Abstract
DNA curvature plays a well-characterized role in many transcriptional regulation mechanisms. We present evidence for the conservation of curvature signals in putative regulatory regions of several archaeal and eubacterial genomes. Genes with highly curved upstream regions were identified in orthologous groups, based on the annotations of the Cluster of Orthologous Groups of proteins (COG) database. COGs possessing a significant number of genes with curvature signals were analyzed, and conserved properties were found in several cases. Curvature signals related to regulatory sites, previously described in single organisms, were located in a broad spectrum of bacterial genomes. Global regulatory proteins, such as HU, IHF and FIS, known to bind to curved DNA and to be autoregulated, were found to present conserved DNA curvature signals in their regulatory regions, emphasizing the fact that structural parameters of the DNA molecule are conserved elements in the process of transcriptional regulation of some systems. It is currently an open question whether these diverse systems are part of an integrated global regulatory response in different microorganisms.
INTRODUCTION
Studies of the relationship between DNA curvature and transcription regulation have been conducted mostly for specific sets of genes and discrete loci. Experimental evidence has demonstrated contributions of DNA curvature in regulating the transcription of several genes, such as those coding for H-NS histone-like protein (1), σs (2), IHF and HU regulatory proteins (3), σ54-dependent glnA (4) and artificial constructs using the T7 virus promoter (5), among others.
Genome-wide analysis of DNA curvature in promoter sequences by our group (6) and others (7) has found these regions to be significantly more curved than coding regions or randomly permuted sequences. Moreover, binding sites for known regulatory proteins were found to present even higher curvature values (7). It was also proposed that mesophilic bacteria, as opposed to hyperthermophilic bacteria and archaea, present a bias to a high DNA curvature content due to a temperature-related adaptation of the transcription regulation mechanisms (8); these data suggest that DNA curvature is not selected because of thermal stability properties. In these previous studies, only average curvature values were considered; therefore, individual genes with significant curvature signals in their regulatory regions were not identified. Recently the curvature profiles of mycobacterial promoters were analyzed, and several genes with similar trends were recognized, suggesting a common transcription mechanism (9).
As far as we know, no attempt has been made to establish that discrete DNA curvature signals are conserved as regulatory features in different organisms. Here we extend the previous studies and address the question of whether static DNA curvature is an element involved in the transcriptional regulation mechanisms within the broad context of 99 available microbial genomes (see Materials and Methods). Using the data compiled in the COG (Cluster of Orthologous Groups of proteins) database (10), and additional orthology data for the genomes not included in it (see Materials and Methods), we demonstrate a significant conservation of curvature signals within the regulatory regions of several clusters spanning a broad spectrum of biological functions. Within this set of orthologous genes, those coding for certain DNA-binding proteins were found to present curved regulatory regions in a large number of genomes. A detailed examination of the most relevant cases is also presented.
MATERIALS AND METHODS
DNA sequence data
DNA sequence was derived from the complete bacterial genomes available in the Entrez Genome Database (fttp://ncbi.nlm.nih.gov/genomes/Bacteria/). We reduced the number of current genomes to a non-redundant collection by eliminating different strains of the same organism, leaving the one with the largest genome. Thus, the analysis contemplated 99 complete archaeal and bacterial genomes. A list of these genomes can be found at http://www.ibt.unam.mx/biocomputo/curvature/genomes.html.
Delimitation of regulatory regions
A 250 nt window, containing 200 bases upstream and 50 bases downstream of the start codon of each coding sequence (CDS), was chosen as our analysis window, since >90% of the regulatory signals are found within this range in Escherichia coli K12 (11). Operons were predicted based on intergenic distances as described by Moreno-Hagelsieb and Collado-Vides (12), and the regulatory region of each gene was considered as the upstream region of the first gene in the operon. This is what we call the minimal upstream regions set (MURs). These MURs are available online (http://www.ibt.unam.mx/biocomputo/curvature/murs/).
Curvature calculations
DNA curvature was calculated using the algorithm BEND (13) and the rotational and translational contribution matrices derived from sequence periodicities in nucleosome core DNA (14). The algorithm and matrices were chosen since they have been compared with five other models of DNA curvature calculation and were found to be the best for predicting experimental curvature data, both A-tract-based and GC-rich-based, as measured by gel retardation, cyclization kinetics and structural data from X-ray crystallography (13); the use of different algorithms gives qualitatively similar results (data not shown). A curvature profile was obtained by assigning to each nucleotide of the genome a curvature value, expressed as a deviation angle from the helical axis per helical turn. Signal-to-noise ratio was minimized by taking the average value of a sliding window of 31 nucleotides (three helical turns), and assigning it to the central nucleotide (13). A frequency histogram was constructed using these values, and the mean and SD were calculated. Since each genome presents a distinctive curvature profile (6,15), a cut-off value of 3 SDs from the genomic curvature mean of each organism was used to identify statistically significant signals in the set of MURs. Each gene selected in this way was collected and sorted into its corresponding orthologous group.
