nakagawa T, Sala C, Sheng M. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature. 2003;424:677–681. doi: 10.1038/nature01781. [DOI] [PubMed] [Google Scholar]
- Perez-Otano I, Lujan R, Tavalin SJ, Plomann M, Modregger J, Liu XB, Jones EG, Heinemann SF, Lo DC, Ehlers MD. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat Neurosci. 2006;9:611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF. Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J Neurosci. 2001;21:1228–1237. doi: 10.1523/JNEUROSCI.21-04-01228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff GJ, Sikela JM. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- Sasaki YF, Rothe T, Premkumar LS, Das S, Cui J, Talantova MV, Wong HK, Gong X, Chan SF, Zhang D, et al. Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J Neurophysiol. 2002;87:2052–2063. doi: 10.1152/jn.00531.2001. [DOI] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci U S A. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M. Molecular organization of the postsynaptic specialization. Proc Natl Acad Sci U S A. 2001;98:7058–7061. doi: 10.1073/pnas.111146298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Song I, Huganir RL. Regulation of AMPA receptors during synaptic plasticity. Trends Neurosci. 2002;25:578–588. doi: 10.1016/s0166-2236(02)02270-1. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Spiess AN, Walther N, Muller N, Balvers M, Hansis C, Ivell R. SPEER--a new family of testis-specific genes from the mouse. Biol Reprod. 2003;68:2044–2054. doi: 10.1095/biolreprod.102.011593. [DOI] [PubMed] [Google Scholar]
- Steigerwald F, Schulz TW, Schenker LT, Kennedy MB, Seeburg PH, Kohr G. C-Terminal truncation of NR2A subunits impairs synaptic but not extrasynaptic localization of NMDA receptors. J Neurosci. 2000;20:4573–4581. doi: 10.1523/JNEUROSCI.20-12-04573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll M, Corneliussen B, Costello CM, Waetzig GH, Mellgard B, Koch WA, Rosenstiel P, Albrecht M, Croucher PJ, Seegert D, et al. Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet. 2004;36:476–480. doi: 10.1038/ng1345. [DOI] [PubMed] [Google Scholar]
- Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, Wu MK, Yuan JP, Jones EG, Lipton SA. Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J Neurosci. 1995;15:6509–6520. doi: 10.1523/JNEUROSCI.15-10-06509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucher NJ, Deitcher DL. PCR and patch-clamp analysis of single neurons. Neuron. 1995;14:1095–1100. doi: 10.1016/0896-6273(95)90257-0. [DOI] [PubMed] [Google Scholar]
- Takumi Y, Ramirez-Leon V, Laake P, Rinvik E, Ottersen OP. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat Neurosci. 1999;2:618–624. doi: 10.1038/10172. [DOI] [PubMed] [Google Scholar]
- Wong HK, Liu XB, Matos MF, Chan SF, Perez-Otano I, Boysen M, Cui J, Nakanishi N, Trimmer JS, Jones EG, et al. Temporal and regional expression of NMDA receptor subunit NR3A in the mammalian brain. J Comp Neurol. 2002;450:303–317. doi: 10.1002/cne.10314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.