Abstract
Gene annotation in viruses often relies upon similarity search methods. These methods possess high specificity but some genes may be missed, either those unique to a particular genome or those highly divergent from known homologs. To identify potentially missing viral genes we have analyzed all complete viral genomes currently available in GenBank with a specialized and augmented version of the gene finding program GeneMarkS. In particular, by implementing genome-specific self-training protocols we have better adjusted the GeneMarkS statistical models to sequences of viral genomes. Hundreds of new genes were identified, some in well studied viral genomes. For example, a new gene predicted in the genome of the Epstein–Barr virus was shown to encode a protein similar to α-herpesvirus minor tegument protein UL14 with heat shock functions. Convincing evidence of this similarity was obtained after only 12 PSI-BLAST iterations. In another example, several iterations of PSI-BLAST were required to demonstrate that a gene predicted in the genome of Alcelaphine herpesvirus 1 encodes a BALF1-like protein which is thought to be involved in apoptosis regulation and, potentially, carcinogenesis. New predictions were used to refine annotations of viral genomes in the RefSeq collection curated by the National Center for Biotechnology Information. Importantly, even in those cases where no sequence similarities were detected, GeneMarkS significantly reduced the number of primary targets for experimental characterization by identifying the most probable candidate genes. The new genome annotations were stored in VIOLIN, an interactive database which provides access to similarity search tools for up-to-date analysis of predicted viral proteins.
INTRODUCTION
Currently, the complete genome of a virus can be sequenced within days. The next step towards understanding the details of a virus life cycle is to identify the whole complement of viral genes and proteins. This information can provide critical insights on many occasions. For instance, for a team working on an antiviral drug design, promising drug targets would be those viral proteins that are basically identical in all major strains of a virus and are significantly different from the proteins in the host, e.g. human.
At the time of this study, the GenBank database (1) contained ∼3000 annotated complete viral genome sequences. In most cases, research groups providing the original annotation are unable to detect and confirm all genes experimentally by the time of submission. Computational approaches have therefore been commonly used since the time of pioneer projects such as the sequencing and annotation of phage λ (2).
There are two major approaches to gene identification, intrinsic and extrinsic (3). The intrinsic approach, which can be also called an ab initio statistical approach, uses statistical patterns of nucleotide frequencies and nucleotide ordering observed in a given genome. These patterns are not the same in protein-coding and non-coding DNA sequences; hence a properly trained intrinsic method can recognize protein-coding regions. Extrinsic methods seek to identify evolutionarily conserved sequences in protein-coding regions. These sequences can be detected by similarity searches. The extrinsic method is thus dependent on external information residing outside the sequence of interest.
Intrinsic and extrinsic methods have complementary strengths. Tests of their predictive power performed with sets of sequences containing known genes show that the intrinsic methods have higher sensitivity than the extrinsic methods which usually have higher specificity. Using intrinsic and extrinsic methods in concert is therefore a worthwhile approach (3).
So far, the use of computational gene identification methods in viral genomes by the groups of researchers submitting genomic data to GenBank was primarily restricted to similarity searches. To reduce the risk of missing real genes, a simple statistics-based rule is frequently applied to take into account the difference in length distributions of real genes and random open-reading frames (ORFs). This rule suggests annotating ‘long enough’ ORFs as genes. For instance, in the rat cytomegalovirus genome any ORF longer than 300 nt not overlapping an adjacent ORF to an extent larger than 60% was annotated as a gene (4). Such a simplistic rule, however, could cause substantial over-annotation, especially in genomes with high G+C content.
Another frequently used simplification is the annotation of a gene start by the ‘longest ORF’ rule (assignment of a gene start to the 5′-most ATG codon). A screening of GenBank identified 26 complete viral genomes with a total of 4400 genes, all annotated using this rule. It was nevertheless shown earlier that the true start may not be pinpointed by this rule in ∼25% of cases (5).
Viral genomes are different from the genomes of their hosts in several aspects that hamper immediate successful application of the gene finding methods developed for their hosts. An important factor is the rather small size of a viral genomic sequence. Currently, the RefSeq collection (19) contains 891 viral genomes shorter than 10 kb with a total of 2900 genes annotated, 169 genomes with lengths between 10 and 100 kb (3500 genes) and 47 genomes longer than 100 kb (7900 genes). A rather short genome size makes it either impossible to apply previously developed training procedures to derive parameters of high order statistical models (for the shortest viral genomes) or significantly limits the accuracy of these models (even in the case of the longest viral genomes). Another important feature of viral genome organization is the high frequency of gene overlaps that occur in viruses of both prokaryotic and eukaryotic hosts. The gene overlaps in viral genomes appear to be considerably longer than those seen in prokaryotic and, much more rarely, eukaryotic genomes. Furthermore, some annotated and experimentally confirmed viral genes are completely overlapped by others. Repetitive DNA may occupy a large portion of a viral genome; for example, in the Epstein–Barr virus genome (NC_001345) repetitive regions amount to ∼30% of the genomic sequence (6), thus making model training more complicated.
In spite of the difficulties mentioned above, several groups have attempted to apply earlier developed statistical gene prediction programs for viral genome annotation. For instance, the GeneMark program (7) was used to identify genes in the genomes of Bovine herpesvirus 4 (8), bacteriophage FKZ of Pseudomonas aeruginosa (9), Mycoplasma virus P1 (10), Mycobacteriophage D29 (11), Stx 2e-encoding phage FP27 (12), coliphage T4 and the marine cyanophage S-PM2 (13), as well as to identify genes in genomes of virulence plasmids in Rhodocuccus equi (14), Shigella felxneri (15) and Escherichia coli (16). Still, these initial attempts did not use a tool developed specifically for the problem in hand (except perhaps the case of T4, where the GeneMark models were adjusted to the genomic T4 sequence).
A significant difference may exist sometimes between the GenBank record and the original publication. For instance, the annotation of the white spot bacilliform virus (GenBank record AF332093) lists 531 protein-coding genes in comparison with only 181 genes mentioned in the original publication (17). On the other hand, only 23 genes are annotated in Rana tigrina ranavirus (GenBank record AF389451), while the original publication (18) describes 105 genes. In order to improve the quality of DNA sequence annotation, the National Center for Biotechnology Information (NCBI) has created the RefSeq collection. While the original GenBank genomic record is maintained as suggested by the authors, the RefSeq record of the same sequence is continuously updated with regard to new relevant data that become available. There were 1191 RefSeq records for complete genomes of viruses of prokaryotic and eukaryotic hosts as of August 2002.
Several attempts have been made to organize data on viral genomes in interactive databases providing tools for analysis of viral genes and proteins (20–22). These projects have been typically focused on specific classes of viruses.
To provide a tool for accurate ab initio gene identification in viral genomes we have modified the earlier developed GeneMarkS program (5) to make it suitable for analysis and gene prediction in viral genomes of different types. As a result of the application of this tool, we have created new annotation records for viral genomes present in GenBank (including its RefSeq part). These records have been compiled in the database VIOLIN (viral genomes online) accessible online at http://opal.biology.gatech.edu/GeneMark/VIOLIN/.
MATERIALS AND METHODS
Materials
A set of 2945 complete viral genome records was downloaded from GenBank. Since several genomic variants (strains, mutants, isolates) were determined for many viral species, many viral genome records had several other almost identical entries. To filter out this redundancy we have specifically focused on the analysis of viral genomes from the RefSeq collection containing 1191 complete genomic records of viruses of eukaryotic (1071) and prokaryotic (120) hosts. RefSeq contains only one record for each virus species. Notably, these 1191 RefSeq viral genome annotations included 86 records that had been updated with the aid of our new predictions. In what follows, these 86 records have been treated differently in terms of comparison of predicted and annotated genes.
Methods
For phage genomes with prokaryotic-type gene organization, computer methods of prokaryotic gene finding could be adjusted rather easily. The prokaryotic version of GeneMark.hmm as well as its self-training version GeneMarkS were previously shown to possess high accuracy both in detecting prokaryotic genes as a whole and in exactly pinpointing gene starts (23,24). Therefore, GeneMarkS was the natural choice as the tool to be applied and adjusted for the analysis of phage genomes. For viruses of eukaryotic hosts, the situation is more complex. Current eukaryotic gene finding algorithms are unable to predict the gene overlaps frequently seen in genomes of viruses of eukaryotic hosts. On the other hand, according to the RefSeq annotation of ∼11 000 genes in 1015 genomes of viruses of eukaryotic hosts, only ∼300 genes have introns. Therefore, use of the program able to predict overlapping genes provides more benefits than the one predicting exon–intron structures. The program suitable for immediate use and further modifications was again the prokaryotic GeneMarkS, which could identify overlapping protein-coding ORFs while rarely occurring exons would be predicted as separate ORFs.
A viral genomic sequence might not provide enough training data to determine parameters of Markov chain models used in GeneMark.hmm. We turned, therefore, to the heuristic training technique described earlier (24), which is able to derive the parameters of the required models from a DNA sequence as short as 400 nt.
