Abstract
The ubiquitous proto-oncogene protein DEK has been found to be associated with chromatin during the entire cell cycle. It changes the topology of DNA in chromatin and protein-free DNA through the introduction of positive supercoils. The sequence and structure specificities of DEK–DNA interactions are not completely understood. The binding of DEK to DNA is not sequence specific, but we describe here that DEK has a clear preference for supercoiled and four-way junction DNA. In the presence of topoisomerase II, DEK stimulates intermolecular catenation of circular DNA molecules. DEK also increases the probability of intermolecular ligation of linear DNA molecules by DNA ligase. These binding properties qualify DEK as an architectural protein.
INTRODUCTION
Chromatin is involved in the regulation of all nuclear processes, most prominently transcription (1–3), replication (4), recombination (5) and repair (6). In recent years, two fundamental principles have emerged that form the basis for interchangeable chromatin structures corresponding to distinct functional states (7). First, the interactions of the histone N-terminal tails can be modulated through post-translational modifications, notably acetylation, phosphorylation and methylation, as well as ubiquitination and ADP-ribosylation (8). These modifications seem to have profound effects on histone–DNA interactions and the binding of regulatory proteins. Secondly, chromatin-remodeling factors alter histone–DNA interactions such that nucleosomal DNA becomes more accessible to interacting proteins (9). In addition, architectural proteins such as the high mobility group (HMG) proteins that typically lack the potential to activate transcription or carry out recombination on their own induce conformational changes in the DNA and facilitate the assembly and activity of multiprotein–DNA complexes (10).
We have identified the proto-oncogene protein DEK as a protein that influences chromatin structure (11,12). The DEK protein was initially identified in a fusion with the CAN nucleoporin in a subtype of acute myeloid leukemias (13). Furthermore, autoantigens to DEK have been detected in human diseases, including systemic lupus erythematosus (14–16), juvenile rheumatoid arthritis (15,17) and sarcoidosis (14,15). Interestingly, DEK has also been linked to ataxia telangiectasia (ATM), as a fragment of dek cDNA reverses the mutagen-sensitive phenotype of cells from ATM patients (18). In spite of these clinical observations, the biological function of DEK remains unclear.
The activity of the DEK protein seems to be regulated by protein–protein interactions as well as through binding to RNA and chromatin. Thus DEK could be involved in tethering different proteins to chromatin. By co-fractionation and co-immunoprecipitation, it was demonstrated that the transcriptional co-repressor hDaxx associates with DEK (19). How ever, the exact function of DEK in hDaxx-mediated repression, however, is not clear. DEK has also been found to be associated with the latency-associated nuclear antigen (LANA) which is constitutively expressed in Kaposi’s sarcoma-associated herpesvirus latent infection (20). The data indicate that LANA is tethered to chromatin through interaction with the DEK protein and the methyl CpG-binding protein MeCP2. In addition, DEK interacts with the celltype-specific transcription factor AP-2α in vitro and stimulates the transactivation activity of AP-2α over the APOE promotor (21).
Two reports suggested that DEK may be involved in RNA metabolism and that DEK occurred in an ∼335 kDa exon junction complex (22,23). However, recent experiments could not confirm these data, and a function for DEK in RNA metabolism remains to be established (24–26).
Our recent data show that the main fraction of DEK is associated with chromatin in vivo. We found that DEK is a constituent of oligonucleosomes, generated by micrococcal nuclease digestion of chromatin in isolated nuclei (27). Association of DEK with metaphase chromosomes has also been reported (28). Purified DEK changes the topology of DNA in viral minichromosomes and reduces the accessibility of chromatin to DNA-binding factors including components of the replication machinery (11). The DEK-induced change in topology is due to the introduction of positive supercoils into the DNA (12).
DEK does not belong to a known family of proteins. Apart from four stretches of acidic amino acids, the only recognizable feature is a homology to a DNA-binding motif, the SAF (scaffold attachment factor) box (29,30) also termed the SAP domain (after SAF-A/B, acinus and PIAS) (31) between amino acids 149 and 183 of the DEK protein. It has been described that DEK binds to DNA and specifically recognizes the peri-ets (pets) sites in the human immunodeficiency virus type 2 (HIV-2) enhancer (32–34). However, Alexiadis et al. (11) have shown that DEK changes the topology of different chromatin templates, independently of the underlying DNA sequence. To further characterize the DEK–DNA interaction, we used substrates with different sequences and structures. We found that DEK binds preferentially to supercoiled and cruciform DNA and that it brings different DNA molecules into close proximity. However, we have no evidence for sequence-specific binding.
MATERIALS AND METHODS
Expression and purification of GST–DEK
The DEK gene was cloned in the pGEX3 vector (Pharmacia) to generate a GST fusion protein. This construct was obtained from G. Grosveld. The GST–DEK fusion protein was expressed in BL21 (DE3) pLys S, and prepared from a 1 l culture in 2× YT medium. Further purification steps were done according to the manufacturer’s protocol (Pharmacia). The experiments were carried out with the uncleaved GST fusion protein.
