Abstract
Accurate measurements of transcript abundance are a prerequisite to understand gene activity in development. Using the NanoString nCounter, an RNA counting device, we measured the prevalence of 172 transcription factors and signaling molecules in early sea urchin development. These measurements show high fidelity over more than five orders of magnitude down to a few transcripts per embryo. Most of the genes included are locally restricted in their spatial expression, and contribute to the divergent regulatory states of cells in the developing embryo. In order to obtain high-resolution expression, profiles from fertilization to late gastrulation samples were collected at hourly intervals. The measured time courses agree well with, and substantially extend, prior relative abundance measurements obtained by quantitative PCR. High temporal resolution permits sequences of successively activated genes to be precisely delineated providing an ancillary tool for assembling maps of gene regulatory networks. The data are available via an interactive website for quick plotting of selected time courses.
Keywords: Transcription factor, Gene expression time course, mRNA prevalence measurement, Embryogenesis
1. Results and discussion
Measurement of transcript prevalence is a direct method to assess gene activity. In conjunction with spatial expression data, it enables a targeted approach to identify genes that contribute to a developmental process. Prevalence data are of particular usefulness if sampling intervals are short enough to yield time courses with high temporal resolution. When genes are activated in close succession such data can reveal the order in which activation occurs. When regulatory genes are profiled these data can be used to establish a very preliminary flow of regulatory information (Materna and Oliveri, 2008). This limits the number of possible regulatory interactions and may directly point at previously unknown regulatory linkages. High resolution expression data may also guide perturbation experiments that directly reveal the regulatory relationships between genes, as accurate knowledge of gene expression profiles allows optimal time points for sampling to be chosen, thus maximizing information gain and robustness of results (Materna and Oliveri, 2008; Revilla-i-Domingo et al., 2007; Smith and Davidson, 2008).
Previous expression profiling efforts in sea urchin were aimed at finding all regulatory genes that are active during early development (Howard-Ashby et al., 2006a, b; Materna et al., 2006; Rizzo et al., 2006; Tu et al., 2006). While these data indicate approximately when genes are being used they do not provide the needed temporal resolution. To provide a set of measurements that could be used for the above purposes we collected sea urchin embryos at hourly intervals and determined transcript prevalence for the majority of regulatory genes that are expressed in a localized manner up to mid-gastrulation (36 hours post fertilization). We counted embryos and added known quantities of external RNA standards to the embryo lysate to yield absolute numbers of transcripts per embryo.
For transcript quantification we made use of the NanoString nCounter (NanoString Technologies, Seattle, WA), a high-throughput RNA molecule counter. The instrument is based on hybridization of gene specific probes to total RNA eliminating the need for a reverse transcription step (Geiss et al., 2008). The instrument visualizes individual hybridized probes, and identifies the transcript species based on a fluorescent bar code. The counts for each transcript species are linearly proportional to its prevalence in the added RNA solution achieving sensitivity comparable to QPCR (Geiss et al., 2008). Using the nCounter, we obtained an extensive collection of high-fidelity expression profiles with high temporal resolution and greatly increased depth.
1.1. Codeset design
The NanoString nCounter identifies and counts RNA species based on a fluorescent barcode attached to a sequence specific hybridization probe (Geiss et al., 2008). Our probe set includes 172 genes covering the majority of active, and spatially restricted regulatory genes in the early Strongylocentrotus purpuratus embryo up to mid-gastrulation (36 hours postfertilization)(Howard-Ashby et al., 2006a, b; Materna et al., 2006; Rizzo et al., 2006; Tu et al., 2006). Table 1 summarizes the gene families represented in the codeset; a full list of genes, accession numbers, and probe sequences is available as Supplementary Table 1. Briefly, the codeset contains probes for 142 mRNAs encoding transcription factors, 20 mRNAs encoding signaling ligands or receptors, and 15 mRNAs used as markers or standards. Of the 142 transcription factors, 115 are known to be localized for at least part of early development whereas only four are expressed ubiquitously. The spatial expression pattern of the remaining 23 is unknown. For two transcription factors (otx, blimp/krox) two splice forms were included by designing probes against unique exons. Of the signaling related genes 14 are known to be spatially restricted while the rest are undetermined. Included in the list of transcription factors are 30 C2H2 zinc finger proteins, only half of which have clear orthologs with known transcriptional regulatory activity (Materna et al., 2006). Probes were designed against confirmed cDNA sequences where available. Gene predictions only were available for 67 genes, but these genes had been previously shown to be expressed by QPCR (Howard-Ashby et al., 2006a, b; Materna et al., 2006; Rizzo et al., 2006; Tu et al., 2006). For these genes, probes were designed in the immediate vicinity of the QPCR amplicon. For all genes a suitable probe was picked by NanoString in house probe design.