For larger viral genomes, the statistical models initially defined by the heuristic procedure could be iteratively refined further by the unsupervised training procedure implemented in GeneMarkS (24). This iterative procedure used simultaneous training and gene prediction to build models of protein-coding and non-coding sequences. For larger phage genomes, GeneMarkS also derived a model for the ribosomal binding site (RBS) and its spacer (the sequence between the rightmost nucleotide of the RBS and the first nucleotide of the start codon). Parameters of both models were determined from the multiple alignment of the nucleotide sequences situated upstream of the predicted gene starts, with the alignment constructed by the Gibbs Motif Sampler (25). For large enough genomes of viruses of eukaryotic hosts, parameters of a model for the Kozak pattern associated with the translational initiation site were determined by GeneMarkS with yet another modification. This GeneMarkS version allowed the use of the Kozak model for gene start prediction. Further modifications were done to adjust the program to different types of viral genome organization.
Since a linear viral genome cannot have a partial coding region at either terminus, a specific restriction imposed at the program initialization stage excluded this possibility. Conversely, an additional post-processing step was implemented for circular viral genomes to detect genes possibly divided by the split point chosen in the original annotation. For the single-stranded RNA (ssRNA) positive strand viruses whose genes are located in one strand only, an additional procedure identified the strand where gene predictions clustered predominantly and the opposing strand was assigned as completely non-coding.
For every viral genome the training procedure had to determine whether the sequence data were only sufficient for obtaining heuristic models or if a full training cycle of GeneMarkS could be initiated. If GeneMark.hmm with the initially defined heuristic models predicted fewer than a certain number of genes, Nr, then the procedure stopped and these initial predictions were not refined further. Otherwise, the full cycle of GeneMarkS training was initiated. The number 50 was assigned as the default Nr number.
In the training process, if several repetitive copies of some predicted protein-coding ORFs were identified, all copies but one were excluded from the training set of protein-coding regions to reduce bias in the protein-coding sequence model. Predicted ORFs longer than 500 nt that appeared in predicted intergenic regions were excluded from the set of non-coding regions to exclude possible ‘contamination’ of the non-coding training set. For viral genomes with a total size of predicted non-coding regions <10 kb, the training set of non-coding regions was augmented with an additional 10 kb sequence generated by the simplest multinomial model, simulating a sequence with the frequencies of the four nucleotides identical to those observed in the native non-coding region (26).
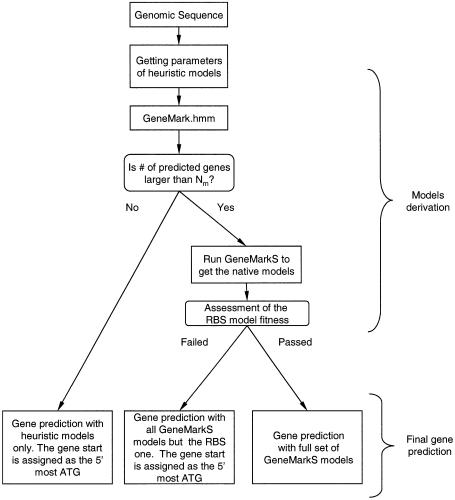
The step-wise diagram of GeneMarkS self-training and gene prediction for the genome of a virus of prokaryotic host is shown in Figure 1. For a virus of a eukaryotic host, a reference to the Kozak model should replace the reference to the RBS model. The evaluation of the RBS model fitness was done by assessing both the variance of the RBS signal localization and the information content of the RBS model derived by the Gibbs Sampler. The Kozak model was evaluated in a similar manner. The self-training procedure was terminated as soon as two subsequent iterations produced the same gene predictions. However, in some cases exact convergence was not achieved due to small cyclic variations observed in subsequent iterations. In these cases the self-training was stopped and the reported sequence parse into coding and non-coding regions was the one with the larger number of predicted genes.
Figure 1.
Flowchart of the statistical gene identification procedure applied to a complete genome of a virus of a prokaryotic host. For viruses of eukaryotic hosts, the Kozak model is used instead of the RBS model.
Assessment of the accuracy of computer gene prediction is a critically important issue. To characterize errors of two sorts, false positive and false negative, we used two parameters of accuracy, sensitivity and specificity. The value of sensitivity (Sn) is defined as the ratio of the number of true predictions to the number of genes in a test set. The fewer the number of false negatives, the higher the sensitivity. The value of specificity (Sp) is defined as the ratio of the number of true predictions to the total number of predictions made. The fewer the number of false positives, the higher the specificity. To determine sensitivity and specificity values for a particular gene prediction method, one needs a test set of nucleotide sequences with experimentally verified genes. To further define the terms we say that a gene is ‘detected’ if its 3′ end coincides with the 3′ end of a verified one. Additionally, a gene is ‘predicted exactly’ if the positions of both ends coincide with the verified gene ends. The accuracy of ‘exact prediction’ in our terms is the same as the accuracy of the ‘gene start prediction’. This value is defined by the fraction of ‘exactly predicted genes’ among ‘detected’ genes.
The BLAST searches used to characterize newly predicted proteins were conducted using standard parameters: BLOSUM62; penalty for gap ‘10’; penalty for gap extension ‘1’; low-complexity filtering ‘on’. In PSI-BLAST searches, the parameters were the same with the exception that the low-complexity filtering was ‘off’.
RESULTS AND DISCUSSION
The overall statistics of the results of our analysis of complete viral genomes from GenBank is shown in Table 1. Our major focus here is on the genomes from the RefSeq collection. Those 86 viral genomes that had previously been reannotated in RefSeq with the aid of our analysis were excluded from our comparisons.
Table 1. Summary of the results of the analysis of viral genomes currently available in GenBank and those viral genomes for which reference sequences (RefSeq collection) have already been created at NCBI.
| GenBanka Total | RefSeq Total | Eukaryotic hosts | Prokaryotic hosts | |
|---|---|---|---|---|
| Database summary | ||||
| Number of viral genomes analyzed | 1750 | 1107 | 1015 | 92 |
| Prediction and annotation comparison | ||||
| Exact match between prediction and annotation | 15703 | 10425 | 8011 | 2414 |
| Predicted gene differs in start location from annotated one | 1479 | 931 | 368 | 563 |
| Predicted gene overlaps with an intron containing annotated gene | 382 | 209 | 190 | 19 |
| Annotated gene was not predicted (possible false negative) | 3885 (25%)b | 2720 (26%)c | 2231 (28%)c | 489 (20%)c |
| Newly predicted genes (possible false positive) | 3520 (22%)b | 1360 (13%)c | 1047 (13%)c | 313 (13%)c |
| Analysis of newly predicted genes | ||||
| Prediction has a BLASTP and CD-Search hit with E-value <0.005 | 622 | 99 | 89 | 10 |
| Prediction has a BLASTP hit with E-value <0.005, no CD-Search hit | 1248 | 336 | 243 | 93 |
| Prediction has a CD-Search hit with E-value <0.005, no BLASTP hit | 35 | 6 | 6 | 0 |
| Prediction has no BLASTP or CD-Search hit with E-value <0.005 | 1615 | 919 | 709 | 210 |
The numbers in the RefSeq columns do not reflect 86 genomes annotated in RefSeq with the aid of the VIOLIN data. Newly predicted genes have been further analyzed by BLASTP and these results are shown in the bottom rows.
aThe GenBank records used in the current analysis did not include RefSeq records; however, the original records for each RefSeq record were included in this GenBank set of genomes.
bThe percentage value is defined with regard to the number of predicted genes exactly matching the annotation in GenBank.
cThe percentage value is defined with regard to the number of predicted genes exactly matching the annotation in RefSeq.
As shown in the RefSeq section of Table 1, 8011 protein-coding genes predicted in 1015 complete genomes of viruses of eukaryotic hosts matched the earlier annotation exactly. However, 1047 gene predictions did not match any previously annotated gene, and for 332 out of these 1047 new predictions, hits to known proteins with E-values <10–5 were found by BLASTP search (27). Interestingly, 135 out of these 332 similarity search supported predictions overlapped with annotated genes but the reading frames were different. A rather large number of 2231 genes in the RefSeq annotated genomes of viruses of eukaryotic hosts were not confirmed by our analysis.
In 92 RefSeq phage genomes, 2414 gene predictions matched the existing annotation exactly. There were 313 entirely new predictions, and 103 of them were corroborated by the BLASTP search with hits to known proteins (E-value <10–5). Again, approximately one-third of predictions corroborated by the similarity search (36 out of 103) overlapped already annotated genes with different reading frames. Our analysis did not confirm 489 genes annotated in phage genomes from the RefSeq collection.
Those 2720 (2231 + 489) genes that were annotated in the RefSeq viral genomes but were not predicted in this study are of a special interest. Subsequent BLASTP searches of these genes protein products against the non-redundant database detected similarity to other known proteins only for 848 out of the 2231 genes annotated in genomes of viruses of eukaryotic hosts and for 137 out of the 489 genes annotated in phages. Overall, we came to the number 985 as the total number of genes not predicted by the ab initio method, though these annotated genes had significant similarity with other known proteins. Therefore, given the whole number of 14 076 genes annotated in 1107 viral genomes, the false negative rate of the ab initio prediction method might be estimated at <10%. Interestingly, in 620 RefSeq viral genomes no annotated gene was missed in predictions.
As is indicated in Table 1, analysis of the original GenBank genomic records produced a larger fraction of newly predicted genes than determined in the genomes from the RefSeq collection. In turn, a larger fraction (28%) of these new genes produced significant BLASTP hits in comparison with the fraction of new genes in RefSeq (10%) supported by BLASTP search.
