Abstract
Human immunodeficiency virus (HIV) RNase H activity is essential for the synthesis of viral DNA by HIV reverse transcriptase (HIV-RT). RNA cleavage by RNase H requires the presence of divalent metal ions, but the role of metal ions in the mechanism of RNA cleavage has not been resolved. We measured HIV RNase H activity associated with HIV-RT protein in the presence of different concentrations of either Mg2+, Mn2+, Co2+ or a combination of these divalent metal ions. Polymerase-independent HIV RNase H was similar to or more active with Mn2+ and Co2+ compared with Mg2+. Activation of RNase H by these metal ions followed sigmoidal dose–response curves suggesting cooperative metal ion binding. Titration of Mg2+-bound HIV RNase H with Mn2+ or Co2+ ions generated bell-shaped activity dose–response curves. Higher activity could be achieved through simultaneous binding of more than one divalent metal ion at intermediate Mn2+ and Co2+ concentrations, and complete replacement of Mg2+ occurred at higher Mn2+ or Co2+ concentrations. These results are consistent with a two-metal ion mechanism of RNA cleavage as previously suggested for a number of polymerase-associated nucleases. In contrast, the structurally highly homologous RNase HI from Escherichia coli is most strongly activated by Mg2+, is significantly inhibited by submillimolar concentrations of Mn2+ and most probably cleaves RNA via a one-metal ion mechanism. Based on this difference in active site structure, a series of small molecule N-hydroxyimides was identified with significant enzyme inhibitory potency and selectivity for HIV RNase H.
INTRODUCTION
Reverse transcription of human immunodeficiency virus (HIV) RNA is an essential early step in the virus life cycle following the entry of HIV into target cells. HIV reverse transcriptase protein (HIV-RT) requires the concerted function of two active sites to generate double-stranded DNA from the viral genomic RNA. The active HIV-RT protein is a heterodimer of a p66 and a p51 subunit. The p66 subunit contains a functional DNA polymerase and a separate RNase H active site. RNase H activity is required to hydrolyse the RNA strand of RNA:DNA heteroduplex molecules that are generated during reverse transcription. RNase H activity is also essential for the progression of two obligatory strand transfer reactions in the reverse transcription process (1,2).
Reverse transcription has been a highly successful anti-retroviral target in HIV therapy in an effort to control the mortality and morbidity of HIV disease. Nucleoside analogues are chain terminators and inhibit DNA synthesis by HIV-RT after incorporation into nascent DNA molecules. Non-nucleoside inhibitors prevent DNA synthesis by HIV-RT after binding to an allosteric site on the polymerase domain of the p66 subunit. In combination, the use of HIV-RT inhibitors has provided treatment options with significant efficacy and durability. However, there is still an urgent need to develop new antiviral agents and find new targets due to the emergence of drug-resistant HIV strains and significant cross-resistance within drug classes. Although RNase H was recognised as an essential viral function over a decade ago (3,4), very few inhibitors of RNA hydrolysis have been described and none of them has been progressed into clinical trials (5,6). Active site binding compounds have been shown to potently inhibit the polymerase-associated, divalent metal ion-dependent influenza RNA endonuclease in vitro and in vivo (7,8). Biochemical studies provided evidence that RNA hydrolysis by influenza endonuclease occurred via a two-metal ion mechanism, similar to the model described for the exonuclease domain of Escherichia coli DNA polymerase I (9,10). Based on a pharmacophore model of inhibitor binding to a two-metal ion active site structure of influenza endonuclease, we recently demonstrated that it was possible to design novel series of influenza endonuclease inhibitors, which competitively bind to the endonuclease active site and do not inhibit divalent metal ion-dependent RNA polymerase activity (11).
The role of divalent metal ions in the mechanism of RNA cleavage by HIV RNase H has remained unclear. Crystallographic analysis of the isolated HIV RNase H domain showed two Mn2+ ions separated by ∼4 Å in the active site and bound to four conserved amino acids (D443, E478, D498 and D549), consistent with the two-metal ion mechanism model of RNA cleavage (12). The measurement of metal ion binding to HIV-RT by solution calorimetry also suggested the binding of two Mn2+ ions to the RNase H domain. However, at a concentration of 0.5 mM, only one Mg2+ was found to bind (13). Mg2+ binding to the HIV RNase H domain has been shown to significantly enhance DNA binding to HIV-RT. Three distinct binding modes were observed, which may correlate with DNA binding in the absence of Mg2+ and binding to one and more than one Mg2+ ion bound at increasing Mg2+ concentrations (14). Evidence for the binding of two Mg2+ ions to HIV RNase H has also been obtained by solution NMR (15).
On the other hand, the data available for the structurally very closely related E.coli RNase HI have been found to be more consistent with a one-metal ion mechanism of RNA cleavage (16,17). Crystallographic analysis showed a single Mg2+ ion in the active site of E.coli RNase HI (18). A single Mg2+-binding site was also observed for E.coli RNase HI using 1H–15N heteronuclear NMR and confirmed by kinetic analysis using 25Mg-NMR (19,20). Two Mn2+ ions were observed to bind to E.coli RNase HI by crystallography, at positions similar to those determined for HIV RNase H (21). Escherichia coli RNase HI is significantly more active in the presence of Mg2+ compared with Mn2+, but RNA hydrolysis is measurable in the presence of Mn2+. In the presence of Mn2+, E.coli RNase HI shows highest activity at concentrations below 5 µM Mn2+, but is inhibited at concentrations above 5 µM Mn2+. Mn2+ could also inhibit Mg2+-bound E.coli RNase HI activity. Based on these results, an activation/attenuation model has been suggested for E.coli RNase HI as an extension to the one-metal ion mechanistic model (22).