Electrophoretic mobility shift assay (EMSA)
A 175 ng aliquot of either SV40 DNA (5243 bp), Litmus™28i (2823 bp, Biolabs) or HK-plasmid (5537 bp, SAF-A cDNA up to base 2580 in bluescript II; Frank Fackelmayer, personal communication) was incubated with increasing amounts of DEK in a reaction volume of 35 µl. DEK was dialyzed before use against nE100 buffer (20 mM HEPES–KOH, pH 7.6, 100 mM NaCl, 10 mM sodium bisulfite, 1 mM EDTA). After 1 h at 37°C, 6× sample buffer was added (0.25% bromophenol blue, 0.25% xylene cyanol, 15% ficoll) and nucleoprotein complexes were separated on 0.6% agarose gels in 0.5× TBE (90 mM Tris, 89 mM boric acid, 2 mM EDTA pH 8) for 16 h at 2 V/cm.
Oligonucleotides corresponding to both strands of the HIV-2 sequence from –162 to –131 were synthesized (MWG Biotech). Wild-type pets sequence, 5′-GATCCAGCTATACTTGGTCAGGGCGAATTCTAACTA-3′; mutated pets sequence, 5′-GATCCAGCTATACTAGATCTGGGCGAATTCTAACTA-3′. Equimolar amounts of the two strands were combined, boiled for 1 min and allowed to cool gradually. The double-stranded oligonucleotide was then radiolabeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP. Labeled oligonucleotides were incubated with DEK (reaction volume 15 µl), separated on 8% native polyacrylamide gels in 1× TBE, and visualized by autoradiography.
Topology assay
DEK assays were performed as described (12). Briefly, purified DEK was dialyzed on Whatman filters (Type VS, pore size 0.025 µm) against buffer nE100 in the presence of 1 µg/µl bovine serum albumin (BSA; Biolabs) for 90 min at 4°C. A 175 ng aliquot of DNA was incubated with the dialyzed DEK protein for 1 h at 37°C in the presence of 1 U of wheat germ topoisomerase I (Promega) or 1 U of human topoisomerase II. Reactions were performed in nE100, containing 0.2 µg/µl BSA in a total volume of 90 µl. After proteinase K digestion, DNA was precipitated and analyzed on 0.8% agarose gels in 0.5× TBE at 2 V/cm for 16 h.
Preparation of duplex and four-way junction DNA
For construction of four-way junction (4WJ) DNA and control duplex DNA, we used oligonucleotides with the following sequences: oligo 1, 5′-CCCTATAACCCCTGCATTGAATTCCAGTCTGATAA-3′; oligo 2, 5′-CTAGTCGTGATAGGTGCAGGGGTTATAGGG-3′; oligo 3, 5′-AACAGTAGC TCTTATTCGAGCTCGCGCCCTATCACGACTA-3′; oligo 4, 5′-TTTATCAGACTGGAATTCAAGCGCGAGCTCGAATAAGAGCTACTGT-3′; and oligo 5, 5′-GTAGTCGTGATAGGGCGCGAGCTCGAATAAGAGCTACTGT-3′. Oligos were annealed as described above and separated on 12% native polyacrylamide gels. 4WJ DNA was cut off the gel, crushed and eluted overnight at room temperature in 900 µl of elution buffer (0.5 mM ammonium acetate, 1 mM EDTA). Gel pieces were separated over glass beads (425–600 µm, Sigma); the supernatant was extracted with phenol/chloroform and precipitated with ethanol.
Ligase assay
A 123 bp marker DNA (Pharmacia) was digested with AvaI and labeled in the presence of [γ-32P]ATP with polynucleotide kinase. A 2 ng aliquot of the 32P-labeled 123 bp fragment was incubated for 30 min at 37°C at different DEK/DNA ratios in ligase buffer (4 mM Tris pH 7.8, 1 mM dithiothreitol, 0.5 mM ATP, 5 mM MgCl2). T4 DNA ligase (1 U) was added and incubated for 10 min at 37°C; the ligase was then inactivated for 15 min at 65°C. To remove linear DNA, samples were incubated for 15 min at 37°C with 50 U of exonuclease III. Samples were digested with proteinase K and analyzed on 7% polyacrylamide gels for 3 h at 10 V/cm.
Electron microscopic analysis
Circular SV40 DNA was incubated with DEK (at molar ratio of 1:90). Unbound protein was removed by gel filtration through a Biogel A-5 column and samples were fixed with glutaraldehyde [0.1% (v/v)] for 15 min at 37°C. Protein–DNA complexes were spread by the BAC (alkyl benzyl dimethyl ammonium chloride) technique of Vollenweider et al. (35). Electron micrographs were taken with a Zeiss EM 900 electron microscope.
RESULTS
DEK binds to and changes the topology of DNA without sequence specificity
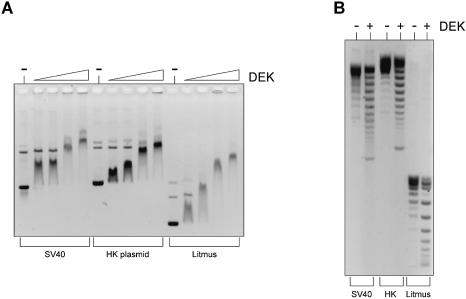
To determine whether DEK binds to different DNA sequences, we used EMSA. SV40 DNA, HK-plasmid DNA and Litmus-plasmid DNA were incubated with increasing amounts of DEK. The resulting nucleoprotein complexes were separated by agarose gel electrophoresis (Fig. 1A). The data of Figure 1 show that DEK binds to all three DNAs with similar efficiency, although with a marked preference for supercoiled DNA (see later). We have also used the different DNAs as substrates in a topology assay incubating supercoiled DNA with DEK and topoisomerase I. The deproteinized DNA was then separated by agarose gel electrophoresis (Fig. 1B). We found that DEK changes the topology of the three templates tested, independent of the underlying DNA sequence.