The gene prediction results for the RefSeq complete viral genomes were grouped together by virus length and type (Tables 2 and 3). Interestingly, a large number of new genes were identified in genomes shorter than 10 kb (892 genomes). For example, in the 8454 nt long genome of single-stranded DNA (ssDNA) enterobacteria phage IF1 (NC_001954) we identified a new 192 nt long gene coding for a homolog of Vibrio cholerae RasR protein. In contrast to all other known genes of this phage, this new gene was located in the DNA strand complementary to the ssDNA present in the virion. The largest numbers of newly identified genes or genes with new start predictions turned out to reside in 193 genomes of double-stranded DNA (dsDNA) viruses and 418 genomes of ssRNA viruses (Table 3).
Table 2. Distribution of the results of the comparative analysis of gene prediction and annotation for viral genomes from the RefSeq collection with the three sets of viruses clustered by genome length.
| a | L < 10 000 ntb (891)c | 10000 <= L <= 100 000 ntb (169)c | L > 100 000 ntb (47)c |
|---|---|---|---|
| Exact match | 1772 | 2493 | 6160 |
| Different start | 225 (12.7%) | 483 (19.4%) | 223 (3.6%) |
| Overlap with interrupted gene | 79 ( 4.5%) | 43 (1.7%) | 87 (1.4%) |
| Annotated gene not predicted | 731 (41.3%) | 499 (20.0%) | 1490 (24.1%) |
| New predictions | 331 (18.7%) | 350 (14.0%) | 679 (11.0%) |
| Analysis of newly predicted genes | |||
| BLASTP and CD-Search hit | 26 | 34 | 39 |
| BLASTP only hit | 51 | 104 | 181 |
| CD-Search only hit | 1 | 0 | 5 |
| No hits | 253 | 212 | 454 |
aThe meaning of the categories in this column is the same as in the left-most column in Table 1.
bThe genome length is designated as L.
cThe number in parentheses designates the number of genomes of a given category.
Table 3. Distribution of the results of the comparative analysis of gene prediction and annotation for viral genomes from the RefSeq collection joined in classes defined by viral classification.
| a | dsDNA (193)b | ssDNA (185)b | dsRNA (127)b | ssRNA positive strand (418)b | ssRNA negative strand (82)b | Retroid (65)b | Satellite (27)b | Virus not classified (6)b | Phage not classified (3)b |
|---|---|---|---|---|---|---|---|---|---|
| Exact match | 8532 | 440 | 142 | 750 | 252 | 151 | 12 | 12 | 132 |
| Different start | 644 | 56 | 5 | 115 | 36 | 32 | 0 | 1 | 42 |
| Overlap with interrupted gene | 125 | 2 | 4 | 51 | 3 | 24 | 0 | 0 | 0 |
| Annotated gene not predicted | 2053 | 275 | 12 | 245 | 32 | 45 | 4 | 6 | 49 |
| New predictions | 1025 | 88 | 54 | 72 | 32 | 53 | 4 | 3 | 29 |
| Analysis of newly predicted genes | |||||||||
| BLASTP and CD-Search hit | 79 | 2 | 0 | 5 | 0 | 13 | 0 | 0 | 0 |
| BLASTP only hit | 279 | 21 | 6 | 12 | 3 | 8 | 1 | 0 | 6 |
| CD-Search only hit | 5 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| No hits | 662 | 65 | 48 | 54 | 29 | 32 | 3 | 3 | 23 |
aThe meaning of the categories in this column is the same as in the left-most column in Table 1.
bThe number in parentheses designates the number of genomes of a given category.
Quite a few new predictions among those that had no BLASTP search support were found to overlap already annotated genes. This occurred 274 times (20% of newly predicted genes) in the RefSeq genomes. In 117 of these cases the product of the annotated gene showed similarity to a protein in another species. Nevertheless, the fact of overlap does not indicate a likely false positive prediction per se. Gene overlap is a quite frequent phenomenon in viral genomes, as 52% of viral genes annotated in RefSeq overlap each other.
Ideally, the characteristics of gene prediction accuracy, sensitivity and specificity (defined in Methods), should be determined for a test set of sequences containing experimentally verified genes. However, any given viral genome, except perhaps several of tiny size, would not have a large fraction of genes annotated experimentally. For this reason, we have compiled sets of so-called ‘trustable’ genes and used them as the test sets. For instance, in nine genomes of human herpesviruses (Table 4) we identified as trustable the genes both annotated and ab initio predicted. Also, we included in this category those genes that were either annotated or predicted and possessed additional ‘extrinsic’ evidence for being a real gene. This could be an experimentally characterized function or statistically significant sequence similarity to previously characterized proteins. For this compiled set of trustable genes of human herpesviruses, we obtained the average values of Sn = 91% and Sp = 84% as the estimates of the accuracy of our method.
Table 4. Gene prediction accuracy assessment for nine human herpesviruses.
| Virus | Number of genes predicted | Number of genes annotated | Number of genes in test set | Number of correct predictions | Prediction sensitivity (%) | Prediction specificity (%) |
|---|---|---|---|---|---|---|
| HHV-1 (HSV-1) | 76 | 73 | 75 | 69 | 92 | 90 |
| HHV-2 (HSV-2) | 77 | 71 | 71 | 65 | 92 | 84 |
| HHV-3 (VZV) | 72 | 71 | 71 | 69 | 97 | 96 |
| HHV-4 (EBV) | 90 | 94 | 78 | 70 | 89 | 78 |
| HHV-5 (HCMV) | 164 | 198 | 148 | 125 | 84 | 76 |
| HHV-6A | 115 | 121 | 119 | 104 | 87 | 90 |
| HHV-6B | 114 | 91 | 85 | 81 | 95 | 71 |
| HHV-7 | 109 | 107 | 104 | 90 | 87 | 83 |
| HHV-8 (KSHV) | 96 | 82 | 88 | 83 | 94 | 86 |
| Total | 913 | 908 | 839 | 767 | 91 | 84 |
The test set was compiled as explained in text.
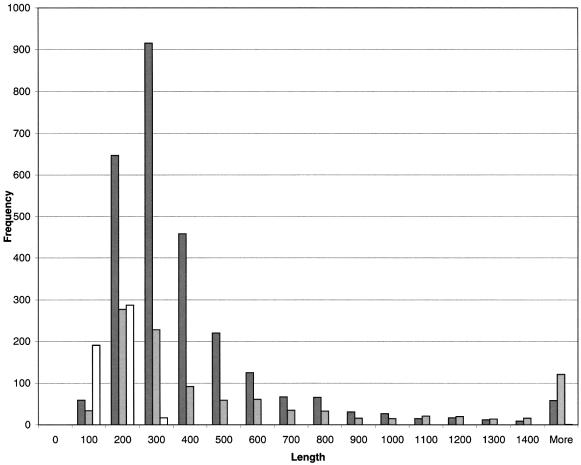
Length comparison between newly predicted genes and genes annotated but missed in predictions indicated that the newly predicted genes tend to be shorter than the ones supposedly real but missed in predictions (Fig. 2). The ratio of newly predicted genes to missed genes decreased from 3.81 for genes shorter than 300 nt to 0.49 for genes longer than 300 nt. This observation seems to be related to a preference in the original records to have longer ORFs annotated as genes. The longer ORFs are generally assumed to be more likely to be real genes while ORFs shorter than 300 nt are difficult to discriminate from random non-coding ORFs and are more risky to annotate as genes. This conventional wisdom could lead to over-annotation of ORFs longer than 300 nt as genes while some short genes could be missed. As Figure 2 shows, many ‘long’ annotated genes were indeed not confirmed while quite a few new ‘short’ genes were predicted.
Figure 2.
Length distributions of several categories of genes predicted or annotated in 1047 RefSeq viral genomes. Dark gray bars are used for genes annotated but not predicted; light gray bars are used for predicted but not annotated genes whose protein products produce BLASTP hits with E-values <10–5; white bars are used for predicted but not annotated genes whose protein products do not produce BLASTP hits with E-values <10–5.
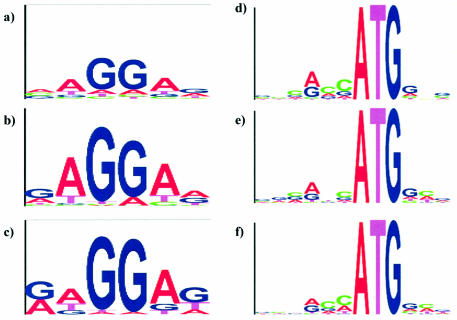
Assessing and improving the gene start prediction accuracy is another important issue. As described above, for more precise gene start prediction we used the RBS model for long enough viruses of prokaryotic hosts and the Kozak model for viruses of eukaryotic hosts. To give an example, the positional frequency matrices of RBS models specific for phage T4 and phage λ are visualized in ‘logo’ images (28) in Figure 3b and c. Notably, these images emphasize the similarity of the nucleotide frequency patterns existing in the RBS of phages to the pattern known for E.coli (Fig. 3a). This observation could be expected given that T4 and λ use the E.coli translational mechanism. While the positional frequency matrix of the RBS model has a fixed length and variable pattern of positional frequencies, the model of the RBS spacer allows for sequences of variable lengths (distances between RBS and start codon) with an invariant positional frequency pattern of the non-coding region.
Figure 3.