The binding of divalent metal ions to RNase H has been shown to significantly affect protein structure and stability (15,23). For the rational design of active site inhibitors of HIV RNase H, it is therefore of great importance to develop a better understanding of these structural effects and potential differences from closely related active site structures. In the current study, we further explored the effect of divalent metal ion binding on HIV-RT-associated RNase H activity. The titration of different divalent metal ions suggested cooperative binding to the enzyme and increased activity in mixed metal ion active sites, consistent with a two-metal ion mechanism of RNA cleavage. Activation of E.coli RNase HI was significantly different, was inhibited by a low concentration of Mn2+ and showed no evidence for cooperativity, in agreement with published results. Based on the previously developed pharmacophore of compound binding to two-metal ion active sites, we were able to identify a novel series of potent HIV RNase H inhibitors, which did not significantly inhibit E.coli RNase HI.
MATERIALS AND METHODS
Materials
HIV-RT protein (p66/p51 dimer) was obtained from Amersham Bioscience or expressed and purified using a previously described protocol, with modifications (24). HXB2 p66 was co-expressed with HIV protease in E.coli, resulting in intracellular processing of p66 into p66/p51 dimers. Analytical ultracentrifugation showed that 86% of p66 was present in heterodimeric form (F.Mueller, personal communication). Cell paste was lysed in 20 mM Tris–HCl pH 8, 1 mM dithiothreitol (DTT), 1 mM EDTA, 100 mM NaCl, 20 mg/ml lysozyme by incubation for 15 min at 4°C and two 30 s sonications. Benzonase (5 U/ml; Novagen) was added and incubated for 15 min at 4°C. The lysate was centrifuged for 30 min using an SS34 Sorvall rotor at 17 000 r.p.m. and the supernatant passed through a Q Sepharose column equilibrated with buffer A (20 mM Tris–HCl pH 8, 1 mM DTT, 1 mM EDTA, 100 mM NaCl). The flow-through was collected and loaded onto a Hi Trap heparin–Sepharose column equilibrated in buffer B (20 mM Tris–HCl pH 8, 1 mM DTT, 1 mM EDTA, 50 mM NaCl). The bound protein was eluted with a 1.5 column volume gradient of buffer B to buffer B with 1 M NaCl. Fractions containing HIV-RT were pooled, and solid ammonium sulfate was added to a final concentration of 1 M. The protein was then loaded onto a phenyl Sepharose column equilibrated in buffer C (20 mM Tris–HCl pH 8, 1 mM DTT, 1 mM EDTA, 1 M ammonium sulfate) and eluted with a 15 column volume gradient from buffer C to buffer B. Fractions containing HIV-RT protein were pooled, dialysed against buffer B, loaded onto a Resource S column equilibrated in buffer B and eluted in a 10 column volume gradient with buffer B with 1 M NaCl. Fractions containing HIV-RT protein were pooled, dialysed against buffer B and stored in aliquots at –80°C. Escherichia coli RNase HI was obtained from Roche Applied Science.
Oligonucleotides were chemically synthesised and gel purified after 5′ labelling. Phosphate-labelled oligonucleotides were generated using T4 polynucleotide kinase from Roche Applied Science and [γ-32P]ATP from Amersham Bioscience. Cap-labelled RNA oligonucleotides were synthesised using a previously published procedure (25). The oligonucleotide-based RNase H substrate GK1 was obtained by annealing 5′-labelled RNA oligoucleotide R1 5′-p*GAAUACUCAAGCUAUGCAUC-3′ with unlabelled DNA oligonucleotide D1 5′-GATGCATAGCTTGAGTATTCTATAGTGAGTCGTATTAA-3′. RNase H substrate CGK1 was obtained by annealing 5′-cap-labelled R1 5′-m7Gppp*GmAAUACUCAAGCUAUGCAUC-3′ with DNA oligonucleotide D1. Tritium-labelled RNase H substrate poly(dC:rG) was synthesised using T7 RNA polymerase and poly(dC) DNA from Amersham Bioscience. Poly(dC) (3.6 U/ml) was incubated with 40 µM [3H]GTP (5 Ci / mmol) and 0.13 U/ml T7 RNA polymerase for 30 min at 37°C. The heteroduplex product was purified by phenol extraction and ethanol precipitation, and unincorporated nucleotides were separated by G50 gel filtration.
Enzyme activity assays
In the standard gel-based HIV RNase assay, 0.1 nM substrate GK1 or CGK1, containing 5′-labelled RNA (specific activity, 50 000–100 000 c.p.m./fmol), was incubated with 1 nM HIV-RT in buffer RH containing 50 mM Tris–HCl pH 7 or 8, 50 mM KCl, 1 mM MgCl2 or 0.1 mM MnCl2 and 0.1 mg/ml bovine serum albumin (BSA) in a total reaction volume of 5 µl for 0–10 min at 30°C. The reaction was stopped by the addition of 5 ml of denaturing gel loading buffer (Ambion). Reaction products were quantified after electrophoresis on 7 M urea, 20% acrylamide gels by phosphorimager analysis (Storm, Amersham Bioscience), using Imagequant 5.2 software. Data analysis was performed using Excel 97 and SigmaPlot 5.0 software.