Figure 1.
EMSA and topology assay with different DNA sequences. (A) EMSA. A 175 ng aliquot of SV40 DNA (lanes 1–5), HK-plasmid (lanes 6–10) and Litmus-plasmid (lanes 11–15) wase incubated for 1 h at 37°C without (–) or with GST–DEK at molar ratios of DEK/DNA of 28, 56, 84 and 112. Nucleoprotein complexes were analyzed on 0.6% agarose gels and visualized by ethidium bromide staining. (B) Topology assay. A 175 ng aliquot of SV40 DNA (lanes 1 and 2), HK-plasmid (lanes 3 and 4) and Litmus-plasmid (lanes 5 and 6) was incubated without (–) or with GST–DEK (at a molar ratio of DEK/DNA of 84) and topoisomerase I for 1 h at 37°C. Deproteinized samples were analyzed by agarose gel electrophoresis and ethidium bromide staining.
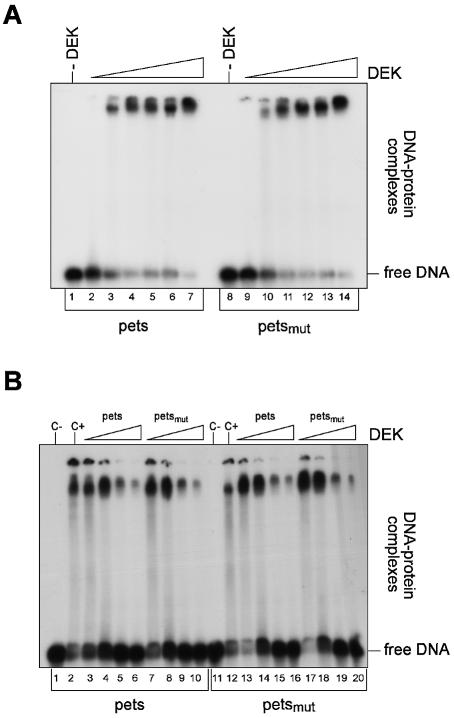
Although no sequence specificity could be detected with these DNAs, it is conceivable that DEK has a higher affinity for the pets sequence as used by Fu et al. (34) and Faulkner et al. (32) compared with a random DNA sequence. The pets sequence is a TG-rich element found between the two Elf-1-binding sites in the HIV-2 enhancer. As a control, we used the same mutated pets sequence that has been used by Fu et al. (34). In this sequence, three bases differ from the wild-type sequence. Both oligonucleotides were labeled and incubated with increasing amounts of purified DEK protein, and nucleoprotein complexes were analyzed on native polyacrylamide gels (Fig. 2A). We found that DEK binds with similar efficiency to the wild-type and the mutated pets sequence. To further investigate these data, we performed competition experiments, and added increasing amounts of either non-labeled wild-type or mutated pets sequence to the standard reaction just shown. Binding to the labeled pets oligonucleotide was competed at an excess of 50-fold competitor DNA of either the pets sequence itself (Fig. 2B lanes 3–6) or the mutated pets sequence (Fig. 2B, lanes 7–10). The same ratio was obtained when we used the mutated pets sequence for DEK binding. Again binding was competed at a 50-fold excess of competitor of both the pets sequence (Fig. 2B, lanes 13–16) and the mutated oligonucleotide (Fig. 2B, lanes 17–20). Identical results were obtained by using GST-tagged or His-tagged DEK protein (data not shown). Thus, these data show that DEK has no preference for the pets sequences of the HIV enhancer. This is clearly in contrast to the data of Fu et al. (33,34), and we presently cannot explain this discrepancy.
Figure 2.
EMSA with pets sequences. (A) EMSA. Radioactively labeled pets or mutated pets oligonucleotides were incubated without (–) or with increasing amounts of GST–DEK for 1 h at 37°C. Nucleoprotein complexes were separated on 8% native polyacrylamide gels and detected by autoradiography. The molar ratios of DEK/DNA were for lanes 1–7 and 8–14: 0, 11, 22, 33, 45, 56 and 90 mol DEK/mol DNA. (B) Competition. Radioactively labeled oligonucleotides were incubated for 1 h at 37°C at a molar ratio of DEK/DNA of 40 with increasing amounts of non-labeled competitor DNA. Lane 1, pets or pets-mut without DEK (c–); lane 2, pets or pets-mut with DEK (c+)
DEK binds preferentially to negatively supercoiled and cruciform DNA
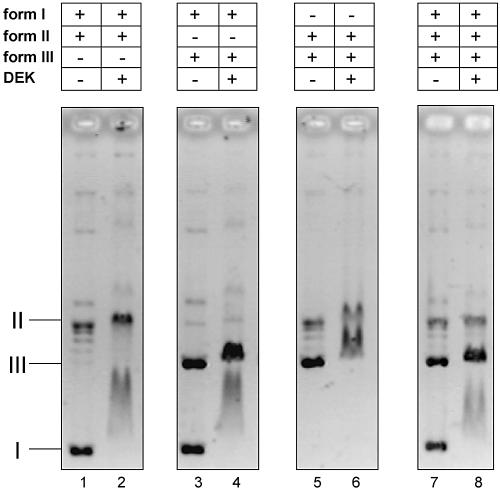
The EMSA experiment in Figure 1A indicated that DEK binds preferentially to negatively supercoiled DNA, provided that relaxed and supercoiled DNA are present within the same sample. To further investigate the binding of DEK to DNAs of different topologies, we incubated DEK with either supercoiled and relaxed DNA, supercoiled and linear DNA or relaxed and linear DNA in the same assay (Fig. 3). The EMSA with supercoiled and relaxed DNA revealed a clear preference of DEK for form I DNA because the entire supercoiled DNA was shifted, whereas no significant shift was observed with relaxed DNA (Fig. 3, lane 2). The same result was obtained when supercoiled and linear DNA was present in the same reaction. Again, the entire supercoiled DNA was shifted by DEK, whereas only a minimal shift was observed with linear DNA (Fig. 3, lane 4). However, when relaxed and linear DNAs were used together, both templates were shifted with similar efficiency (Fig. 3, lane 6). The clear preference for supercoiled DNA was also obvious when all three templates were present in the same reaction. In this case, the entire supercoiled DNA was shifted, but none of the relaxed and linear DNA (Fig. 3, lane 8).