The positional nucleotide frequency patterns of the GeneMarkS models of the RBS pattern for phage T4 (b) and phage λ (c) are shown in the logo form (27), as compared with the RBS pattern of E.coli shown in (a). Similarly, the Kozak pattern for human herpesvirus 4 (e) and human herpesvirus 8 (f) are shown in the logo form, with the Kozak pattern for human genes shown in (d).
The logos for the Kozak model determined for the Epstein–Barr virus (HHV4) and for Kaposi’s sarcoma herpesvirus (HHV8) shown in Figure 3e and f clearly indicate that the information content of these signals is lower than that of RBS. However, the Kozak patterns observed in these viruses are still similar to the Kozak pattern known for the genome of the human host (Fig. 3d). Accurate evaluation of the gene start prediction accuracy requires a set of genes with experimentally verified gene starts. Evaluation of GeneMarkS performance was done earlier on the test set of E.coli genes with 5′ ends verified by sequencing of N-terminals of encoded proteins (29). In this test the accuracy of start prediction was observed to be as high as 94% (5). A comparison of predictions for phage T4 both with and without the use of the RBS model was carried out (Supplementary Material, Table 1). This comparison showed that predictions made with the use of the RBS model made an almost 10% better match with the annotation, which we consider sufficiently accurate for this well studied phage genome.
Considering viruses of eukaryotic hosts, we compiled a set of genes from nine human herpesviruses with translation starts confirmed by similarity search on a protein level. The 5′ end of the protein having the highest BLASTP hit (excluding one or several self hits) was compared with the 5′ end of the query protein to assess the accuracy of the gene start prediction. After selection of the most unambiguous cases, we obtained an estimate of the accuracy of start prediction as 85% (Supplementary Material, Table 2).
The whole set of newly predicted genes was used further to search for similarity and reconstruct possible orthologous relationships. A database of 1360 newly predicted proteins was compiled and was cross-searched using BLASTP. We found that 237 predicted proteins had some similarity to other members in the database and could be further grouped into 106 protein clusters (Supplementary Material, Table 3). Some of these clusters show highly conserved regions; for instance, a cluster of protein products of new genes identified in poxviruses.
Now we take a closer look at several individual gene predictions. In the well studied genome of Bacteriophage λ (JO2459) we identified as many as five new genes. These genes have already been included in the RefSeq version of the phage λ annotation (NC_001416). Two genes, coding for a putative envelope protein (NP_597781) and Bor protein precursor (NP_597780), are similar to genes in prophage CP-933X, being a part of the E.coli O157 genome (NC_002655). A gene for superinfection exclusion protein B (NP_597779) must have been known for some time since its protein product had been included into the PIR database (P03762). The other two genes were classified as hypothetical.
Our predictions of 16 new genes in Porcine adenovirus A (NC_001997) were corroborated by similarity search. For instance, the protein encoded by predicted ORF6 is a member of a family of DNA polymerases present in 39 other adenoviruses.
A potentially important finding was a gene located in positions 10443–11138 of the genome of Alcelaphine herpesvirus 1 (NC_002531) coding for a 231 amino acid long putative protein (NP_597933). Initially, the new protein was shown to be similar to the uncharacterized putative protein ORF E4 (NP_042601, AAC13792) of unclassified γ-herpesvirus Equine herpesvirus 2. A subsequent PSI-BLAST search revealed a striking similarity between these two proteins and recently discovered antagonists of the lymphocryptovirus antiapoptotic BCL-2 proteins (30). Later, the sequence of a third non-lymphocryptovirus protein, hypothetical v-BCL2 of another unclassified γ-herpesvirus (Porcine lymphotropic herpesvirus 1) was released (31) and we have found its sequence to be very similar to the newly identified protein (NP_597933). The PSI-BLAST search profile built from the three proteins further identified similarity with ORF1 protein of Callitrichine herpesvirus 3 (a lymphocryptovirus BALF1-like BCL-2 like protein) and with the BALF1 protein (AAK01916) of Allitrichine herpesvirus 3 (a lymphocryptovirus) with E-values of 8 × 10–4 and 0.007, respectively. This range of E-values has been characterized as being indicative of significant sequence similarity (32,33). The output of the third iteration of PSI-BLAST included all the BALF1-like proteins at the top of the list. Human GRS protein and other BCL-2-like non-viral proteins were also present in the list at a substantial score distance.
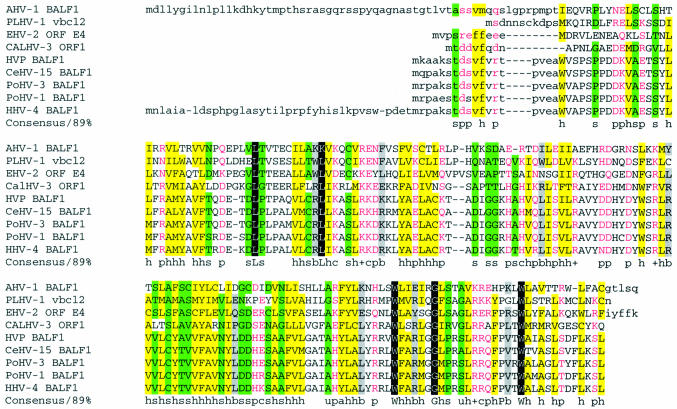
In the next round of analysis, the RPS-BLAST (the NCBI program comparing protein sequences with the Conserved Domain Database) readily detected a BCL motif in all three non-lymphocryptovirus proteins. Moreover, multiple alignment by hierarchical clustering (34) of the newly predicted protein (NP_597933) with proteins NP_042601, AAM22111 and all the lymphocryptovirus BALF1 proteins (Fig. 4) further supported the probable functional significance of the observed pairwise similarity by making evident the patterns of amino acids conserved in all sequences. Interestingly enough, a TBLASTN search failed to reveal additional un-annotated homologs of NP_597933. It is tempting to speculate that, given the function of BALF1 (30), the newly identified BALF1-like protein may be involved in a complex regulation of the host cell apoptosis, presumably as an antagonist of the herpesvirus antiapoptotic BCL-2 proteins, and, perhaps, as a part of a gene network involved in carcinogenesis.
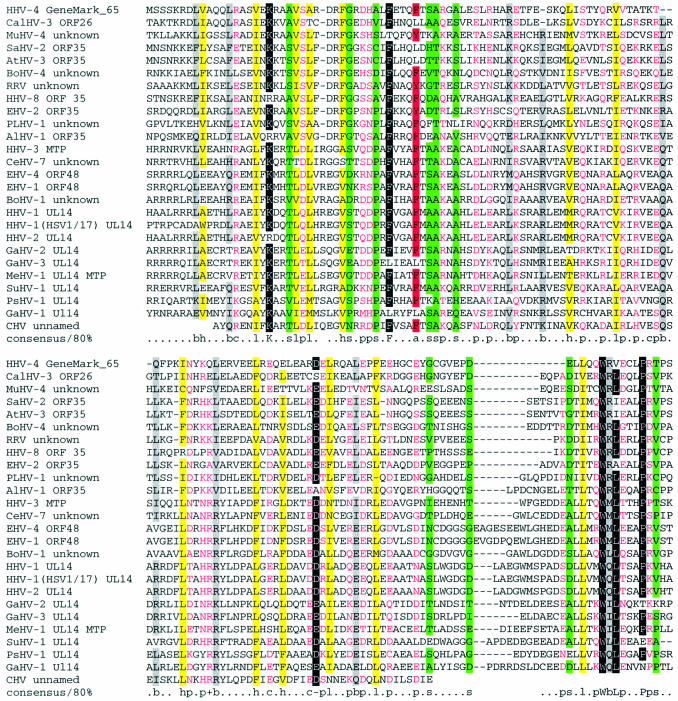
Figure 4.
MultAlin alignment of (putative) BALF1-like proteins (33). The variable N- and C-termini are shown in lower case. Protein names are abbreviated as follows: AHV-1 BALF1, BALF1 homolog (NP_597933) predicted by GeneMarkS in the genome of Alcelaphine herpesvirus 1 (NC_002531); PLHV-1 vbcl2, Porcine lymphotropic herpesvirus 1 hypothetical v-bcl2 (AAM22111); CALHV-3 ORF1, Callitrichine herpesvirus 3 ORF1 (AAK38208); HVP BALF1, Herpesvirus papio BALF1 (AAK01916); PoHV-3 BALF1, Pongine herpesvirus 3 BALF1 (AAK60342); HHV-4 BALF1, Human herpesvirus 4 BALF1 (NP_039912); PoHV-1 BALF1, Pongine herpesvirus 1 (AAK01917); CeHV-15 BALF1, Cercopithicine herpesvirus 15 (AAK95480); EHV-2 ORF E4, Equine herpesvirus 2 ORF E4 protein (NP_042601). The conserved positions are color coded based on the type of amino acid residue as indicated in the consensus line, where h and a stand for hydrophobic residues (A, C, F, I, L, M, V, W, Y: yellow background in alignment) and for aromatic residues (F, Y, W), respectively; b stands for ‘large ’ residues (E, K, R, I, L, M, F, Y, W: gray background); p stands for polar residues (D, E, H, K, N, Q, R, S, T: shown in pink); s and u stand for small residues (A, C, S, T, D, N, V, G, P: green background) and tiny residues (G, A, S), respectively; c and + stand for charged residues (K, R, D, E, H: shown in pink) and positively charged residues (K, R), respectively. Invariant amino acid residues (in 85% or more sequences) are highlighted with black background.