The filtration-based RNase H assay combined 0.5 nM HIV-RT or 0.01 U/µl E.coli RNase HI with 0.1 mU/ml (OD260) poly(dC:rG) substrate in buffer RH containing 10% dimethylsulfoxide (DMSO) in a total volume of 50 µl. After incubation at 37°C for 15 min, the reaction products were precipitated by adding 25 ml of cold trichloroacetic acid (TCA) and incubated at 4°C for 30 min before filtration through glass fibre filters on Millipore NOB filterplates. The precipitates were washed five times with 10% TCA and once with 70% ethanol, and quantified by scintillation counting. For the analysis of RNase H inhibitor potencies, control reactions were included in the absence of inhibitor and corresponding activity was defined as 100% RNase H activity or 0% inhibition. Control reactions in the absence of protein were defined as 0% activity or 100% inhibition.
HIV-RT DNA polymerase activity was measured after incubating 1–4 nM HIV-RT protein, 5 µg/ml poly(rA) RNA, 2.5 µg/ml oligo(dT16) DNA, 5 µM TTP, 0.15 µCi of [3H]TTP in 50 mM Tris–HCl pH 8, 6 mM MgCl2, 50 mM NaCl, 1 mM EDTA, 10% DMSO at 37°C for 30 min. Product DNA was quantified by TCA precipitation and scintillation counting as described above.
RESULTS
Cleavage pattern and metal ion preference of HIV RNase H
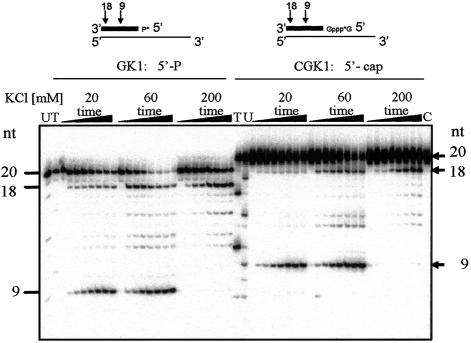
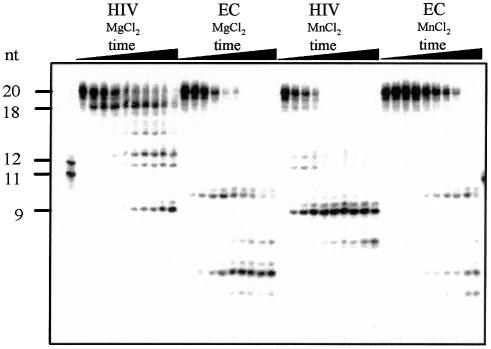
From structural and functional studies, the distance between the polymerase and RNase H active sites on HIV-RT has been determined as 18 bp on RNA:DNA heteroduplex substrates (26–30). On substrates longer than 18 bp, HIV RNase H preferentially cleaves at positions 18 and 8–9 nt from the 5′ end of RNA on substrates containing 3′-DNA overhangs. These cleavage reactions occur independently of each other with different rates, and the 18mer can serve as a substrate for the formation of the 9mer product (31,32). HIV RNase H generated the expected 18 and 9 nt major cleavage products from substrate GK1, which contains a 5′-phosphorylated, 20 nt RNA hybridised to a 49 nt DNA oligonucleotide with a 3′-DNA overhang (Fig. 1). This preferred cleavage pattern was also observed when the RNA oligonucleotide was blocked at the 5′ end by adding a guanosyl-5′-5′-guanosine triphosphate cap structure (substrate CGK1, Fig. 1). In agreement with previous observations, cleavage rates observed on capped and phosphorylated substrates were similar (33). The formation of the 9mer cleavage product was significantly slower than for the 18mer product and was strongly inhibited in the presence of 200 mM KCl (Fig. 1). The formation of the primary 18mer product was more resistant to high salt, and the 18mer could also serve as a substrate for the formation of 9mer product (Fig. 1). Escherichia coli RNase HI showed a significantly different cleavage pattern on CGK1 substrate compared with HIV RNase H (Fig. 2). Substrate CGK1 is therefore a useful probe for the differential analysis of HIV and E.coli RNase HI activities. Figure 2 shows that HIV RNase H activity was significantly higher in the presence of Mn2+ compared with Mg2+ at pH 7. At pH 8, HIV RNase H activity was similar with Mg2+, Mn2+ and Co2+ (Fig. 3). Reduced activity with Mg2+ at lower pH compared with Mn2+ is consistent with a role for the catalytic metal ion in increasing the concentration of the nucleophile OH– at the active site according to the respective pKa values of the metal hydrates (9). On the other hand, E.coli RNase HI activity was low in the presence of Mn2+ and highest in the presence of Mg2+ (Fig. 2). These results confirmed the previously published preference of E.coli RNase HI for Mg2+ and show a significant difference in metal ion activation for the two structurally related enzymes.
Figure 1.
Polymerase-independent HIV RNase H activity. Comparative analysis of 5′-phosphorylated and 5′-capped RNA substrates at different concentrations of KCl as indicated. RNase H products were analysed by denaturing acrylamide gel electrophoresis. HIV RNase H reactions were incubated for 0, 0.5, 1, 2, 4, 6, 8 and 10 min with 1 mM MgCl2. T, U, RNA sequencing reactions. T, RNase T1; U, RNase U2; C, annealed substrate incubated for 10 min. The positions of the 20mer RNA substrate and the major 18mer and 9mer cleavage products are indicated. Sequencing reactions with RNases U2 and T1 generate products with 3′-phosphate groups, which migrate slightly faster than the corresponding products of HIV RNase H, which generates 3′-OH ends.
Figure 2.
Cleavage pattern differences between HIV and E.coli RNases H. RNase H products were analysed by denaturing acrylamide gel electrophoresis. HIV RNase H reactions were incubated for 0, 0.5, 1, 2, 4, 6, 8, 10, 15 and 30 min with substrate CGK1 and 1 mM MgCl2 or 0.1 mM MnCl2 at pH 7 as indicated. The positions of the 20mer RNA substrate, the major 18mer and 9mer cleavage products and the 11mer and 12mer size marker are indicated on the left of the gel. HIV, HIV RNase H; EC, E.coli RNase H.