Figure 3.
Binding of DEK to different DNA structures. A 100 ng aliquot of each of form I and II DNA (lanes 1 and 2), form I and III DNA (lanes 3 and 4), form II and III (lanes 5 and 6) and form I, II and III (lanes 7 and 8) were incubated with DEK at a molar ratio of 15 mol DEK/mol DNA (lanes 2, 4 and 6) or 10 mol DEK/mol DNA (lane 8). Nucleoprotein complexes were analyzed by agarose gel electrophoresis and ethidium bromide staining. I, supercoiled; II, relaxed, closed circular and nicked DNA; III, linear DNA.
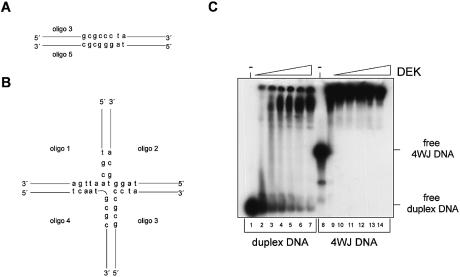
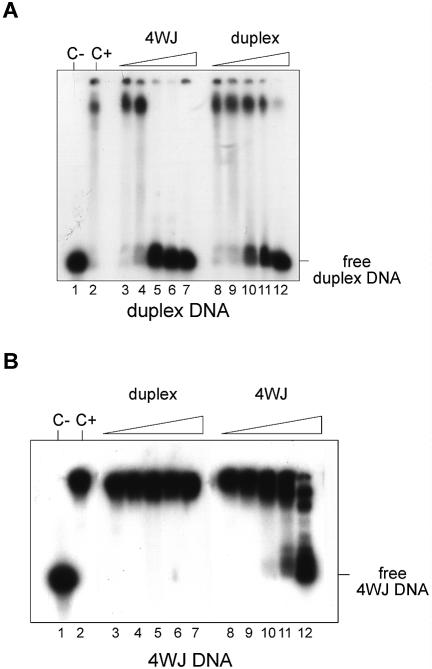
The preferential binding of DEK to supercoiled DNA could be an indication that DEK recognizes unusual DNA forms such as cruciform DNA. The classical experiment to test this is the binding to 4WJ DNA as found in the Holliday junction DNA of intermediates in homologous genetic recombination (36). Accordingly, we used five different single-stranded oligonucleotides which were annealed to form either duplex DNA or cruciform DNA (Fig. 4A and B) (37). The 32P-labeled DNA substrates were incubated with increasing amounts of DEK and analyzed on a native polyacrylamide gel (Fig. 4C). We found that DEK binds to both DNA substrates although with different efficiencies. Low DEK/DNA ratios were sufficient to shift the entire 4WJ DNA, whereas at least 4-fold higher DEK/DNA ratios were required to shift the duplex DNA.
Figure 4.
EMSA with duplex and 4WJ DNA. (A) Model and sequence of the duplex DNA. (B) Model and sequence of the cruciform DNA. Oligonucleotides 1–4 are partially complementary to each other and, when annealed, assemble into the cruciform molecule indicated. (C) Bandshift with duplex and cruciform DNA. Oligonucleotides were annealed, purified on native polyacrylamide gels and then labeled. Labeled duplex DNA (lanes 1–7) and 4WJ DNA (lanes 8–14) were incubated for 1 h at 37°C with increasing amounts of GST–DEK and analyzed on an 8% polyacrylamide gel. Bands were visualized by autoradiography. The molar ratios of DEK/DNA were as follows: lanes 1–7 and lanes 8–14, 0, 11, 22, 33, 45, 56 and 90.
To confirm these data, we performed competition experiments. Binding to labeled duplex DNA was competed with increasing amounts of either unlabeled 4WJ DNA or duplex DNA (Fig. 5A). Whereas binding to duplex DNA was competed already at a 50-fold excess of 4WJ DNA as competitor, almost a 1000-fold excess of duplex DNA was necessary to eliminate binding to the labeled duplex DNA. When labeled 4WJ DNA was used as substrate, binding could not be competed even with a 1000-fold excess of cold duplex DNA and was significantly reduced at a 500-fold excess of competing 4WJ DNA (Fig. 5B). These data demonstrate that DEK binds with higher preference to cruciform DNA compared with duplex DNA.