Table 1.
Contents of the NanoString nCounter codeset listed by gene family. A full list is given as supplementary online material.
| Family | No. of Genes |
|---|---|
| bHLH | 15 |
| bZIP | 4 |
| C2H2 zinc finger | 30 |
| ETS | 7 |
| Forkhead | 15 |
| Homeobox | 39 |
| Nuclear Hormone Receptor | 6 |
| SOX/HMG | 7 |
| T-Box | 4 |
| Miscellaneous TFs | 15 |
| Signaling - Ligands and antagonists | 16 |
| Signaling - Receptors | 3 |
| Markers/Standards | 11 |
1.2. Prevalence measurements with the NanoString nCounter
To determine the dynamic range of nCounter quantification assays we spiked 100 ng of sea urchin total RNA with different amounts of in vitro transcribed GFP and RFP RNA spanning a range of several orders of magnitude in concentration. As has been reported previously, the NanoString counts show a linear proportional relationship to transcript abundance in the hybridization reaction over more than five orders of magnitude (Geiss et al., 2008). With RNA from 200 embryos added to the hybridization reactions for the time course measurements below, no gene in our codeset reached a transcript level high enough to exceed the linear range. We determined the technical error associated with our measurements and confirmed that variation between technical replicates is minimal even for low counts as reported previously (Geiss et al., 2008).
The background resulting from non-specific interactions between capture probes and detection probes was determined in four runs without RNA and was found to be insignificant. Across the probe set the median of the probe specific background amounts to just three counts. It reaches double digits for only 10% of genes (Fig. S1). Using Poisson statistics with λ = 5 the probability of observing 11 or more counts is <0.01. For comparison, 50 NanoString counts correspond to 25 transcripts per embryo, a transcript level that is exceeded by even very lowly expressed genes such as scl and snail. Only six probes have a background count of 17 or higher, with 77 counts being the highest background observed (nfe2, see Fig. S1). Even for these genes the background is negligible compared to their expression level that in all cases reaches several thousand transcripts per embryo. In conclusion, with 200 embryos per hybridization reaction the detection limit lies at a few transcripts per embryo for virtually all genes included in the codeset.
All quantification of nucleic acids based on hybridization of sequence specific probes is susceptible to slight variation in hybridization efficiencies. According to the manufacturer NanoString probes may exhibit variation in hybridization efficiencies of up to two-fold, but this may be exceeded by a few outliers. Although we have not tested the hybridization efficiency for each probe, the good agreement with prior QPCR data (see next section) underlines the validity of our measurements. To illustrate this point: For the time course measurements described in the next section 107 GFP and RFP molecules were added as an external standard. Based on the reported recovery efficiency of ~1% (Geiss et al., 2008), GFP and RFP were expected to produce about 100,000 counts. Indeed, GFP and RFP together produce a mean of almost exactly 100,000 counts after adjusting for hybridization efficiency. However, the counts for GFP are about 20% higher, while those for RFP are lower by roughly the same margin. Thus, even though very low abundance transcripts can be detected reproducibly with high fidelity, the absolute numbers of transcripts per embryo have to be viewed as an approximation.
1.3. NanoString nCounter expression profiling with high temporal resolution
The probe set described above was used to collect expression data from fertilization to late gastrulation (48 hours postfertilization). Samples were drawn at hourly intervals to capture in detail the dynamics of gene expression throughout early development. The relatively slow rate of transcription at 15°C in Strongylocentrotus purpuratus (Davidson, 1986) causes a considerable lag in subsequent gene activation events (Ben-Tabou de-Leon and Davidson, 2009; Bolouri and Davidson, 2003). Thus, this sampling interval is sufficient to reveal the order in which genes are activated even where genes become transcribed in close succession. The counts obtained in the NanoString nCounter assay were normalized using the average counts for the external standards GFP and RFP to yield absolute numbers of transcripts per embryo (see Materials and methods). Examples of the measured time courses are shown in Fig. 1, 2 and 3; all 172 expression profiles are presented in Fig. S2.