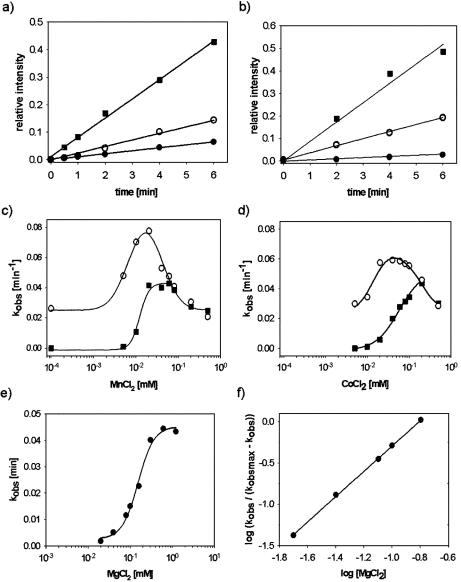
Figure 3.
Activation of HIV RNase H by divalent metal ions. (a) Time courses of 9mer formation from CGK1 substrate were determined in the presence of 0.02 mM MnCl2 (filled circles), 0.2 mM MgCl2 (open circles) or 0.02 mM MnCl2 plus 0.2 mM MgCl2 (filled squares). (b) Time courses of 9mer formation from CGK1 substrate were determined in the presence of 0.02 mM CoCl2 (filled circles), 0.2 mM MgCl2 (open circles) or 0.02 mM CoCl2 plus 0.2 mM MgCl2 (filled squares). (c) Rates of 9mer formation were determined from time courses in the presence of increasing concentrations of MnCl2 alone (filled squares) or 0.2 mM MgCl2 (open circles). The dose–response curve of RNA cleavage activation by MnCl2 (filled squares) fitted to a sigmoidal equation with a Hill coefficient of nH = 3.1. (d) As (c) in the presence of increasing concentrations of CoCl2 alone (filled squares) or 0.2 mM MgCl2 (open circles). The dose–response curve of RNA cleavage activation by CoCl2 (filled squares) fitted to a sigmoidal equation with a Hill coefficient of nH = 1.6. (e) Rates of 9mer formation were determined from time courses in the presence of increasing concentrations of MgCl2. The dose–response curve of RNA cleavage activation by MgCl2 (filled squares) fitted to a sigmoidal equation with a Hill coefficient of nH = 2.0. (f) Hill plot of 9mer formation kinetics in the presence of MgCl2. Determination of the slope provided a Hill coefficient of nH = 1.8. All reactions were performed as described in Materials and Methods and at pH 8.
Cooperative binding of divalent metal ions to activate HIV RNase H
Previous evidence from structural studies indicated that HIV RNase H can accommodate two divalent metal ions in the active site. Using substrate CGK1, the rates of formation of the HIV-specific 9mer cleavage product were determined at different concentrations of either Mg2+, Mn2+ or Co2+ as activating divalent metal ions. About 10-fold higher concentrations of Mg2+ were required to achieve maximal cleavage rates compared with Mn2+ or Co2+. Maximal cleavage rates were similar for the three metal ions at pH 8 (Fig. 3). Interestingly, dose–response curves of metal ion titration experiments showed slopes greater than unity, and Hill plots of the dose–response curves consistently showed Hill coefficients larger than 1 for the activation of HIV RNase H (Fig. 3c–f). These results suggested cooperative binding of more than one divalent metal ion to activate the generation of 9mer product from substrate CGK1 by HIV-RT.
One important implication of a two-metal ion mechanism of RNA cleavage is that stepwise titration should occur when a pre-bound divalent metal ion (M1–M1) is exchanged with increasing concentrations of a second type of divalent metal ion (M2) to form first a mixed active site (M1–M2), followed by a homogeneous two-metal ion active site (M2–M2) at higher concentrations of M2. It has previously been observed with influenza RNA endonuclease and the hammerhead ribozyme RNA hydrolysis activities that mixed M1–M2 active sites could have higher activities than homogeneous M1–M1 sites under certain conditions (9,34). Stepwise titration of metal ions will lead to bell-shaped titration curves under these conditions. HIV RNase H also showed more than an additive increase in RNA cleavage activity when low concentrations of either Mn2+ or Co2+ were combined with Mg2+-bound HIV-RT (Fig. 3a and b). Titration of Mg2+-bound HIV-RT with either Mn2+ or Co2+ generated bell-shaped titration curves (Fig. 3c and d). At concentrations above 100 µM Mn2+ or Co2+, HIV RNase H activity was identical when measured with Mn2+ or Co2+ alone or when Mn2+ or Co2+ had been added to an Mg2+-bound protein (Fig. 3c and d), indicating complete replacement of Mg2+ at these higher concentrations.