Figure 5.
Competition with duplex and 4WJ DNA. Labeled DNA fragments were incubated for 1 h at 37°C with DEK (at a molar ratio of DEK/DNA of 40) and increasing amounts of non-labeled competitor DNA. (A) Com petition of duplex binding. Lane 1, duplex DNA without DEK (c–); lane 2, duplex DNA with DEK (c+); lanes 3–7 and 8–12 were incubated at the following ratios of the indicated competitor DNA: 1:1, 1:10, 1:50, 1:500 and 1:1000. (B) Competition of 4WJ DNA binding. Lane 1, 4WJ DNA without DEK (c–); lane 2, 4WJ DNA with DEK (c+); lanes 3–7 and 8–12 were incubated at the following ratios of the indicated competitor DNA: 1:1, 1:10, 1:50, 1:500 and 1:1000.
DEK stimulates intermolecular interactions
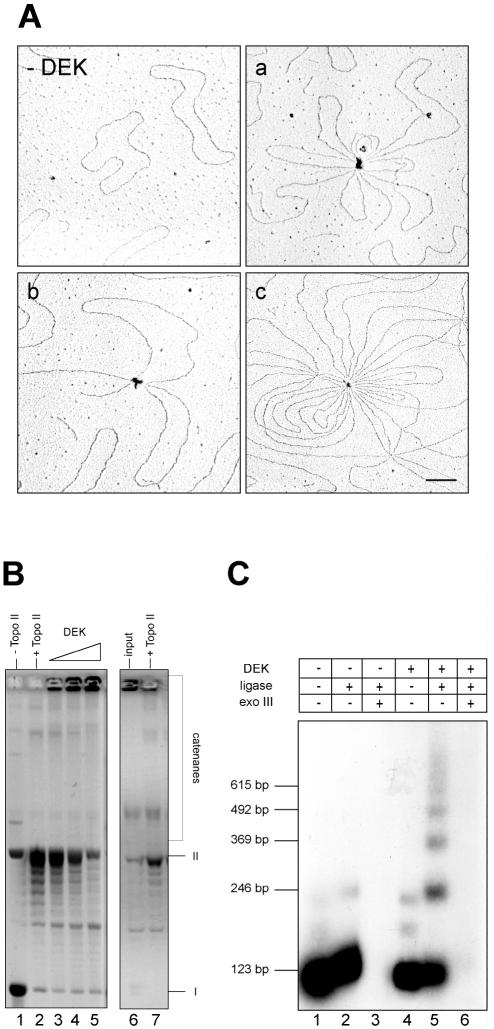
The binding behavior of DEK to DNA resembles that of architectural proteins such as the HMG proteins (38). For example, the HMGB1 protein preferentially binds to supercoiled over linear or nicked closed circular DNA (39) and is also able to manipulate DNA by looping, compaction and changes in DNA topology (40–42). To gain further insights into DEK–DNA interactions, we investigated the binding of DEK to circular DNA by electron microscopy (Fig. 6A). We detected a population of molecules (∼5%) containing two or more DNA molecules with multiple DNA loops connected at a protein bead in addition to large complexes with many looped DNA molecules of variable size. These data indicate that DEK brings different DNA molecules into close proximity, and thereby increases the local concentration of DNA.
Figure 6.
DEK directs intermolecular interactions. (A) Electron microscopic analysis. Circular SV40 DNA was incubated in the absence (–DEK) or presence of DEK (a, b and c) (at a molar ratio of DEK to DNA of 90:1) and visualized by electron microscopy. Bar = 100 nm. (B) A 175 ng aliquot of SV40 DNA was incubated with increasing amounts of DEK in the absence (– topo II) or presence of topoisomerase II (+ topo II). Deproteinized DNA was analyzed by agarose gel electrophoresis and ethidium bromide staining. Molar DEK/DNA ratios were in lane 3, 28; in lane 4, 56; and in lane 5, 112 mol DEK/mol DNA. I, supercoiled; II, relaxed, closed circular and nicked DNA. Deproteinized DNA of reaction 5 (input) was treated with topoisomerase II and fractionated on an agarose gel. (C) A 123 bp 32P-labeled DNA fragment (8 ng) with AvaI sticky ends was incubated in the absence (lanes 1–3) or presence (lanes 4–6) of DEK. Protein/DNA molar ratios used were 10:1 (lanes 4–6). T4 DNA ligase was added to the reactions as indicated and incubated at 37°C for 10 min. Samples in lanes 3 and 6 were subsequently incubated with exonuclease III to remove linear ligation products. Reaction products were electrophoresed on an 8% polyacrylamide gel and visualized by autoradiography. The positions of the 123 bp multimers are indicated.
To test whether DEK stimulates intermolecular interactions between circular DNA molecules, we incubated supercoiled SV40 DNA with increasing amounts of DEK and topoisomerase II (Fig. 6B). Topoisomerase II is known to induce double-strand breaks (43), and could induce the formation of catenated DNA from closely adjacent DNA circles. We found indeed that topoisomerase II converted DNA circles into large structures that were not able to enter the gel (Fig. 6B, lanes 3–5). These large DNA structures were most probably catenanes because topoisomerase II could resolve them into circular monomers (Fig. 6B, lane 7).