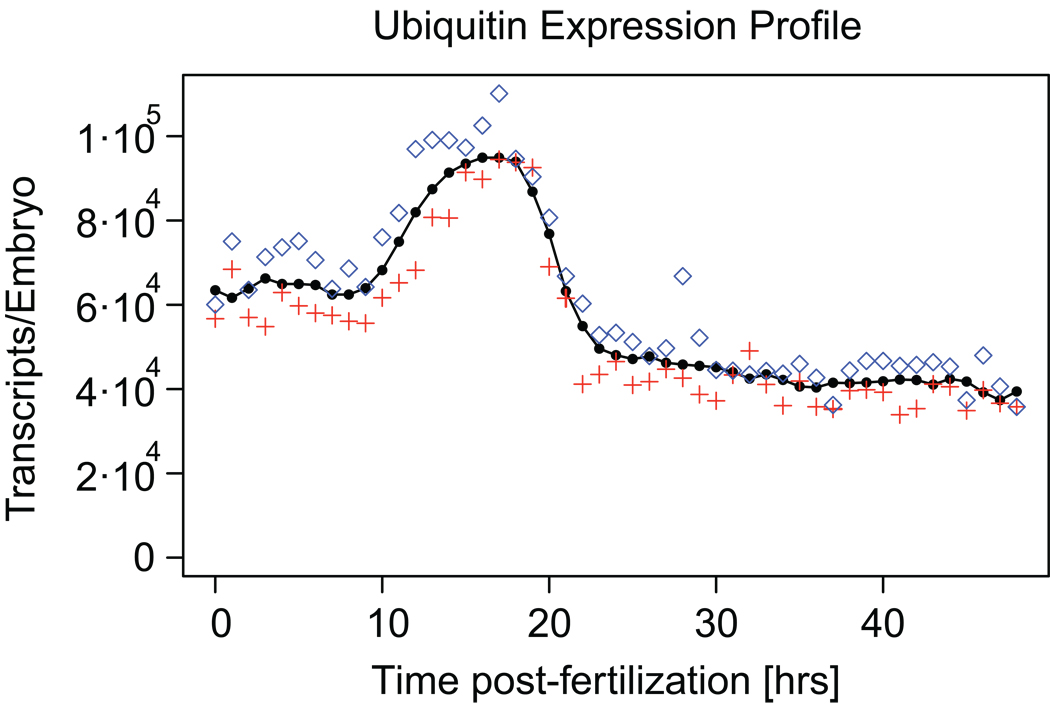
Fig. 1.
Expression profile for the ubiquitin domain. This domain has been used as an internal standard in previous expression profiling efforts (Howard-Ashby et al., 2006a, b; Materna et al., 2006; Rizzo et al., 2006; Tu et al., 2006). The average prevalence is shown in black, data for each of the two embryo batches are shown in red and blue.
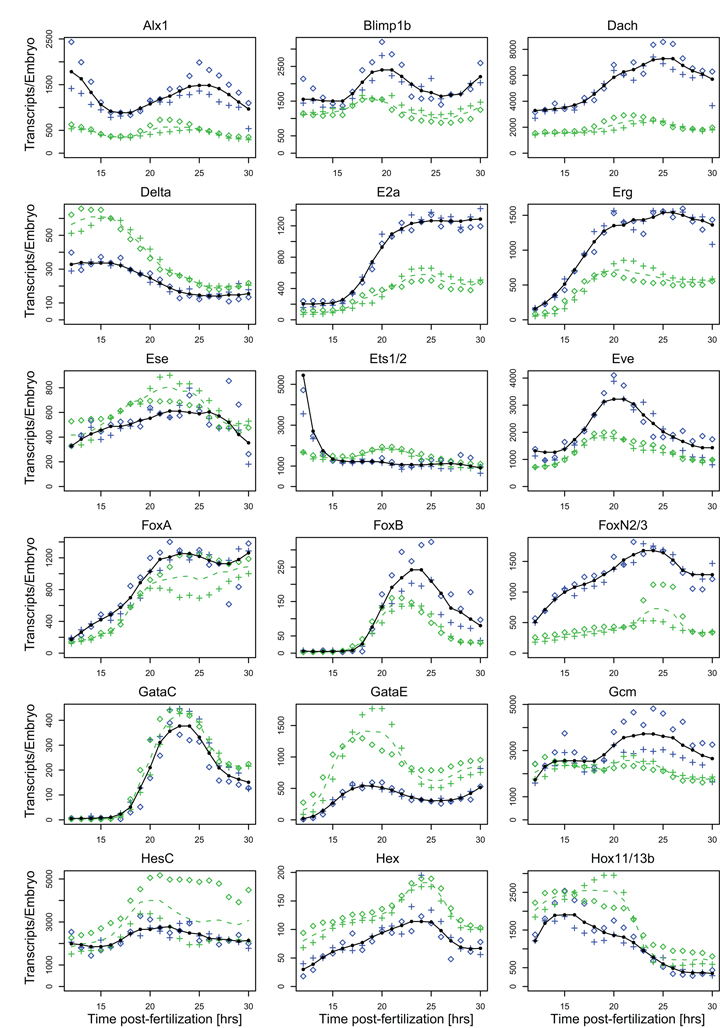
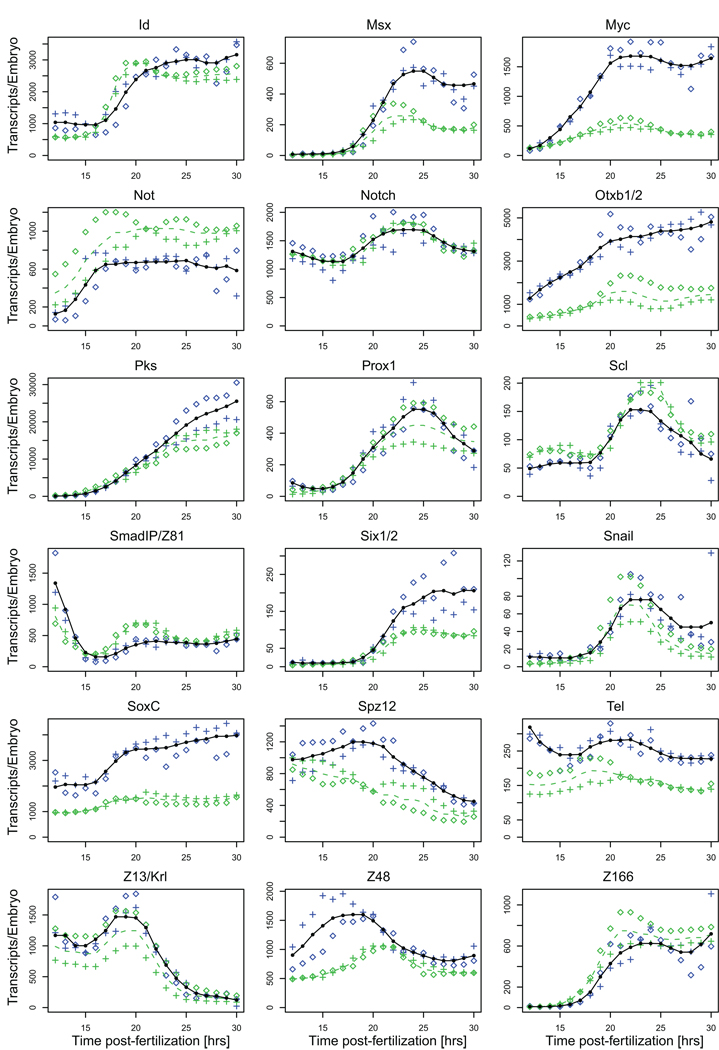
Fig. 2.