Another interesting new finding was a gene (ORF65) predicted in the genome of Epstein–Barr virus (HHV-4, NC_001345). Initially, the protein product of this gene was found to be significantly similar (with an E-value of <10–5) to uncharacterized ORF26/ORF35 proteins of other γ-herpesviridae. The subsequent PSI-BLAST search revealed after four iterations a similarity (with an E-value of 6 × 10–4) to the ORF26/ORF35 protein family and the ORF48 protein of Equine herpesvirus 4, an α-herpesvirus. The ORF48 protein belongs to the UL14 family of proteins which are present in a minor component of the virion tegument and possess heat shock protein-like functions (35). Eight further PSI-BLAST iterations brought up all the members of this family. Multiple alignment of the ORF26/ORF35 and UL14-like protein sequences (Fig. 5) highlights common features that could not be readily seen in pair-wise alignments, particularly, similar patterns of distribution of charged residues. The observed sequence similarity strongly indicates a common function which remains to be determined by direct experiments. It is likely that these proteins play an important role since the members of the ORF26/ORF35 protein family are now confirmed to be present in all complete genomes of γ-herpesviruses. Interestingly, none of the β-herpesviruses genomes has a TBLASTN detectable homolog of ORF26/ORF35 or UL14, which indicates that ORF26/ORF35 proteins are likely to fulfill a subfamily-level function.
Figure 5.
Alignment of the sequences of ORF26/ORF35 and UL14-like proteins. For most sequences, the N- and C-termini are not shown. The coloring is as in Figure 4. The protein gi numbers and the organism names are: HHV-4 GeneMark_65 prediction (positions 1–139) (Human herpesvirus 4); SaHV-2 ORF35 (1–147), 9625991 (Saimiriine herpesvirus 2); HHV-3 MTP (minor tegument protein, positions 11–159), 9625920 (Human herpesvirus 3); CeHV-7 unknown (11–159), 13242439 (Cercopithecine herpesvirus 7); AtHV-3 ORF35 (1–147), 9631227 (Ateline herpesvirus 3); EHV-4 ORF48 (7–155), 9629775 (Equine herpesvirus 4); BoHV-4 unknown (4–150), 13095612 (Bovine herpesvirus 4); RRV unknown (3–146), 18653842 (Rhesus rhadinovirus, Macaca mulatta rhadinovirus); HHV-1 UL14 (7–151), 9629394 (Human herpesvirus 1); HHV-2 UL14 (7–155), 9629283 (Human herpesvirus 2); HHV-1 (HSV1/17) UL14 (3–155), 136823 [Herpes simplex virus (type 1/strain 17)]; EHV-1 ORF48 (7–155), 9626785 (Equine herpesvirus 1); EHV-2 ORF35 (5–150), 9628038 (Equine herpesvirus 2); CalHV-3 ORF26 (3–148), 13676668 (Callitrichine herpesvirus 3); HHV-8 ORF35 (3–147), 18846002 (Human herpesvirus 8); PLHV-1 unknown (3–149), 20453822 (Porcine lymphotropic herpesvirus 1); AlHV-1 ORF35 (2–148), 10140956 (Alcelaphine herpesvirus 1); GaHV-2 UL14 (19–161), 9635049 (Gallid herpesvirus 2); MeHV-1 UL14 MTP (13–156), 12084842 (Meleagrid herpesvirus 1); GaHV-3 UL14 (8–156), 10834883 (Gallid herpesvirus 3); MuHV-4 unknown (3–149), 9629576 (murid herpesvirus 4); PsHV-1 UL14 (15–163), 13094667 (Psittacid herpesvirus 1); BoHV-1 unknown (18–170), 9629861 (Bovine herpesvirus 1); GaHV-1 Ul14 (62–210), 5708112 (Gallid herpesvirus 1); CHV unnamed (1–112, the entire sequence; appears to be incomplete), 1066253 (Canine herpesvirus); SuHV-1 UL14 (6–159, end of sequence), 267201 [Suid herpesvirus 1 (strain NIA-3)].
Some coding regions in viral genomes were missed in the earlier annotation because of their unusual organization. For instance, some viral genes contain a weak, read-through stop codon, which in the original annotation is considered the end of the gene; thus, a part of the real gene (and protein) is missed. In Barmah Forest virus a GeneMarkS prediction (ORF2), recovers the second part of the non-structural polyprotein gene in positions 5679–7298, missed in the original record U73745. Only after combining together these two parts, the protein (NC_001786) shows full-length similarity to the complete polyprotein encoded, for instance, in Ross River virus.
The vast majority of genes in viral genomes have no introns. There are, however, a few genes with introns and even some with whole separate genes located inside introns, such as an IE glycoprotein gene, HCMVUL37, in Human herpesvirus 5 (NC_001347). Genes interrupted by introns were identified by GeneMarkS as series of separate protein-coding ORFs. For instance, in Enterobacteria phage T4 (introns may appear not only in viruses of eukaryotic hosts but in phages as well) a gene for DNA topoisomerase small subunit protein (NC_000866) consists of two exons both predicted by GeneMarkS as separate ORFs. Developing an ab initio approach for exact prediction of introns in viral genes is a challenging problem. However, quite frequently the combination of data obtained by intrinsic and extrinsic methods becomes easily amenable to further delineation of exon–intron structure by expert analysis. For instance, in the complete genome of Human adenovirus D (Human adenovirus type 17), GeneMarkS revealed 32 potential genes or gene fragments missed in the original annotation (AF108105). Only 11 of them appeared to be complete genes while the other 21 predicted coding regions were manually assembled into nine genes in the RefSeq record (NC_002067).
The above discussed examples of confirmation and functional characterization of new ab initio predictions by subsequent application of an extrinsic method make it quite plausible that many not yet confirmed ab initio predictions will be supported extrinsically as more DNA and protein data become available. Still, the absence of similarity to known proteins may also indicate the uniqueness of the protein whose expression and function might be established only by direct experiments.
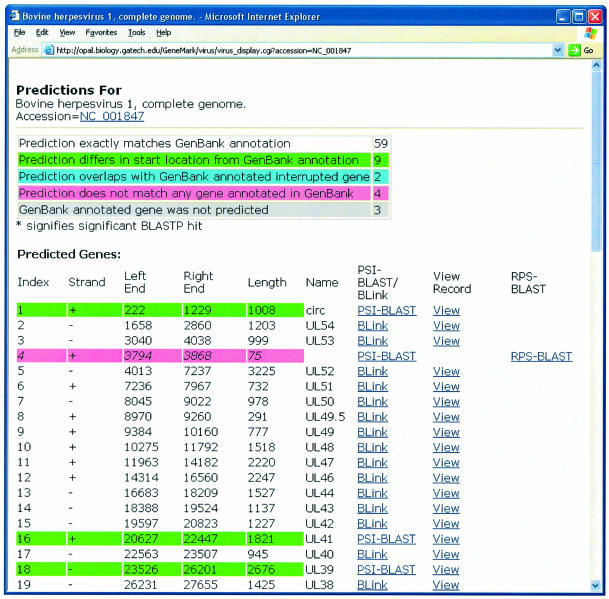
The VIOLIN database
Newly defined genome annotations were compiled in the VIOLIN database http://opal.biology.gatech.edu/GeneMark/VIOLIN/. This database currently has flat text file architecture. Differences between the VIOLIN and GenBank annotations are visualized by color codes (Fig. 6). The VIOLIN web site provides hypertext links to the NCBI similarity search programs directly from a genome annotation record. For a gene exactly matching an already known one, the line citing its coordinates is linked to the original gene record in GenBank as well as to the BLink program providing up-to-date information on the protein product (the BLink program, ‘BLAST Link’, displays the prerecorded results of BLAST searches that have been done for every protein sequence in the Entrez proteins data domain). For a predicted gene with no exact or partial match to the previous annotation, links to the programs PSI-BLAST and RPS-BLAST allow one to proceed with further up-to-date characterization of the putative protein. Genes annotated in a GenBank record but not confirmed by our analysis are shown at the bottom of the VIOLIN record with links to the BLink, PSI-BLAST and RPS-BLAST programs to help re-analyze the previously annotated genes.
Figure 6.
Snapshot of a sample viral genome record as it appears at the VIOLIN web site.
VIOLIN has been regularly used by the NCBI curators to improve the annotation of viral genomes in the RefSeq collection (36). Gene predictions have been subjected to additional analysis and manual curation by NCBI staff for quality control and functional assignment. Some of the new findings that originally appeared in VIOLIN and that are now included into annotations of 86 viral genomes in the RefSeq collection are shown in Table 5. For example, in Fowl adenovirus D (NC_000899) 14 proteins have been added to 15 existing in the original GenBank record AF083975. This was a particularly difficult case because many of the newly added genes were disrupted by frameshifts that likely resulted from sequencing errors. The new tentative protein sequences were assembled from fragments predicted by GeneMarkS using the ORF Finder (R. Tatusov and T. Tatusova, unpublished results), and BLASTP searches. In another example, in Lymphocystis disease virus (NC_001824) 110 coding regions were identified while the original GenBank record (AF083975) contained only one gene for a major capsid protein.