We finally show that DEK stimulates the intermolecular end-to-end joining of linear DNA molecules by DNA ligase. For this purpose 32P-labeled 123 bp DNA fragments with cohesive ends were incubated with increasing amounts of DEK and treated with T4 DNA ligase. Increasing concentrations of DEK favored the formation of linear multimers (Fig. 6C). To discriminate between linear and circular DNA, the deproteinized DNA was digested with exonuclease III (Fig. 6C, lanes 3 and 6) (44). All reaction products were sensitive to exonuclease III, indicating that DEK does not bend the DNA fragment but stimulates intermolecular end-to-end joining.
DISCUSSION
We found that DEK exhibits distinctive structure-specific rather than sequence-specific DNA binding properties. In contrast to published data (32–34), our results do not support a sequence-specific binding mode of DEK. We found that DEK is able to bind to and change the topology of different DNA substrates. More specifically, we show that DEK binds with similar efficiency to the pets sequence and the mutated pets sequence. Binding to the pets sequences could be competed equally well with both the specific pets sequence and the mutated sequence, and vice versa. These results contradict the data of Fu et al. (33,34) who claimed a specific binding of DEK to the pets sequences. The reasons for this discrepancy are not clear; however, we believe that their competition experiment with the labeled pets sequence competed with the unlabeled pets oligonucleotide or an unrelated HIV-2 κB oligonucleotide was inconclusive [fig. 7 of Fu and Markovitz (33)]. Our present data agree with the previous findings of Alexiadis et al. (11), who had shown that changes in chromatin topology occur independently of the DNA sequence. Thus, we conclude that the underlying DNA sequence is not a critical determinant for DEK–DNA interaction.
In fact, the structure of the DNA could be a more important parameter that determines the interaction of DEK with DNA. In support of this, we report here that DEK has a strong preference for negatively supercoiled DNA compared with relaxed or linear DNA, and that the element recognized by DEK could be a 4WJ DNA. The 4WJ (Holliday junction) DNA was initially proposed as the central intermediate in homologous genetic recombination; it is also involved in replication-related events. Furthermore, binding to 4WJ DNA seems to be a common property of architectural proteins (38) as shown, for example, for the HMG proteins (45,46), the linker histone H1 (47) or the structural maintenance of chromosomes (SMC) proteins (48). It has also been reported that the chromatin-remodeling complex SWI/SNF preferentially binds to 4WJ DNA (49). Binding and competition experiments demonstrated that DEK binds preferentially to 4WJ DNA compared with duplex DNA.
At least some of the binding properties of DEK are reminiscent of the HMGB1 and HMGB2 proteins [old name: HMG 1 and 2 (50)]. They bind DNA without sequence specificity but have a high affinity for bent or distorted DNA such as 4WJs (46). They also bind to crossovers in supercoiled DNA and can cause looping of linear DNA. HMGB1 and 2 play important architectural roles in the assembly of nucleoprotein complexes in a variety of biological processes, e.g. V(D)J recombination and the initiation of transcription. A function in DNA repair has also been reported (51).
We have observed that DEK stimulates the intermolecular end-to-end joining of linear DNA molecules in the presence of DNA ligase (Fig. 6C). Increasing concentrations of DEK favored the formation of linear multimers but did not induce circularization of the substrate, as observed with the HMG protein (44). We also found that DEK promoted the formation of catenated DNA molecules in the presence of topoisomerase II (Fig. 6B). The same properties were observed for the cohesin complex, which consists of a heterodimeric pair of SMC proteins and at least two non-SMC subunits (52). Cohesin functions as a molecular cross-linker and holds sister chromatids together (53). Whether DEK is involved in such a kind of chromatin organization has yet to be investigated.
It has been postulated that proteins which bind to 4WJs all perform a particular kind of architectural role in vivo (38). In most cases studied, the non-specific binders induce DNA looping and change the topology of circular closed DNA molecules, in both a negative and positive orientation. These are precisely the properties of DEK: it promotes the looping of DNA (Fig. 6A) and induces positive supercoils into protein-free DNA (12). The mechanism of DEK-induced positive supercoiling is not known. Electron microscopic analysis demonstrated that DEK caused no shortening of DNA molecules, excluding a wrapping of the DNA around DEK (12). Another possibility is that protein–protein interactions between DNA-bound DEK molecules cause a change in helical twist by bending the DNA and thus introducing positive supercoils (see Fig. 6A). Further studies are necessary to clarify the exact mechanism.
The function of DEK in vivo is not yet known, but its structure-specific binding strongly resembles those of architectural proteins. The DNA entering and exiting from the nucleosome, which shows structural similarity to 4WJ DNAs (54), provides possible DEK-binding sites. Whatever the precise function of DEK may be, it seems to be required in many physiological states, because we see no significant changes in the amount of chromatin-bound DEK throughout the cell cycle (27).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Rolf Knippers, Ferdinand Kappes and Ingo Scholten for stimulating discussions and critical reading of the manuscript. The GST–DEK expression vector was a kind gift from Gerald Grosveld; human topoisomerase II was kindly provided by Fritz Boege. This work was supported by grants from the DFG to C.G.