Comparison of NanoString prevalence measurements and QPCR data. The average transcript abundance as measured with the NanoString nCounter is shown in black, data for the two embryo batches as blue crosses and diamonds. Average prevalence data collected in QPCR assays are shown as dashed green lines, data for the two embryo batches used in these assays as green crosses or diamonds. Absolute transcript numbers were computed using the ubiquitin domain numbers obtained in the nCounter assays (Fig. 1).
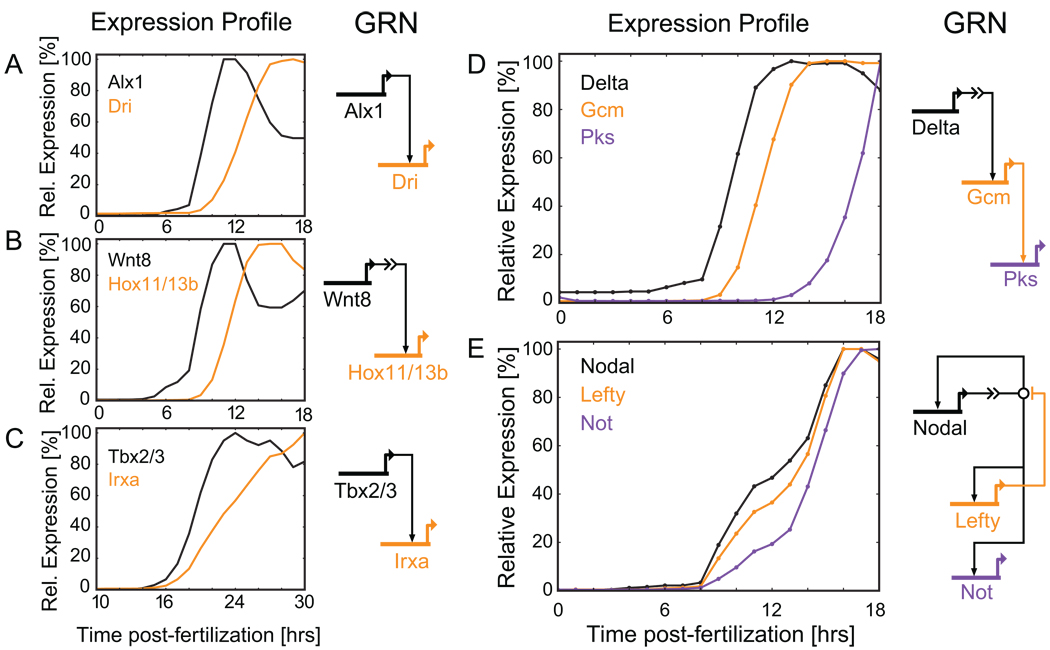
Fig. 3.