Inhibition of HIV RNAse H by N-hydroxyimides
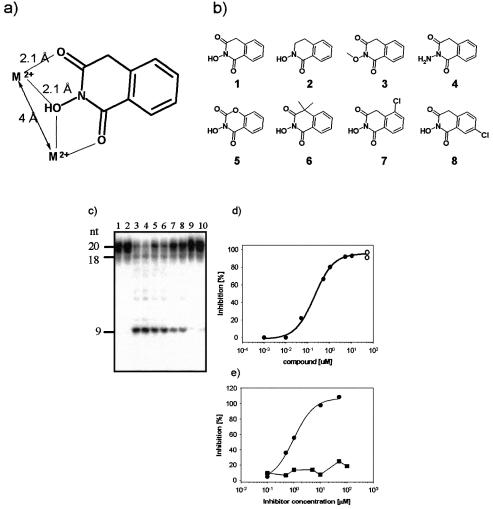
The results described above were most consistent with a two-metal ion mechanism for the formation of 9mer product from substrate CGK1 by HIV RNase H, but were difficult to reconcile with a one-metal ion or activation/attenuation mechanism. We have previously developed a pharmacophore model for the binding of compounds to the two-metal ion active site of influenza endonuclease. The major feature of the pharmacophore is a specific arrangement of three oxygen atoms to mimic the metal ion-mediated protein interaction with substrate, nucleophile and leaving group oxygens in the enzyme active site (11). In this model, the catalytic divalent metal ions are bound at a distance of 4–5 Å in the active site, and each metal ion interacts with two out of three compound oxygen atoms at 2–2.5 Å distance (Fig. 4a). Compound 1 is a prototype structure to present the oxygen atoms for optimal metal ion interaction with two metal ions bound at 4–5 Å distance (Fig. 4). As predicted, compound 1 inhibited the generation of 9mer product from substrate CGK1 (Fig. 4c and d). Complete inhibition was observed at 10 µM, with an IC50 of 0.6 µM (Table 1). Similarly, in a filtration assay format, using poly(dC:rG) as a template, compound 1 inhibited HIV RNase H activity with an IC50 of 1 µM (Fig. 4e). Under the same conditions, compound 1 did not show significant inhibition of E.coli RNase HI activity (Fig. 4e, Table 1). Inhibition of HIV-RT DNA polymerase activity was detectable with an IC50 of 40 µM. Compounds 2–8 were used to characterise structural features of HIV RNase H inhibition by this class of compounds (Fig. 4b). In particular, the requirement for the oxygen ligands for inhibitory potency was investigated. The N-hydroxyl group, which represents the bridging ligand for both metal ions, was found to be essential. RNase H inhibitory potency was lost when the hydroxyl group was replaced by a methoxy or an amino group (compounds 3 and 4). Substitutions on the phenyl moiety could further modulate inhibitory potency and selectivity in this series (compounds 7 and 8).
Figure 4.
Design and inhibitory activity of a chemical probe to interact with two-metal ion active sites. (a) N-hydroxyimides present three oxygen atoms at positions compatible with the interaction of two divalent metal ions at a distance of 4 Å. Each metal ion can undergo two liganding interactions with oxygen atoms at equal distances of 2.1 Å. (b) Structures of the N-hydroxyimides described. Inhibition of enzyme activity was determined and the results are shown in Table 1. (c) Compound 1 inhibited the formation of 9mer from substrate CGK1 in a dose-dependent manner in the presence of 1 mM MgCl2. Polyacrylamide gel analysis of HIV RNase H activity. Lanes 1 and 2, control reactions in the absence of protein; lanes 3–10, RNase H reactions in the presence of 0, 0.01, 0.05, 0.1, 0.5, 1, 5 and 10 µM compound 1. (d) The dose–response curve obtained with compound 1 on the gel-based assay was fitted to a hyperbolic, single site interaction equation. IC50 = 0.6 µM. (e) Compound 1 inhibited HIV RNase H-directed degradation of poly(dC:rG) in the presence of 1 mM MgCl2. IC50 = 1.0 µM (filled circles). Escherichia coli RNase H activity was not affected under identical conditions (filled squares).
Table 1. In vitro inhibition of HIV RNase H activity by N-hydroxyimides.
| Compounda | HIV-RH poly(dC:rG)b IC50 [µM] | HIV-RH G20 RNAc IC50 [µM] | EC-RH poly(dC:rG)b IC50 [µM] | EC-RH G20 RNAc IC50 [µM] | HIV-RTd IC50 [µM] |
|---|---|---|---|---|---|
| 1 | 1.0 | 0.6 | >50 | >10 | 41 |
| 7 | 0.3 | nd | nd | nd | 83 |
| 8 | 7 | nd | nd | nd | nd |
aCompound numbers are according to Figure 4b. All other compounds shown in Figure 4b (compounds 2–6) did not significantly inhibit HIV RNase H activity in vitro (IC50 > 50 µM).
bIC50 values determined using poly(dC:rG) double-stranded DNA:RNA hybrid substrate (nd, not determined).
cIC50 values determined using G20 RNA hybridised to D1 DNA double-stranded DNA:RNA hybrid substrate.
dIC50 values determined using a filtration-based DNA polymerase assay with poly(rA):oligo(dT16) substrate.
DISCUSSION
Metal ion binding to the active site of HIV RNase H is a critical factor to consider in the design of inhibitors targeting this essential viral function. In particular, a one-metal or a two-metal ion active site will provide significantly different binding environments for small molecules. As shown before, cross-titration of divalent metal ions with different physicochemical properties can generate characteristic bell-shaped titration curves, due to the fact that mixed metal active sites can have higher activities. In contrast, one-metal ion active sites will show typical hyperbolic titration curves due to single metal ion exchange in the active site (9,34).