REFERENCES
- 1.Armstrong J.A. and Emerson,B.M. (1998) Transcription of chromatin: these are complex times. Curr. Opin. Genet. Dev., 8, 165–172. [DOI] [PubMed] [Google Scholar]
- 2.Pollard K.J. and Peterson,C.L. (1998) Chromatin remodeling: a marriage between two families? Bioessays, 20, 771–780. [DOI] [PubMed] [Google Scholar]
- 3.Urnov F.D. and Wolffe,A.P. (2001) Chromatin remodeling and transcriptional activation: the cast (in order of appearance). Oncogene, 20, 2991–3006. [DOI] [PubMed] [Google Scholar]
- 4.Demeret C., Vassetzky,Y. and Mechali,M. (2001) Chromatin remodelling and DNA replication: from nucleosomes to loop domains. Oncogene, 20, 3086–3093. [DOI] [PubMed] [Google Scholar]
- 5.Smerdon M.J. and Conconi,A. (1999) Modulation of DNA damage and DNA repair in chromatin. Prog. Nucleic Acid Res. Mol. Biol., 62, 227–255. [DOI] [PubMed] [Google Scholar]
- 6.Green C.M. and Almouzni,G. (2002) When repair meets chromatin. EMBO Rep., 3, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- 8.Cheung P., Allis,C.D. and Sassone-Corsi,P. (2000) Signaling to chromatin through histone modifications. Cell, 103, 263–271. [DOI] [PubMed] [Google Scholar]
- 9.Aalfs J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. [DOI] [PubMed] [Google Scholar]
- 10.Bewley C.A., Gronenborn,A.M. and Clore,G.M. (1998) Minor groove-binding architectural proteins: structure, function and DNA recognition. Annu. Rev. Biophys Biomol. Struct., 27, 105–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexiadis V., Waldmann,T., Andersen,J., Mann,M., Knippers,R. and Gruss,C. (2000) The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the replication efficiency of DNA replication in a chromatin-specific manner. Genes Dev., 14, 1308–1312. [PMC free article] [PubMed] [Google Scholar]
- 12.Waldmann T., Eckerich,C., Baack,M. and Gruss,C. (2002) The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J. Biol. Chem., 277, 24988–24994. [DOI] [PubMed] [Google Scholar]
- 13.von Lindern M., Fornerod,M., van Baal,S., Jaegle,M., de Wit,T., Buijs,A. and Grosveld,G. (1992) The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol. Cell. Biol., 12, 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong X., Michelis,M.A., Wang,J., Bose,R., DeLange,T. and Reeves,W.H. (1998) Autoantibodies to DEK oncoprotein in a patient with systemic lupus erythematosus and sarcoidosis. Arthritis Rheum., 41, 1505–1510. [DOI] [PubMed] [Google Scholar]
- 15.Dong X., Wang,J., Kabir,F.N., Shaw,M., Reed,A.M., Stein,L., Andrade,L.E., Trevisani,V.F., Miller,M.L., Fujii,T. et al. (2000) Autoantibodies to DEK oncoprotein in human inflammatory disease. Arthritis Rheum., 43, 85–93. [DOI] [PubMed] [Google Scholar]
- 16.Wichmann I., Respaldiza,N., Garcia-Lozano,J.R., Montes,M., Sanchez-Roman,J. and Nunez-Roldan,A. (2000) Autoantibodies to DEK oncoprotein in systemic lupus erythematosus (SLE). Clin. Exp. Immunol., 119, 530–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sierakowska H., Williams,K.R., Szer,I.S. and Szer,W. (1993) The putative oncoprotein DEK, part of a chimera protein associated with acute myeloid leukaemia, is an autoantigen in juvenile rheumatoid arthritis. Clin. Exp. Immunol., 94, 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyn M.S., Lu-Kuo,J.M. and Herzing,L.B.K. (1993) Expression cloning of multiple human cDNAs that complement the phenotypic defects of ataxia-telangiectasia group D fibroblasts. Am. J. Hum. Genet., 53, 1206–1216. [PMC free article] [PubMed] [Google Scholar]
- 19.Hollenbach A.D., McPherson,C.J., Mientjes,E.J., Iyengar,R. and Grosveld,G. (2002) Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci., 115, 3319–3330. [DOI] [PubMed] [Google Scholar]
- 20.Krithivas A., Fujimuro,M., Weidner,M., Young,D.B. and Hayward,S.D. (2002) Protein interactions targeting the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus to cell chromosomes. J. Virol., 76, 11596–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campillos M., García,M.A., Valdivieso,F. and Vázquez,J. (2003) Transcriptional activation by AP-2a is modulated by the oncogene DEK. Nucleic Acids Res., 31, 1571–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGarvey T., Rosonina,E., McCracken,S., Li,Q., Arnaout,R., Mientjes,E., Nickerson,J.A., Awrey,D., Greenblatt,J., Grosveld,G. et al. (2000) The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon–product complexes. J. Cell Biol., 150, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lejeune F., Ishigaki,Y., Li,X. and Maquat,L.E. (2002) The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J., 21, 3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science, 293, 1836–1839. [DOI] [PubMed] [Google Scholar]
- 26.Reichert V.L., Le Hir,H., Jurica,M.S. and Moore,M.J. (2002) 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev., 16, 2778–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kappes F., Burger,K., Baack,M., Fackelmayer,F.O. and Gruss,C. (2001) Subcellular localization of the human proto-oncogene protein DEK. J. Biol. Chem., 276, 26217–26223. [DOI] [PubMed] [Google Scholar]