Expression dynamics of genes with close regulatory relationships. (A-C) Downstream targets often show a visible lag after their main activator first becomes transcribed. (A) Alx drives dri transcription in the skeletogenic cells of the sea urchin (Oliveri et al., 2008). (B) Wnt8 signaling leads to accumulation of Tcf1 in the nucleus of endomesodermal cells resulting in transcription of hox11 (Peter and Davidson, 2010). (C) Tbx2/3 activates irxa transcription in the aboral ectoderm of the early sea urchin embryo (Su et al., 2009). (D) Delta/Notch signaling from the skeletogenic cells activates gcm in the neighboring endomesodermal cells. Gcm in turn is a direct activator of pks (Ransick and Davidson, 2006; Calestani and Rogers, 2010). (E) More complex regulatory relationships are reflected in the expression dynamics. Nodal activates its own transcription, lefty, and not. Lefty is an antagonist of Nodal signaling (Duboc et al., 2008; Nam et al., 2007; Range et al., 2007).
The poly-ubiquitin gene has been used in several earlier studies as an internal reference and has been assumed to be expressed at a constant level of about 88,000 ubiquitin domain copies per embryo, present in several copies in each transcript (Howard-Ashby et al., 2006a, b; Materna et al., 2006; Nemer et al., 1991). A graph showing the prevalence of ubiquitin domain copies as determined with the nCounter is presented in Fig. 1. The time course reveals a roughly twofold rise in prevalence between 10 hours and 22 hours postfertilization with a peak expression of about 105 domain copies per embryo. Thereafter it remains more or less constant throughout gastrulation at just under 50,000 ubiquitin domain copies per embryo, an altogether minor difference from the previous assumption.
The prevalence of the spz12 message had been measured previously by RNA probe excess titration (Wang et al., 1995). Four time points of this earlier study fall into the period covered by our measurements and show the same overall expression profile. However, the absolute numbers we obtain are about 1.5 to two-fold lower. RNA probe excess titration is highly sensitive and not susceptible to variation in hybridization efficiency, but the absolute values obtained depend directly on the accuracy of the probe specific activity estimation (Davidson, 1986). Absolute prevalence as determined by this method may vary by a factor of two. The confidence intervals for the RNA probe excess titration measurements of spz12 and our nCounter data overlap, confirming the fidelity of the present measurements.
Batch to batch variation of gene expression in the nCounter assays is apparent for about 40% of genes included in the codeset – a significant fraction (Fig. S2). The remaining genes have essentially identical expression profiles in both batches. These differences indicate polymorphisms among the four genomes included in the comparison which affect synthesis or turnover rates for specific transcripts, not surprising given the very high level of genomic polymorphism characteristic of this species (Sodergren et al., 2006). Polymorphic differences are usually limited to given times, and only few genes show significant discrepancies throughout. The magnitude of variation generally does not exceed three-fold. Many of the most noticeable differences are found very early, up to about ten hours post-fertilization. This is likely due to differences in maternal transcript accumulation (e.g. ets1/2, z48, Fig. S2). In addition to polymorphisms that affect RNA synthesis or turnover, variation in maternal deposition may contribute to variation of zygotically expressed genes downstream. Other common differences include expression peaks that reach different levels in different batches or the timing of a ramping step. For the latter, differences usually persist until a peak or plateau has been reached.
We compared a subset of the expression profiles collected in the nCounter assays to independently gathered high resolution QPCR data (Fig. 2). Again two independent batches of fertilized eggs were assayed. In these QPCR assays prevalence was measured relative to the ubiquitin domain transcript as the internal standard and converted to absolute transcript numbers per embryo using the prevalence data for the ubiquitin domain obtained with the nCounter (Fig. 1). In general, prevalence and overall shape of the expression profile are in excellent agreement. As might be expected from the fact that there are now eight genomes rather than four in the dataset, some genes that display no batch to batch variation in the nCounter assays display variation between the batches used for QPCR (e.g. foxA, gataE, hesC, Fig. 2). Vice versa, there are genes that show batch to batch variation in the QPCR but not the nCounter assays (e.g. six1/2, z48, Fig. 2).