The assay system described here to measure HIV RNase H activity is based on substrate GK1, a 20mer RNA hybridized to a 49mer DNA with a 3′-DNA overhang. The length of this substrate is expected to span the nucleic acid-binding site on HIV-RT from the polymerase to the RNase H active site (26). As demonstrated before, this nucleic acid structure will direct binding of the polymerase domain of HIV-RT to the RNA 5′ end, leading to a preferential fast primary cleavage site 18 nt downstream of the RNA 5′ end and a preferential slow secondary cleavage site 9 nt downstream of the RNA 5′ end (31,32). The 9mer formation occurs independently of the 18mer formation and constitutes an essential polymerase-independent RNA cleavage activity that is easily monitored in time course measurements. It is not known if HIV-RT undergoes a conformational change to perform 18mer and 9mer cleavages, or if a rigid HIV-RT protein slides along the substrate to cleave the RNA. A hinge region involved in a conformational change during this process may constitute a potential new drug-binding site. If HIV-RT went through a simple, rigid sliding motion along substrate GK1, then the polymerase active site would have to release the double-stranded nucleic acid interaction in order for RNA cleavage to occur at the 9mer site. This should be associated with a lower affinity substrate interaction leading to higher sensitivity to high salt conditions. Interestingly, RNA cleavage rates decreased with increasing ionic strength of the reaction. In particular, there was no 9mer cleavage product detectable when the reaction was performed in the presence of 200 mM KCl. The disappearance of 9mer is unlikely to be due to an overall reduction in cleavage rates, because the reduction of cleavage rate is significantly higher for 9mer production compared with 18mer production (Fig. 1). This finding therefore supports a protein sliding model, by which HIV-RT generates preferential cleavages at 18mer and 9mer positions.
We found that adding a cap structure to the 5′ end of the RNA molecule generated a substrate, CGK1, which showed significantly different signature cleavages for HIV RNase H compared with E.coli RNase HI. The RNA cleavage preference of HIV RNase H as well as the cleavage rates were identical on 5′-phosphorylated and 5′-capped substrates. This may reflect an adaptation of HIV RNA to allow the complete digestion of 5′-capped HIV RNA during reverse transcription, as has been suggested previously (33). Interestingly, the cleavage site choice was not affected by the addition of a cap structure to the RNA substrate, although capping resulted in the presence of an additional nucleotide at the 5′ end of the RNA. A previous study suggested that on RNase H substrates with recessed RNA 5′ ends, the polymerase active site was located at the RNA 5′ end, leading to preferential RNA cleavage at a position 18 nucleotides downstream (31,32). Further studies are required to map the position of the polymerase active site relative to the RNase H active site on capped RNA substrates. The difference in cleavage site preference and divalent metal ion preference between E.coli and HIV RNases H confirmed the absence of E.coli RNase HI contamination in the preparation of HIV-RT and suggested a significant difference in active site structure and substrate interaction. HIV RNase H activity was strongly activated by Mn2+ compared with Mg2+, whereas E.coli RNase HI was only weakly active in 0.1 mM MnCl2. Both enzyme active sites have shown the presence of two Mn2+ ions in the active site of crystal structures. Therefore, these results are consistent with a model in which binding of two metal ions to E.coli RNase HI is possible, but inhibitory (activation/attenuation model) (22). In contrast, HIV RNase H is highly active under conditions where two Mn2+ ions are likely to bind to the active site (13).
HIV RNase H could be activated by a combination of Mg2+ and Mn2+ or by a combination of Mg2+ and Co2+ ions to levels higher than that observed with each metal ion separately. In addition, the titration of Mg2+-bound enzyme with increasing concentrations of either Mn2+ or Co2+ generated bell-shaped dose–response curves, indicating a two-step titration process. The impact on activity by Co2+ was lower than that achieved with Mn2+, which was similar to results obtained with influenza endonuclease and could be related to increased loss of Co2+ due to the formation of insoluble hydroxide (9). The RNase H activities at the highest concentrations of Mn2+ and Co2+ were unchanged when measured in the presence or absence of Mg2+. This indicated the complete replacement of Mg2+ in a biphasic process, consistent with a two-metal ion mechanism of RNA cleavage.
Compounds forming the N-hydroxyimide series of RNase inhibitors were established as probes for the interaction of two metal ions presented at a distance of 4–5 Å with three oxygen atoms. The N-hydroxyl group could function as a bridging ligand for both metal ions, with each metal ion undergoing an additional interaction with two carboxyl groups on the molecule. NMR studies with the isolated HIV RNase H domain confirmed the interaction of N-hydroxyimide 1 with the protein, which occurred in the presence, but not in the absence of Mg2+ (G.Williams, unpublished results). In addition, compounds which were inactive as RNase H inhibitors did not interact with the isolated HIV RNase H domain. Compound interaction with the protein in the presence of Mg2+ caused a spectral change consistent with enol formation. This could explain the fact that compounds 5 and 6 were inactive, as they have lost the ability to form the enol tautomer. N-Hydroxyimides were found to potently inhibit HIV RNase H, but not E.coli RNase HI activity. To our knowledge, this is the first demonstration of an inhibitor that could differentially inhibit HIV, but not E.coli RNase HI. In addition, the active compounds also inhibited RNase H activity of the isolated HIV RNase H domain with similar potency, consistent with binding to the RNase H active site (J.Q.Hang, S.Rajendran, Y.Yang, Y.Li, P.Wong Kai In, H.Overton, K.E.B.Parkes, N.Cammack, J.A.Martin and K.Klumpp, submitted). Inhibition of influenza virus RNA endonuclease was reported previously (11). However, the potency of compound 1 was ∼15-fold lower with influenza endonuclease compared with HIV RNase H. This could reflect the higher affinity of influenza endonuclease for substrate RNA compared with HIV-RT, because compound 1 competes with substrate binding to the active site (11).