- 28.Fornerod M., Boer,J., van Baal,S., Jaegle,M., von Lindern,M., Murti,K.G., Davis,D., Bonten,J., Buijs,A. and Grosveld,G. (1995) Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene, 10, 1739–1748. [PubMed] [Google Scholar]
- 29.Göhring F., Schwab,B.L., Nicotera,P., Leist,M. and Fackelmayer,F.O. (1997) The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J., 16, 7361–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kipp M., Schwab,B.L., Przybylski,M., Nicotera,P. and Fackelmayer,F.O. (2000) Apoptotic cleavage of scaffold attachment factor A (SAF-A) by caspase-3 occurs at a noncanonical cleavage site. J. Biol. Chem., 275, 5031–5036. [DOI] [PubMed] [Google Scholar]
- 31.Aravind L. and Koonin,E.V. (2000) SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci., 25, 112–114. [DOI] [PubMed] [Google Scholar]
- 32.Faulkner N.E., Hilfinger,J.M. and Markovitz,D.M. (2001) Protein phosphatase 2A (PP2A) activates the HIV-2 promoter through enhancer elements which include the Pets site. J. Biol. Chem., 24, 24. [DOI] [PubMed] [Google Scholar]
- 33.Fu G.K. and Markovitz,D.M. (1996) Purification of the pets factor. A nuclear protein that binds to the inducible TG-rich element of the human immunodeficiency virus type 2 enhancer. J. Biol. Chem., 271, 19599–19605. [DOI] [PubMed] [Google Scholar]
- 34.Fu G.K., Grosveld,G. and Markovitz,D.M. (1997) DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc. Natl Acad. Sci. USA, 94, 1811–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vollenweider H.J., Sogo,J. and Koller,T. (1975) A routine method for protein-free spreading of double- and single-stranded DNA molecules. Proc. Natl Acad. Sci. USA, 72, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lilley D.M. and Clegg,R.M. (1993) The structure of the four-way junction in DNA. Annu. Rev. Biophys Biomol. Struct., 22, 299–328. [DOI] [PubMed] [Google Scholar]
- 37.Bianchi M.E. (1988) Interaction of a protein from rat liver nuclei with cruciform DNA. EMBO J., 7, 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zlatanova J. and van Holde,K. (1998) Binding to four-way junction DNA: a common property of architectural proteins? FASEB J., 12, 421–431. [PubMed] [Google Scholar]
- 39.Sheflin L.G., Fucile,N.W. and Spaulding,S.W. (1993) The specific interactions of HMG 1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry, 32, 3238–3248. [DOI] [PubMed] [Google Scholar]
- 40.Javaherian K., Liu,J.F. and Wang,J.C. (1978) Nonhistone proteins HMG1 and HMG2 change the DNA helical structure. Science, 199, 1345–1346. [DOI] [PubMed] [Google Scholar]
- 41.Stros M., Stokrova,J. and Thomas,J.O. (1994) DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res., 22, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stros M., Reich,J. and Kolibalova,A. (1994) Calcium binding to HMG1 protein induces DNA looping by the HMG-box domains. FEBS Lett., 344, 201–206. [DOI] [PubMed] [Google Scholar]
- 43.Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- 44.Paull T.T., Haykinson,M.J. and Johnson,R.C. (1993) The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev., 7, 1521–1534. [DOI] [PubMed] [Google Scholar]
- 45.Pöhler J., Norman,D.G., Bramham,J., Bianchi,M.E. and Lilley,D.M. (1998) HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J., 17, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas J.O. (2001) HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans, 29, 395–401. [DOI] [PubMed] [Google Scholar]
- 47.Varga-Weisz P., van Holde,K. and Zlatanova,J. (1993) Preferential binding of histone H1 to four-way helical junction DNA. J. Biol. Chem., 268, 20699–20700. [PubMed] [Google Scholar]
- 48.Akhmedov A.T., Frei,C., Tsai-Pflugfelder,M., Kemper,B., Gasser,S.M. and Jessberger,R. (1998) Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J. Biol. Chem., 273, 24088–24094. [DOI] [PubMed] [Google Scholar]
- 49.Quinn J., Fyrberg,A.M., Ganster,R.W., Schmidt,M.C. and Peterson,C.L. (1996) DNA-binding properties of the yeast SWI/SNF complex. Nature, 379, 844–847. [DOI] [PubMed] [Google Scholar]
- 50.Bustin M. (2001) Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci., 26, 152–153. [DOI] [PubMed] [Google Scholar]
- 51.Birger Y., West,K.L., Postnikov,Y.V., Lim,J.H., Furusawa,T., Wagner,J.P., Laufer,C.S., Kraemer,K.H. and Bustin,M. (2003) Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J., 22, 1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Losada A. and Hirano,T. (2001) Intermolecular DNA interactions stimulated by the cohesin complex in vitro: implications for sister chromatid cohesion. Curr. Biol., 11, 268–272. [DOI] [PubMed] [Google Scholar]
- 53.Hirano T. (1999) SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev., 13, 11–19. [DOI] [PubMed] [Google Scholar]
- 54.Allan J., Hartman,P.G., Crane-Robinson,C. and Aviles,F.X. (1980) The structure of histone H1 and its location in chromatin. Nature, 288, 675–679. [DOI] [PubMed] [Google Scholar]