Prominent differences between expression profiles collected in QPCR and nCounter assays were observed for eight out of 36 genes. In these cases the nCounter prevalence data are usually higher than those obtained with QPCR and they remain higher by a constant factor throughout their expression (Fig. 2: alx1, dach, e2a, erg, foxn2/3, myc, otxb1/2, soxc). Absolute values aside, the expression profiles are more or less identical regardless of the quantification method. In addition, almost no batch to batch variation is observed for these genes between the embryo batches in either QPCR or nCounter assays. This indicates that the discrepancies are likely due to differences in hybridization efficiencies of NanoString probe and QPCR primers. Since QPCR values are lower than those obtained with the nCounter it is most likely that the QPCR primers were suboptimal.
Only two transcripts showed significantly higher prevalence in QPCR compared to nCounter assays (Fig. 2: delta, gataE). But these genes, gataE in particular, display significant batch to batch differences in the QPCR assays, indicating that this is the likely cause for the discrepancy. Together these findings suggest that nCounter assays are less prone to the minor quantitative problems affecting absolute transcript determinations by QPCR, such as low amplification efficiency in the first few cycles due to secondary structures in the cDNA, and polymorphisms that result in poor primer hybridization (Geiss et al., 2008; Wong and Medrano, 2005).
1.4. High resolution expression data as a tool
Gene expression data are a useful ancillary tool for assembling maps of gene regulatory networks. High temporal resolution produces a detailed picture of the expression dynamics for the genes under study and can indicate which genes could be linked at the regulatory level. However, they are no substitute for perturbation experiments, the effects of which directly reveal causal regulatory relationships among genes. High resolution expression data can, however, augment and guide perturbation experiments and expedite the discovery of unknown linkages (Materna and Oliveri, 2008).
Where a regulatory gene serves as the main driver of a downstream gene, the target is often expressed with a visible delay after the activator is first being transcribed. This is due to the time needed to produce a sufficient amount of protein for activation to occur. Kinetic analyses using the transcription, turnover and protein synthesis rates for S. purpuratus have demonstrated a typical lag of about two hours between activation of an upstream gene and activation of its direct target (Ben-Tabou de-Leon and Davidson, 2009; Bolouri and Davidson, 2003). This is illustrated by the gene pairs in Fig. 3A-C. The activators in Fig. 3A and 3C are likely direct inputs into their downstream target genes, although some of the connections have yet to be confirmed at the cis-regulatory level (Nam et al., 2007; Oliveri et al., 2008; Ransick and Davidson, 2006; Su et al., 2009). The activation in Fig. 3B is indirect: Wnt8 is a signaling factor. Signal reception causes accumulation of a positively active form of the transcription factor Tcf1 in the nucleus to which the target gene hox11/13b responds (Peter and Davidson, 2010, C. Theodoris, J. Smith, and E. Davidson, unpublished data; for kinetic analysis: Bolouri and Davidson, 2009). Figure 3D shows an example of three genes linked in a rather simple activation cascade. The signaling ligand Delta is presented on the surface of the skeletogenic cells, and via its effector, Suppressor of Hairless, activates gcm in the neighboring endomesodermal cells (Ransick and Davidson, 2006). Gcm in turn directly activates pks (Calestani and Rogers, 2010). However, activation of pks may require genes in addition to gcm as indicated by the relatively long period between gcm and pks activation.
Despite these clear-cut examples, the connection between expression dynamics and how genes are linked at the regulatory level might become clear only in hindsight. After all, gene activation is combinatorial and often requires several activating inputs acting in concert for a target gene to turn on. In addition, the flow of regulatory information is often not as unidirectional as in the examples above. Regulatory genes are frequently part of feedback loops and influence each other’s expression, as in the example in Fig. 3E: The signaling ligand Nodal is initially activated by bZIP factors but, once expressed, strongly activates itself (Nam et al., 2007; Range et al., 2007). Nodal also directly activates lefty transcription. Lefty, in turn, antagonizes the activity of Nodal thus effectively limiting its transcription (Duboc et al., 2008). not, a second downstream target of Nodal (S.C.Materna., E. Li and E.H.Davidson, unpublished data) has a similar expression profile. The two feedback loops between Nodal and Lefty make it impossible to infer the linkage pattern from timecourse data alone. However, perturbation experiments and cis-regulatory analysis have clearly established their relationship (Duboc et al., 2008; Nam et al., 2007; Range et al., 2007; Su et al., 2009). The kinetic aspescts of these interactions have been discussed elsewhere (Bolouri and Davidson, 2009).
In conclusion, we have acquired a set of high quality prevalence data for the majority of spatially restricted regulatory genes in early sea urchin development. The data describe in detail the expression dynamics of the genes included. This information will be useful for unraveling the gene regulatory network underlying early sea urchin development.