Influenza RNA polymerase and T7 RNA polymerase activities were not affected by compound 1, although measurable inhibition was observed with HIV DNA polymerase. There is significant evidence suggesting that polymerase active sites contain only a single high affinity metal ion-binding site, even though they catalyse nucleotide incorporation via a two-metal ion mechanism. The second metal ion is most probably carried into the active site as a complex with the nucleotide triphosphate substrate (35–37). Therefore, the results of enzyme inhibition are consistent with N-hydroxyimides showing significant selectivity towards pre-formed two-metal ion compared with one-metal ion-binding sites. Preliminary structure–activity relationship (SAR) suggested the potential to increase potency and selectivity of these compounds by substitutions on the phenyl moiety.
Taken together, the results from single and multiple divalent metal ion titration and the successful design of two-metal ion-selective enzyme inhibitors have provided strong evidence to suggest that the essential polymerase-independent HIV RNase H activity is provided by a two-metal ion active site structure. There was no indication of metal ion cooperativity for E.coli RNase HI, suggesting that a different mechanism of RNA cleavage may operate in the two closely related enzymes. The current data do not exclude the possibility that substrate structure or sequence may influence the binding of metal ions, resulting in different active site structures catalysing functionally different RNase H activities. Also it is not known if the binding of metal ions to the polymerase active site may affect the RNase H activity of HIV-RT. Therefore, further experiments are aimed at the role of divalent metal ions in the polymerase-dependent, as well as the fast, primary cleavage reactions by HIV RNase H.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Glyn Williams, Astex Technology, Cambridge, UK, for the communication of results prior to publication, Dr Kenneth Straub, Roche Palo Alto for N-terminal sequencing and peptide mapping of HIV-RT and HIV RNase H domain protein preparations, and John Merrett, Roche Welwyn, for the chemical synthesis of reference compounds.
REFERENCES
- 1.Freed E.O. and Martin,M.A. (2001) HIVs and their replication. In Knipe,D. and Howley,P. (eds), Fields Virology. Lippincott Williams & Wilkins, Philadelphia, PA, Vol. 2, pp. 1971–2042. [Google Scholar]
- 2.Telesnitsky A. and Goff,S.P. (1997) Reverse transcription and the generation of retroviral DNA. In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 121–160. [PubMed] [Google Scholar]
- 3.Schatz O., Cromme,F.V., Nass,T. and Lindemann,D. (1989) Inactivation of RNaseH domain of HIV-1 reverse transcriptase blocks viral infectivity. In Papas,T.S. (ed.), Gene Regulation and AIDS: Transcriptional Activation, Retroviruses and Pathogenesis. Gulf Publishing Company, Houston, TX, pp. 293–303. [Google Scholar]
- 4.Tisdale M., Schulze,T., Larder,B.A. and Moelling,K. (1991) Mutations within the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase abolish virus infectivity. J. Gen. Virol., 72, 59–66. [DOI] [PubMed] [Google Scholar]
- 5.Andreola M.L., De Soultrait,V.R., Fournier,M., Parissi,V., Desjobert,C. and Litvak,S. (2002) HIV-1 integrase and RNase H activities as therapeutic targets. Expert Opin. Ther. Targets, 6, 433–446. [DOI] [PubMed] [Google Scholar]
- 6.Shaw-Reid C.A., Munshi,V., Graham,P., Wolfe,A., Witmer,M., Danzeisen,R., Olsen,D.B., Carroll,S.S., Embrey,M., Wai,J.S., Miller,M.D., Cole,J.L. and Hazuda,D.J. (2003) Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem., 278, 2777–2780. [DOI] [PubMed] [Google Scholar]
- 7.Tomassini J., Selnick,H., Davies,M.E., Armstrong,M.E., Baldwin,J., Bourgeois,M., Hastings,J., Hazuda,D., Lewis,J., McClements,W., Ponticello,G., Radzilowski,E., Smith,G., Tebben,A. and Wolfe,A. (1994) Inhibition of cap (m7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2,4-dioxobutanoic acid compounds. Antimicrob. Agents Chemother., 38, 2827–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hastings J.C., Selnick,H., Wolanski,B. and Tomassini,J.E. (1996) Anti-influenza virus activities of 4-substituted 2,4-dioxobutanoic acid inhibitors. Antimicrob. Agents Chemother., 40, 1304–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doan L., Handa,B., Roberts,N.A. and Klumpp,K. (1999) Metal ion catalysis of RNA cleavage by the influenza virus endonuclease. Biochemistry, 38, 5612–5619. [DOI] [PubMed] [Google Scholar]
- 10.Beese L.S. and Steitz,T.A. (1991) Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J., 10, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkes K.E., Ermert,P., Fassler,J., Ives,J., Martin,J.A., Merrett,J.H., Obrecht,D., Williams,G. and Klumpp,K. (2003) Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J. Med. Chem., 46, 1153–1164. [DOI] [PubMed] [Google Scholar]
- 12.Davies J.F. II, Hostomska,Z., Hostomsky,Z., Jordan,S.R. and Matthews,D.A. (1991) Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science, 252, 88–95. [DOI] [PubMed] [Google Scholar]
- 13.Cowan J.A., Ohyama,T., Howard,K., Rausch,J.W., Cowan,S.M. and Le Grice,S.F. (2000) Metal-ion stoichiometry of the HIV-1 RT ribonuclease H domain: evidence for two mutually exclusive sites leads to new mechanistic insights on metal-mediated hydrolysis in nucleic acid biochemistry. J. Biol. Inorg. Chem., 5, 67–74. [DOI] [PubMed] [Google Scholar]