Table 5. Sample of the newly added RefSeq genes identified by the statistical gene finding methods described in this work.
| Group | Prediction | Predicted length | Best BLASTP hit | BLASTP length | Score | E-value | Annotated function |
|---|---|---|---|---|---|---|---|
| dsDNA | Alcelaphine herpesvirus 1 | NC_002531 | |||||
| 10443–11138 | 231 | gi|9628007| | 183 | 66.3 | 4.00E–10 | Putative BALF1 homolog | |
| Amsacta moorei entomopoxvirus | NC_002520 | ||||||
| complement (114621–114773) | 50 | gi|9629968| | 52 | 65.6 | 9.00E–11 | Conotoxin-like protein | |
| Ateline herpesvirus 3 | NC_001987 | ||||||
| 73911–75053 | 380 | gi|331012| | 384 | 603 | 1.00E–171 | Immediate-early phosphoprotein (transactivator) | |
| Avian adenovirus CELO | NC_001720 | ||||||
| 26793–27119 | 108 | gi|9633186| | 302 | 95.6 | 2.00E–19 | Late 33 kDa protein | |
| Bovine adenovirus 2 | NC_002513 | ||||||
| 10583–12295 | 570 | gi|13487865| | 573 | 755 | 0 | Peripentonal hexon-associated protein | |
| 12347–13783 | 478 | gi|13487866| | 471 | 793 | 0 | Penton protein | |
| 15888–16382 | 164 | gi|13487870| | 233 | 201 | 4.00E–51 | Minor capsid protein VI precursor | |
| 16628–19324 | 898 | gi|13487871| | 910 | 1546 | 0 | Hexon protein | |
| 21366–23579 | 737 | gi|13487873| | 722 | 1004 | 0 | Hexon assembly-associated 100 kDa protein | |
| complement (30406–30735) | 109 | gi|13487881| | 245 | 101 | 3.00E–21 | 245R protein homolog | |
| complement (30823–31383) | 186 | gi|13487880| | 253 | 188 | 5.00E–47 | 253R protein homolog | |
| Deer papillomavirus | NC_001523 | ||||||
| 3914–4048 | 44 | gi|137747| | 44 | 85.4 | 9.00E–17 | E5 transforming protein | |
| Equine herpesvirus 1 | NC_001491 | ||||||
| complement (112994–113785) | 263 | gi|15235673| | 608 | 179 | 5.00E–44 | Glycine-rich protein | |
| Fowl adenovirus 8 | NC_000899 | ||||||
| 14583–16211 | 542 | gi|9628848| | 575 | 799 | 0 | Peripentonal hexon associated protein | |
| complement (38665–40446) | 593 | gi|3845680| | 195 | 381 | 1.00E–104 | Glycine-rich protien | |
| Fowlpox virus | NC_002188 | ||||||
| 52914–54572 | 552 | gi|1083970| | 552 | 1122 | 0 | Rifampicin resistance N3L protein | |
| Human adenovirus type 2 | NC_001405 | ||||||
| 30444–30830 | 128 | gi|119063| | 128 | 264 | 5.00E–70 | Early E3B protein | |
| complement (30852–31019) | 55 | gi|9626584| | 53 | 143 | 4.00E–09 | U protein | |
| complement (35146–35532) | 128 | gi|119716| | 283 | 246 | 1.00E–64 | E4 protein | |
| Human adenovirus type 12 | NC_001460 | ||||||
| 25202–25558 | 118 | gi|9626562| | 211 | 135 | 2.00E–31 | 33 kDa phosphoprotein | |
| complement (31183–31407) | 74 | gi|93525| | 74 | 154 | 1.00E–37 | Early E4 17 kDa protein | |
| Human adenovirus type 17 | NC_002067 | ||||||
| 560–1138 | 192 | gi|4323354| | 251 | 316 | 1.00E–85 | Early E1A protein | |
| 1491–2117 | 208 | gi|4323357| | 182 | 377 | 1.00E–104 | Small T-antigen fragment | |
| 2165–2533 | 122 | gi|4323358| | 495 | 214 | 5.00E–55 | Small T-antigen fragment | |
| 2530–2976 | 148 | gi|4323358| | 495 | 301 | 5.00E–81 | Small T-antigen fragment | |
| 3033–3359 | 108 | gi|4323358| | 495 | 227 | 4.00E–59 | Small T-antigen fragment | |
| complement (3888–4499) | 203 | gi|130244| | 448 | 408 | 1.00E–113 | IVa2 maturation protein | |
| complement (4501–4935) | 144 | gi|130244| | 448 | 250 | 8.00E–66 | IVa2 maturation protein | |
| 15724–15960 | 78 | gi|9626191| | 368 | 74.5 | 2.00E–13 | V minor core protein | |
| 16177–16713 | 178 | gi|9626570| | 358 | 148 | 4.00E–35 | V minor core protein | |
| 16798–16953 | 51 | gi|9626571| | 70 | 74.5 | 2.00E–13 | L2 protein mu precursor | |
| 17754–18065 | 103 | gi|780528| | 947 | 161 | 3.00E–39 | Hexon capsid protein | |
| 18068–20617 | 849 | gi|780528| | 947 | 1595 | 0 | Hexon capsid protein | |
| complement (21293–21745) | 150 | gi|118737| | 517 | 238 | 3.00E–62 | E2A DNA binding protein | |
| complement (21724–22503) | 259 | gi|118735| | 512 | 341 | 6.00E–93 | E2A DNA binding protein | |
| 23513–23779 | 88 | gi|209871| | 652 | 99.8 | 7.00E–21 | Hexon assembly-associated protein | |
| 23799–24956 | 385 | gi|9626180| | 805 | 331 | 1.00E–89 | Hexon assembly-associated protein | |
| 25472–25774 | 100 | gi|9626578| | 233 | 129 | 9.00E–30 | pVIII protein | |
| 27021–27494 | 157 | gi|1279435| | 166 | 314 | 7.00E–85 | HLA-binding protein | |
| 29892–30287 | 131 | gi|6940696| | 130 | 264 | 4.00E–70 | E3B protein | |
| 30280–30672 | 130 | gi|6940697| | 130 | 272 | 1.00E–72 | E3B protein | |
| complement (30770–30919) | 49 | gi|9626584| | 53 | 54.3 | 2.00E–07 | U protein | |
| complement (32308–32970) | 220 | gi|3913555| | 292 | 464 | 1.00E–130 | E4 protein | |
| complement (33116–33478) | 120 | gi|1699394| | 120 | 259 | 2.00E–68 | E4 protein | |
| complement (33481–33834) | 117 | gi|1699393| | 117 | 243 | 7.00E–64 | E4 protein | |
| complement (33831–34058) | 75 | gi|1699392| | 130 | 142 | 5.00E–34 | E4 protein | |
| complement (34266–34463) | 65 | gi|1699391| | 125 | 132 | 7.00E–31 | E4 protein | |
| Human herpesvirus 3 | NC_001348 | ||||||
| 10678–10905 | 75 | gi|13242466| | 87 | 112 | 9.00E–25 | Membrane protein | |
| Human herpesvirus 4 | NC_001345 | ||||||
| 503–805 | 100 | gi|330387| | 365 | 160 | 5.00E–39 | Latent membrane protein | |
| 1546–1680 | 44 | gi|330387| | 365 | 85.8 | 7.00E–17 | Latent membrane protein | |
| 166576–166920 | 114 | gi|126379| | 497 | 257 | 4.00E–68 | Latent membrane protein | |
| complement (169031–169474) | 147 | gi|126373| | 386 | 224 | 6.00E–58 | Latent membrane protein | |
| Human herpesvirus 5 | NC_001347 | ||||||
| 160003–160173 | 56 | gi|7542409| | 176 | 97.1 | 3.00E–20 | Interleukin-10-like protein | |
| Human herpesvirus 6B | NC_000898 | ||||||
| 23343–23774 | 143 | gi|11346494| | 305 | 300 | 1.00E–80 | G-protein coupled receptor | |
| Human herpesvirus 7 | NC_001716 | ||||||
| 129708–129848 | 46 | gi|2746315| | 153 | 101 | 2.00E–21 | Membrane glycoprotein | |
| Human papillomavirus type 1a | NC_001356 | ||||||
| 812–2650 | 612 | gi|137646| | 612 | 1251 | 0 | Replication protein E1 | |
| Human papillomavirus type 53 | NC_001593 | ||||||
| 892–1140 | 82 | gi|9627323| | 631 | 125 | 1.00E–28 | Replication protein E1 | |
| 1391–1591 | 66 | gi|9627323| | 631 | 104 | 2.00E–22 | Replication protein E1 | |
| Human papillomavirus type 56 | NC_001594 | ||||||
| 895–1149 | 84 | gi|9628585| | 630 | 112 | 1.00E–24 | Replication protein E1 | |
| 1395–2804 | 469 | gi|9628585| | 630 | 927 | 0 | Replication protein E1 | |
| Human papillomavirus type 71 | NC_002644 | ||||||
| 559–828 | 89 | gi|1491685| | 100 | 99.8 | 8.00E–21 | Transforming protein E7 | |
| 3004–3858 | 284 | gi|9626037| | 383 | 264 | 1.00E–69 | Regulatory protein E2 | |
| 4443–5783 | 446 | gi|13186281| | 524 | 583 | 1.00E–165 | Minor capsid protein L2 | |
| 5776–7341 | 521 | gi|3845719| | 505 | 689 | 0 | Late major capsid protein L1 | |
| Macaca mulatta rhadinovirus | NC_003401 | ||||||
| 70403–70888 | 161 | gi|13506781| | 234 | 279 | 2.00E–74 | bZIP transcription factor | |
| 71468–72160 | 230 | gi|13506783| | 275 | 292 | 4.00E–78 | Glycoprotein R8.1 | |
| Murine adenovirus type 1 | NC_000942 | ||||||
| 2897–3175 | 92 | gi|209749| | 97 | 187 | 2.00E–47 | Early E1A protein | |
| complement (29726–30076) | 116 | gi|9800520| | 810 | 67.9 | 6.00E–11 | Tropoelastin | |
| Ovine papillomavirus 1 | NC_001789 | ||||||