1.5. Data availability
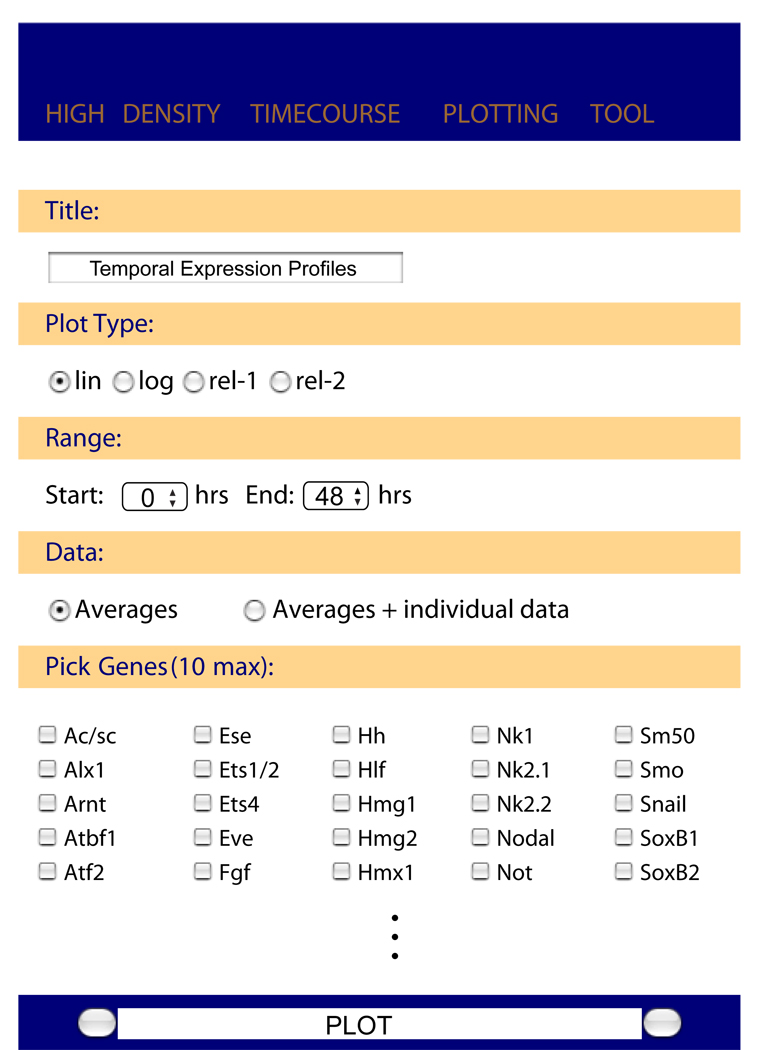
In order to allow quick access to the expression data for all genes we have created a simple visualization tool that is available via http://sugp.caltech.edu/endomes/. A screenshot is shown in Fig. 4. The website enables easy viewing of multiple time courses covering the whole 48 hours of early development or fractions thereof as determined by the user. The data can be plotted either on a linear or logarithmic scale, to accommodate genes with different expression levels. Two options for plotting relative expression levels are available: The first relates the gene expression level to the maximum measured for this gene during the whole time period covered, regardless of the period chosen for plotting. The second option sets the maximum expression during the plotting period to 100%. These options are useful when the timing of gene expression matters more than the absolute transcript levels – as is often the case when examining regulatory linkages. The data for the two individuals sampled can be plotted if desired. With this tool all graphs displaying nCounter data in this paper can essentially be recreated in print quality. A table with all expression data is available on request.
Fig. 4.
Screen shot of the online plotting tool. This website enables easy plotting of all expression profiles. What genes are to be plotted and the details of the presentation can be specified by the user. The website is available via http://sugp.caltech.edu/endomes/.
2. Materials and methods
2.1. Embryo culture and RNA extraction
Sea urchin embryos were fertilized in filtered seawater and washed repeatedly to remove excess sperm. Embryos were cultured at low density at 15°C and closely monitored for proper development. For the time course measurements 200 embryos were counted for each time point. Samples were collected hourly just prior to lysis in 350 µl RLT buffer from the Qiagen RNeasy Micro Kit (Qiagen, Hilden, Germany). Embryo lysates were immediately stored at −70°C until use. gfp and rfp genes were transcribed in vitro and 1x107 transcripts of both GFP and RFP were added to each lysate after thawing. RNA was extracted according to manufacturers instructions but, to maximize recovery, RNA was eluted with 50 µl nuclease free water. The samples were ethanol precipitated and resuspended in 5 µl nuclease free water all of which was used in the following NanoString hybridization.
2.2. NanoString nCounter assays
For each individual and time point transcript prevalence was measured using the NanoString nCounter. Hybridization reactions were performed according to manufacturer’s instructions with 5 µl RNA solution. Care was taken to minimize the time after addition of capture probeset in order to minimize background due to nonspecific interactions between detection probes and capture probes. All hybridization reactions were incubated at 65°C for a minimum of 18 h. Hybridized probes were recovered with the NanoString Prep Station and immediately evaluated with the NanoString nCounter. For each reaction 600 fields of view were counted.