- 14.Cristofaro J.V., Rausch,J.W., Le Grice,S.F. and DeStefano,J.J. (2002) Mutations in the ribonuclease H active site of HIV-RT reveal a role for this site in stabilizing enzyme–primer–template binding. Biochemistry, 41, 10968–10975. [DOI] [PubMed] [Google Scholar]
- 15.Pari K., Mueller,G.A., DeRose,E.F., Kirby,T.W. and London,R.E. (2003) Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry, 42, 639–650. [DOI] [PubMed] [Google Scholar]
- 16.Kanaya S., Oobatake,M. and Liu,Y. (1996) Thermal stability of Escherichia coli ribonuclease HI and its active site mutants in the presence and absence of the Mg2+ ion. Proposal of a novel catalytic role for Glu48. J. Biol. Chem., 271, 32729–32736. [DOI] [PubMed] [Google Scholar]
- 17.Haruki M., Tsunaka,Y., Morikawa,M., Iwai,S. and Kanaya,S. (2000) Catalysis by Escherichia coli ribonuclease HI is facilitated by a phosphate group of the substrate. Biochemistry, 39, 13939–13944. [DOI] [PubMed] [Google Scholar]
- 18.Katayanagi K., Okumura,M. and Morikawa,K. (1993) Crystal structure of Escherichia coli RNase HI in complex with Mg2+ at 2.8 Å resolution: proof for a single Mg(2+)-binding site. Proteins, 17, 337–346. [DOI] [PubMed] [Google Scholar]
- 19.Oda Y., Nakamura,H., Kanaya,S. and Ikehara,M. (1991) Binding of metal ions to E.coli RNase HI observed by 1H–15N heteronuclear 2D NMR. J. Biomol. NMR, 1, 247–255. [DOI] [PubMed] [Google Scholar]
- 20.Huang H.W. and Cowan,J.A. (1994) Metallobiochemistry of the magnesium ion. Characterization of the essential metal-binding site in Escherichia coli ribonuclease H. Eur. J. Biochem., 219, 253–260. [DOI] [PubMed] [Google Scholar]
- 21.Goedken E.R. and Marqusee,S. (2001) Co-crystal of Escherichia coli RNase HI with Mn2+ ions reveals two divalent metals bound in the active site. J. Biol. Chem., 276, 7266–7271. [DOI] [PubMed] [Google Scholar]
- 22.Keck J.L., Goedken,E.R. and Marqusee,S. (1998) Activation/attenuation model for RNase H. A one-metal mechanism with second-metal inhibition. J. Biol. Chem., 273, 34128–34133. [DOI] [PubMed] [Google Scholar]
- 23.Goedken E.R., Keck,J.L., Berger,J.M. and Marqusee,S. (2000) Divalent metal cofactor binding in the kinetic folding trajectory of Escherichia coli ribonuclease HI. Protein Sci., 9, 1914–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeGrice S.F. and Gruninger-Leitch,F. (1990) Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem., 187, 307–314. [DOI] [PubMed] [Google Scholar]
- 25.Klumpp K., Hooker,L. and Handa,B. (2001) Influenza virus endoribonuclease. Methods Enzymol., 342, 451–466. [DOI] [PubMed] [Google Scholar]
- 26.Sarafianos S.G., Das,K., Tantillo,C., Clark,A.D.,Jr, Ding,J., Whitcomb,J.M., Boyer,P.L., Hughes,S.H. and Arnold,E. (2001) Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J., 20, 1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schatz O., Mous,J. and Le Grice,S.F. (1990) HIV-1 RT-associated ribonuclease H displays both endonuclease and 3′–5′ exonuclease activity. EMBO J., 9, 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wohrl B.M. and Moelling,K. (1990) Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA–DNA hybrids. Biochemistry, 29, 10141–10147. [DOI] [PubMed] [Google Scholar]
- 29.Gopalakrishnan V., Peliska,J.A. and Benkovic,S.J. (1992) Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl Acad. Sci. USA, 89, 10763–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gotte M., Maier,G., Gross,H.J. and Heumann,H. (1998) Localization of the active site of HIV-1 reverse transcriptase-associated RNase H domain on a DNA template using site-specific generated hydroxyl radicals. J. Biol. Chem., 273, 10139–10146. [DOI] [PubMed] [Google Scholar]
- 31.Palaniappan C., Fuentes,G.M., Rodriguez-Rodriguez,L., Fay,P.J. and Bambara,R.A. (1996) Helix structure and ends of RNA–DNA hybrids direct the cleavage specificity of HIV-1 reverse transcriptase RNase H. J. Biol. Chem., 271, 2063–2070. [PubMed] [Google Scholar]
- 32.Wisniewski M., Balakrishnan,M., Palaniappan,C., Fay,P.J. and Bambara,R.A. (2000) Unique progressive cleavage mechanism of HIV reverse transcriptase RNase H. Proc. Natl Acad. Sci. USA, 97, 11978–11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisniewski M., Chen,Y., Balakrishnan,M., Palaniappan,C., Roques,B.P., Fay,P.J. and Bambara,R.A. (2002) Substrate requirements for secondary cleavage by HIV-1 reverse transcriptase RNase H. J. Biol. Chem., 277, 28400–28410. [DOI] [PubMed] [Google Scholar]
- 34.Lott W.B., Pontius,B.W. and von Hippel,P.H. (1998) A two-metal ion mechanism operates in the hammerhead ribozyme-mediated cleavage of an RNA substrate. Proc. Natl Acad. Sci. USA, 95, 542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Korolev,S. and Waksman,G. (1998) Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J., 17, 7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steitz T.A. (1998) A mechanism for all polymerases. Nature, 391, 231–232. [DOI] [PubMed] [Google Scholar]
- 37.Sosunov V., Sosunova,E., Mustaev,A., Bass,I., Nikiforov,V. and Goldfarb,A. (2003) Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J., 22, 2234–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]