| 747–2624 | 625 | gi|9627078| | 611 | 744 | 0 | Replication protein E1 | |
| 2611–3780 | 389 | gi|9627069| | 416 | 379 | 1.00E–104 | Regulatory protein E2 | |
| 3780–3941 | 53 | gi|137747| | 44 | 66.3 | 5.00E–11 | Transforming protein E5 | |
| 4268–5623 | 451 | gi|9627086| | 447 | 445 | 1.00E–124 | Minor capsid protein L2 | |
| Ovine papillomavirus 2 | NC_001790 | ||||||
| 745–2628 | 627 | gi|9627078| | 611 | 753 | 0 | Replication protein E1 | |
| 2615–3778 | 387 | gi|9627069| | 416 | 369 | 1.00E–101 | Regulatory protein E2 | |
| 3778–3930 | 50 | gi|137747| | 44 | 65.2 | 1.00E–10 | E5 protein | |
| 4122–5615 | 497 | gi|9627086| | 477 | 525 | 1.00E–148 | Minor capsid protein L2 | |
| Tupaia herpesvirus | NC_002794 | ||||||
| complement (60731–61684) | 317 | gi|9845327| | 478 | 120 | 2.00E–26 | US22 family protein | |
| Vaccinia virus | NC_001559 | ||||||
| complement (5422–5526) | 34 | gi|3096964| | 351 | 67.1 | 3.00E–11 | TNF receptor II | |
| complement (6231–6377) | 48 | gi|3096965| | 586 | 96.7 | 4.00E–20 | K1R protein (ankyrin repeat protein) | |
| 76530–76721 | 63 | gi|11346541| | 63 | 130 | 3.00E–30 | RNA polymerase | |
| 162151–162264 | 37 | gi|401315| | 193 | 58.9 | 9.00E–09 | Guanylate kinase | |
| 183524–183640 | 38 | gi|3096966| | 672 | 66.7 | 4.00E–11 | D4L protein (ankyrin repeat protein) | |
| 185397–185507 | 36 | gi|3096965| | 586 | 70.6 | 3.00E–12 | K1R protein (ankyrin repeat protein) | |
| 186212–186316 | 34 | gi|3096964| | 351 | 67.1 | 3.00E–11 | TNF receptor II | |
| ssDNA | Chloris striate mosaic virus | NC_001466 | |||||
| complement (1864–2376) | 170 | gi|137410| | 295 | 348 | 3.00E–95 | Replication-associated protein | |
| Periplaneta fuliginosa densovirus | NC_000936 | ||||||
| complement (5134–5388) | 84 | gi|5689346| | 291 | 83.1 | 6.00E–16 | Structural protein | |
| Phage | Bacteriophage bIL311 | NC_002670 | |||||
| 2252–2464 | 70 | gi|15673928| | 68 | 139 | 4.00E–33 | ps3 protein 14-like transcriptional regulator | |
| Bacteriophage L5 | NC_003695 | ||||||
| 2–340 | 112 | gi|4098413| | 348 | 216 | 8.00E–56 | Integrase | |
| Bacteriophage lambda | NC_001416 | ||||||
| 34482–35036 | 184 | gi|140702| | 183 | 374 | 1.00E–103 | Superinfection exclusion protein B | |
| complement (46459–46752) | 97 | gi|137520| | 97 | 196 | 5.00E–50 | Bor protein precursor | |
| complement (47042–47575) | 177 | gi|16128541| | 150 | 309 | 2.00E–83 | Putative envelope protein | |
| Bacteriophage VT2-Sa provirus | NC_000902 | ||||||
| complement (11467–11595) | 42 | gi|15830439| | 217 | 59.7 | 5.00E–09 | c1 repressor protein | |
| Chlamydia phage phiCPAR39 | NC_002180 | ||||||
| 1–147 | 48 | gi|9634956| | 84 | 104 | 2.00E–22 | Non-structural protein | |
| 4425–4532 | 35 | gi|9634956| | 84 | 75.3 | 1.00E–13 | Non-structural protein | |
| Enterobacteria phage HK022 virion | NC_002166 | ||||||
| 19015–20130 | 371 | gi|9634179| | 321 | 270 | 2.00E–71 | Tail fiber protein | |
| complement (26155–26307) | 50 | gi|9634191| | 50 | 104 | 1.00E–22 | kil protein | |
| 32436–33047 | 203 | gi|15832758| | 188 | 106 | 3.00E–22 | Endonuclease | |
| 33876–34316 | 146 | gi|9910800| | 146 | 294 | 4.00E–79 | Protein Nin B | |
| 35667–36029 | 120 | gi|9634210| | 120 | 244 | 4.00E–64 | Holiday-junction resolvase | |
| Enterobacteria phage Mu | NC_000929 | ||||||
| complement (33531–34064) | 177 | gi|96899| | 177 | 360 | 8.00E–99 | Tail fiber assembly protein | |
| complement (34067–35053) | 328 | gi|96901| | 536 | 678 | 0 | Tail fiber | |
| Roseophage SIO1 | NC_002519 | ||||||
| complement (39527–39826) | 99 | gi|9964612| | 271 | 124 | 3.00E–28 | gp5-like protein | |
| Streptococcus thermophilus bacteriophage 7201 | NC_002185 | ||||||
| 3148–3330 | 60 | gi|9634634| | 218 | 116 | 3.00E–26 | Erf protein | |
| Streptococcus thermophilus bacteriophage Sfi21 | NC_000872 | ||||||
| 37175–37687 | 170 | gi|9635004| | 167 | 317 | 6.00E–86 | DNA binding protein | |
| Sulfolobus Virus 1 | NC_001338 | ||||||
| 12585–13001 | 138 | gi|75696| | 144 | 270 | 7.00E–72 | Structural protein VP1 | |
| Retroid | Abelson murine leukemia virus | NC_001499 | |||||
| 4425–4580 | 51 | gi|332031| | 636 | 104 | 2.00E–22 | env polyprotein | |
| Feline immunodeficiency virus | NC_001482 | ||||||
| 9006–9170 | 54 | gi|128015| | 122 | 118 | 1.00E–26 | nef protein | |
| Friend spleen focus-forming virus | NC_001500 | ||||||
| 2173–2292 | 39 | gi|11120675| | 1733 | 79.6 | 5.00E–15 | gag polyprotein | |
| 2289–2543 | 84 | gi|510896| | 538 | 168 | 2.00E–41 | gag polyprotein | |
| Human foamy virus | NC_001736 | ||||||
| 11054–11827 | 257 | gi|227764| | 356 | 562 | 1.00E–159 | bel-2 protein | |
| Human T-cell lymphotropic virus type 2 | NC_001488 | ||||||
| 6–119 | 37 | gi|6539751| | 48 | 77.6 | 2.00E–14 | tax protein | |
| Moloney murine sarcoma virus | NC_001502 | ||||||
| 2485–2967 | 160 | gi|9626961| | 1737 | 271 | 5.00E–72 | pol polyprotein | |
| 2945–3388 | 147 | gi|9626961| | 1737 | 293 | 1.00E–78 | pol polyprotein | |
| 4563–4718 | 51 | gi|332031| | 636 | 102 | 5.00E–22 | Envelope protein | |
| Murine osteosarcoma virus | NC_001506 | ||||||
| complement (2305–2706) | 133 | gi|15822914| | 137 | 250 | 4.00E–66 | Ubiquitin-like protein | |
| Murine sarcoma virus | NC_001363 | ||||||
| 2970–3452 | 160 | gi|9626961| | 1737 | 271 | 5.00E–72 | pol polyprotein | |
| 3430–3873 | 147 | gi|9626961| | 1737 | 293 | 1.00E–78 | pol polyprotein | |
| 5048–5203 | 51 | gi|332031| | 636 | 102 | 5.00E–22 | spike protein | |
| Simian foamy virus | NC_001364 | ||||||
| 3–377 | 124 | gi|9626108| | 417 | 279 | 1.00E–74 | bet protein | |
| Simian immunodeficiency virus | NC_001549 | ||||||
| 3–335 | 110 | gi|9627209| | 223 | 247 | 5.00E–65 | nef protein | |
| Simian type D virus 1 | NC_001551 | ||||||
| 5194–5973 | 259 | gi|9627214| | 1771 | 450 | 1.00E–125 | pol polyprotein | |
| Y73 sarcoma virus | NC_001404 | ||||||
| 2865–3194 | 109 | gi|13508442| | 611 | 206 | 7.00E–53 | Transmembrane envelope protein | |
| Barmah Forest virus | NC_001786 | ||||||
| 5679–7298 | 539 | gi|7444406| | 2493 | 816 | 0 | Non-structural polyprotein | |
| ssRNA(+) | Northern cereal mosaic virus | NC_002251 | |||||
| 6740–12916 | 2058 | gi|2961429| | 1967 | 536 | 1.00E–150 | Polymerase |
CONCLUDING REMARKS
We have demonstrated that GeneMarkS, the ab initio gene finding method can be adjusted for analysis of viral genomes of different types and can generate useful information. In small viral genomes, any single missed gene could be of significant interest and the reliable identification of a narrow set of putative proteins to work with by extrinsic and experimental methods saves a considerable amount of time and effort. As the never ending discovery of new viruses brings about new names such as Mimivirus (37) or SARS (38), accurate ab initio computer methods for viral gene identification will remain of great value.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Yiming Bao for valuable comments on his experience of using the GeneMarkS program for annotating viral genomes. We are grateful to Dr John Besemer, Dr Dwight Hall and Dr Chris Klausmeier for useful remarks on the manuscript. M.B., A.L. and R.M. were supported in part by grant HG00783 from the US National Institutes of Health as well as by a grant jointly awarded by Georgia Tech and the US Centers for Disease Control and Prevention.