The resulting counts were normalized using the mean of GFP and RFP counts for each time point and converted to transcript numbers per embryo by using the known numbers of GFP/RFP transcripts and embryos. For each gene and time point the arithmetic mean was calculated for the expression data from the two individuals sampled. To discard outliers in the average time course, a running average was calculated over five time points, discarding the minimum and maximum value and computing the arithmetic mean of the three remaining values.
2.3. QPCR assays
For quantitative PCR assays RNA was collected from embryos of two different females. RNA was extracted with the Qiagen RNeasy Mini kit. 1 µg of total RNA was converted to cDNA using the BioRad iScript cDNA synthesis kit (BioRad, Carlsbad, CA). QPCR was performed with the BioRad SYBR Green reagent on an AB 7900 HT instrument (Applied Biosystems, Foster City, CA). Data were evaluated with the dCt method using poly-ubiquitin as the reference gene (Materna and Oliveri, 2008). dCt values were converted to transcripts per embryo by applying the transcript numbers for poly-ubiquitin obtained with the NanoString nCounter. Data were averaged as above.
Supplementary Material
Acknowledgements
We would like to thank the NanoString team, and in particular Stephen Jackson and Sean Ferree, for their tireless support. Research was supported by NIH grant HD 37105. Most of the funding for the purchase of the nCounter was a gift from the Ahmanson Research & Equipment Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ben-Tabou de-Leon S, Davidson EH. Modeling the dynamics of transcriptional gene regulatory networks for animal development. Dev Biol. 2009;325:317–328. doi: 10.1016/j.ydbio.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci U S A. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. The gene regulatory network basis of the "community effect," and analysis of a sea urchin embryo example. Dev. Biol. 2009 doi: 10.1016/j.ydbio.2009.06.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calestani C, Rodgers D. Cis-regulatory analysis of the sea urchin pigment cell gene polyketide synthase. Dev. Biol. 2010 doi: 10.1016/j.ydbio.2010.01.026. in press. [DOI] [PubMed] [Google Scholar]
- Duboc V, Lapraz F, Besnardeau L, Lepage T. Lefty acts as an essential modulator of Nodal activity during sea urchin oral-aboral axis formation. Dev Biol. 2008;320:49–59. doi: 10.1016/j.ydbio.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Geiss G, Bumgarner R, Birditt B, Dahl T, Dowidar N, Dunaway D, Fell H, Ferree S, George R, Grogan T, James J, Maysuria M, Mitton J, Oliveri P, Osborn J, Peng T, Ratcliffe A, Webster P, Davidson E, Hood L. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev Biol. 2006a;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev Biol. 2006b;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Dev Biol. 2006;300:108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Materna SC, Oliveri P. A protocol for unraveling gene regulatory networks. Nat Protoc. 2008;3:1876–1887. doi: 10.1038/nprot.2008.187. [DOI] [PubMed] [Google Scholar]
- Nam J, Su Y-H, Lee PY, Robertson AJ, Coffman JA, Davidson EH. Cis-regulatory control of the nodal gene, initiator of the sea urchin oral ectoderm gene network. Dev Biol. 2007;306:860–869. doi: 10.1016/j.ydbio.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemer M, Rondinelli E, Infante D, Infante AA. Polyubiquitin RNA characteristics and conditional induction in sea urchin embryos. Dev Biol. 1991;145:255–265. doi: 10.1016/0012-1606(91)90124-l. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2010 doi: 10.1016/j.ydbio.2009.10.037. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Range R, Lapraz F, Quirin M, Marro S, Besnardeau L, Lepage T. Cis-regulatory analysis of nodal and maternal control of dorsal-ventral axis formation by Univin, a TGF-beta related to Vg1. Development. 2007;134:3649–3664. doi: 10.1242/dev.007799. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci U S A. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus) Dev Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Smith J, Davidson EH. Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc Natl Acad Sci U S A. 2008;105:20089–20094. doi: 10.1073/pnas.0806442105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sea Urchin Genome Consortium. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y-H, Li E, Geiss GK, Longabaugh WJR, Krämer A, Davidson EH. A perturbation model of the gene regulatory network for oral and aboral ectoderm specification in the sea urchin embryo. Dev Biol. 2009;329:410–421. doi: 10.1016/j.ydbio.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Wang DG, Kirchhamer CV, Britten RJ, Davidson EH. SpZ12-1, a negative regulator required for spatial control of the territory-specific CyIIIa gene in the sea urchin embryo. Development. 1995;121:1111–1122. doi: 10.1242/dev.121.4.1111. [DOI] [PubMed] [Google Scholar]
- Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.