REFERENCES
- 1.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2002) GenBank. Nucleic Acids Res., 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanger F., Coulson,A.R., Hong,G.F., Hill,D.F. and Peterson,G.B. (1982) Nucleotide sequence of bacteriophage λ DNA. J. Mol. Biol., 162, 729–773. [DOI] [PubMed] [Google Scholar]
- 3.Borodovsky M., Rudd,K.E. and Koonin,E.V. (1994) Intrinsic and extrinsic approaches for detecting genes in a bacterial genome. Nucleic Acids Res., 22, 4756–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vink C., Beuken,E. and Bruggeman,C.A. (2000) Complete DNA sequence of the rat cytomegalovirus genome. J. Virol., 74, 7656–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besemer J., Lomsadze,A. and Borodovsky,M. (2001) GeneMarkS: a self-training method for predicition of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res., 29, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieff E. and Rickinson,A.B. (2001) Epstein–Barr virus and its replication. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincott Williams and Wilkins, Philadelphia, PA, Vol. 2, pp. 2511–2672. [Google Scholar]
- 7.Borodovsky M.Y. and McIninch,J.D. (1993) GeneMark: parallel gene recognition for both DNA strands. Comput. Chem., 17, 123–153. [Google Scholar]
- 8.Zimmerman W., Broll,H, Ehlers,B., Buhk,H., Rosenthal,A. and Goltz,M. (2001) Genome sequence of bovine herpesvirus 4, a bovine Rhadinovirus and identification of an origin of DNA replication. J. Virol., 75, 1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesyanzhinov V.V., Robben,R., Grymonprez,B., Kostyuchenko,V.A., Bourkaltseva,M.V., Sykilinda,N.N., Krylog,V.N. and Volckaert,G. (2002) The genome of bacteriophage ΦKZ of Pseudomonas aeruginosa. J. Mol. Biol., 317, 1–19. [DOI] [PubMed] [Google Scholar]
- 10.Tu A.T., Voelker,L.L., Shen,X. and Dybvig,K. (2001) Complete nucleotide sequence of the Mycoplasma virus P1 genome. Plasmid, 45, 122–126. [DOI] [PubMed] [Google Scholar]
- 11.Ford M.E., Sarkis,G.J., Belanger,A.E., Hendrix,R.W. and Hatfull,G.F. (1998) Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol., 279, 143–164. [DOI] [PubMed] [Google Scholar]
- 12.Recketenwald J. and Schmidt,H. (2002) The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage ΦP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun., 70, 1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hambly E., Tetart,F., Desplats,C., Wilson,W.H., Krisch,H.M. and Mann,N.H. (2001) A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl Acad. Sci. USA, 98, 11411–11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takai S., Hines,S.A., Sekizaki,T., Nicholson,V.M., Alperin,D.A., Osaki,M., Takamatsu,D., Nakamura,M., Suzuki,K., Ogino,N., Kakuda,T., Dan,H. and Prescott,J.F. (2000) DNA sequence and comparison of virulence plasmids from Rhodocuccus equi ATCC 33701 and 103. Infect. Immun., 68, 6840–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatesan M.M., Goldberg,M.B., Rose,D.J., Grotbeck,E.J., Burland,V. and Blattner,F.R. (2001) Complete DNA sequence and analysis of the large virulence plasmid of Shigella felxneri. Infect. Immun., 69, 3271–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burland V., Shao,Y., Perna,N.T., Plunkett,G., Sofia,H.J. and Blattner,F.R. (1998) The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res., 26, 4196–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F., He,J., Lin,X., Li,Q., Pan,D., Zhang,X. and Xu,X. (2001) Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol., 75, 11811–11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He J.G., L,L., Deng,M., He,H.H., Weng,S.P., Wang,X.H., Zhou,S.Y., Long,Q.Z., Wang,X.Z. and Chan,S.M. (2002) Sequence analysis of the complete genome of an iridovirus isolated from the tiger frog. Virology, 292, 185–197. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2001) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 29, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farmer A.D., Calef,C.E., Millman,K. and Myers,G.L. (1995) The Human Papillomavirus Database. J. Biomed. Sci., 2, 90–104. [DOI] [PubMed] [Google Scholar]
- 21.Hiscock D. and Upton,C. (2000) Viral Genome DataBase: storing and analyzing genes and proteins from complete viral genomes. Bioinformatics, 16, 484–485. [DOI] [PubMed] [Google Scholar]
- 22.Mar Albà M., Lee,D., Pearl,F.M.G., Shepherd,A.J., Martin,N., Orengo,C.A. and Kellam,P. (2001) VIDA: a virus database system for the organisation of virus genome open reading frames. Nucleic Acids Res., 29, 133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukashin A. and Borodovsky,M. (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res., 26, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Besemer J. and Borodovsky,M. (1999) Heuristic approach to deriving models for gene finding. Nucleic Acids Res., 27, 3911–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wootton,J.C. (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 26.Durbin S., Eddy,A., Krogh,G. and Mitchison,G. (1998) Biological Sequence Analysis. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 27.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 26, 3986–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider T.D. and Stephens,R.M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Link A.J., Robison,K. and Church,G.M. (1997) Comparing the predicted and observed properties of proteins encoded in the genome of Escherchia coli K-12. Electrophoresis, 18, 1259–1313. [DOI] [PubMed] [Google Scholar]
- 30.Bellows D.S., Howell,M., Pearson,C., Hazlewood,S.A. and Hardwick,J.M. (2002) Epstein-Barr virus BALF1 is a BCL-2-like antagonist of the herpesvirus antiapoptotic BCL-2 proteins. J. Virol., 76, 2469–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goltz M., Ericsson,T., Patience,C., Huang,C.A., Noack,S., Sachs,D.H. and Ehlers,B. (2002) Sequence analysis of the genome of porcine lymphotropic herpesvirus 1 and gene expression during post-transplant lymphoproliferative disease of pigs. Virology, 294, 383–393. [DOI] [PubMed] [Google Scholar]
- 32.Schaffer A.A., Aravind,L., Madden,T.L., Shavirin,S., Spouge,J.L., Wolf,Y.I., Koonin,E.V. and Altschul,S.F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res., 29, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul S.F., Bundschuh,R., Olsen,R. and Hwa,T. (2001) The estimation of statistical parameters for local alignment score distributions. Nucleic Acids Res., 29, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamauchi Y., Wada,K., Goshima,F., Daikoku,T., Ohtsuka,K. and Nishiyama,Y. (2002) Herpes simplex virus type 2 UL14 gene product has heat shock protein (HSP)-like functions. J. Cell Sci., 115, 2517–2527. [DOI] [PubMed] [Google Scholar]
- 36.Tatusova T.A., Karsch-Mizrachi,I. and Ostell,J.A. (1999) Complete genomes in WWW Entrez: data representation and analysis. Bioinformatics, 15, 536–543. [DOI] [PubMed] [Google Scholar]
- 37.Scola B.L, Audic,S., Robert,C., Jungang,L., de Lamballerie,X., Drancourt,M., Birtles,R., Claverie,J. and Raoult,D. (2003) A giant virus in Amoebae. Science, 299, 2033. [DOI] [PubMed] [Google Scholar]
- 38.Marra M.A., Jones,S.J., Astell,C.R., Holt,R.A., Brooks-Wilson,A., Butterfield,Y.S., Khattra,J., Asano,J.K., Barber,S.A., Chan,S.Y., Cloutier,A., Coughlin,S.M., Freeman,D., Girn,N., Griffith,O.L., Leach,S.R., Mayo,M., McDonald,H., Montgomery,S.B., Pandoh,P.K., Petrescu,A.S., Robertson,A.G., Schein,J.E., Siddiqui,A., Smailus,D.E., Stott,J.M., Yang,G.S., Plummer,F. andonov,A., Artsob,H., Bastien,N., Bernard,K., Booth,T.F., Bowness,D., Czub,M., Drebot,M., Fernando,L., Flick,R., Garbutt,M., Gray,M., Grolla,A., Jones,S., Feldmann,H., Meyers,A., Kabani,A., Li,Y., Normand,S., Stroher,U., Tipples,G.A., Tyler,S., Vogrig,R., Ward,D., Watson,B., Brunham,R.C., Krajden,M., Petric,M., Skowronski,D.M., Upton,C. and Roper,R.L. (2003) The Genome sequence of the SARS-associated coronavirus. Science, 300, 1399–1404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.