Clustering of orthologs and identification of COGs with a statistically significant number of curvature signals
Our orthologous groups were mainly those found in the COGs database (10). Orthologs not included in this database were identified using the bi-directional best-hit criterion, as implemented elsewhere (12). In order to evaluate the statistical significance of the number of genes with curved DNA signals in their regulatory regions in a given COG, the following procedure was used: (i) a database of Monte Carlo permutations of the MURs within each complete genome was generated; (ii) for every permutation, the number of genes within each COG that presented a curvature signal in its MUR was counted; (iii) the previous two steps were repeated 1000 times to find the mean and SD of the number of signals for each COG; and (iv) a statistical significance value was given to every COG based on the distance between the number of observed signals within the COG and the mean of the number of signals obtained in the Monte Carlo permutations (expected mean) expressed in SD units (Table 1). COGs with proteins from less than five organisms represented were excluded from our analysis. The nucleotide sequences of the MURs with significant curvature signals, grouped by COGs, are avail able at http://www.ibt.unam.mx/biocomputo/curvature/curved-cogs/. Most COGs presented a broad range of organisms; the phylogenetical data of significant COGs is available at http://www.ibt.unam.mx/biocomputo/curvature/tree/.
Table 1. COGs with the most significant conserved curvature signals in their regulatory regions.
| SD | COG | Genes/organisms | Function | |
|---|---|---|---|---|
| Observed | Total | |||
| Nucleotide transport and metabolism | ||||
| 4.17 | COG0299 | 25/25 | 64/62 | Phosphoribosylglycinamide formyltransferase |
| 3.95 | COG3072 | 8/8 | 12/12 | Adenylate cyclase |
| Signal transduction mechanisms | ||||
| 3.38 | COG1217 | 22/21 | 62/61 | GTPase |
| 3.09 | COG3275 | 10/9 | 23/21 | Two-component sensor histidine kinase |
| Cell motility | ||||
| 3.21 | COG1344 | 38/20 | 115/36 | Flagellin |
| 3.08 | COG1360 | 22/19 | 60/36 | Flagellar protein MotB |
| Transcription | ||||
| 4.19 | COG4977 | 31/15 | 61/18 | Transcriptional regulator araC family |
| 3.81 | COG0085 | 32/31 | 98/90 | RNA polymerase β subunit |
| 3.28 | COG0553 | 32/24 | 98/59 | SNF2 RNA helicase |
| 3.16 | COG1522 | 98/36 | 336/53 | Transcriptional regulator asnC/lrp family |
| Amino acid transport and metabolism | ||||
| 4.66 | COG4992 | 38/29 | 96/65 | PLP-dependent aminotransferases |
| 3.36 | COG0253 | 24/24 | 68/64 | Diaminopimelate epimerase |
| 3.30 | COG0703 | 25/25 | 72/66 | Shikimate kinase |
| 3.03 | COG0174 | 46/31 | 142/74 | Glutamine synthetase |
| Defense mechanisms | ||||
| 3.67 | COG2746 | 8/6 | 13/10 | Aminoglycoside N3′-acetyl transferase |
| Cell wall/membrane/envelope biogenesis | ||||
| 5.37 | COG3637 | 29/12 | 65/16 | Outer membrane protein x precursor |
| 5.30 | COG0275 | 31/31 | 74/74 | SAM-dependent methyltransferase |
| 4.24 | COG0768 | 52/36 | 158/69 | Penicillin-binding protein 2 |
| 4.10 | COG2821 | 14/14 | 27/26 | Membrane-bound lytic murein transglycosylase A precursor |
| 3.86 | COG0472 | 42/37 | 133/77 | Phospho-N-acetylmuramoyl-pentapeptide-transferase |
| 3.33 | COG0797 | 20/17 | 52/37 | Rare lipoprotein A |
| 3.04 | COG1212 | 15/15 | 36/36 | 3-Deoxy-manno-octulosonate cytidylyltransferase |
| 3.01 | COG0770 | 23/23 | 72/72 | UDP-N-acetylmuramoylalanyl-d-glutamyl-2,6-diaminopimelate–d-alanyl-d-alanyl ligase |
| Replication, recombination and repair | ||||
| 6.81 | COG0776 | 64/43 | 150/71 | DNA-binding protein HU-α |
| 4.57 | COG0188 | 45/37 | 134/80 | DNA gyrase subunit A |
| 4.23 | COG0187 | 45/38 | 131/80 | DNA gyrase subunit B |
| 4.15 | COG3385 | 38/6 | 109/10 | Transposase |
| 4.08 | COG1604 | 5/5 | 8/7 | Hypothetical protein |
| 3.13 | COG0468 | 30/30 | 102/87 | RecA protein |
| 3.04 | COG3611 | 7/7 | 17/17 | Chromosome replication protein |
| Translation, ribosomal structure and biogenesis | ||||
| 3.55 | COG0268 | 23/23 | 64/64 | 30S ribosomal protein S20 |
| 3.50 | COG0173 | 25/25 | 76/75 | Aspartyl-tRNA synthetase |
| 3.28 | COG0012 | 29/29 | 95/90 | GTP-binding protein |
| 3.22 | COG4108 | 18/18 | 48/48 | Peptide chain release factor 3 |
| 3.04 | COG5256 | 7/7 | 14/14 | Translation elongation factor EF-1, α chain |
| Post-translational modification, protein turnover, chaperones | ||||
| 3.27 | COG0719 | 37/24 | 125/65 | ABC transporter permease |
| Inorganic ion transport and metabolism | ||||
| 4.19 | COG1392 | 19/18 | 43/37 | Pit accessory protein |
| 3.47 | COG1553 | 12/9 | 28/22 | Hypothetical protein, involved in intracellular sulfur reduction |
| Cell cycle control, cell division, chromosome partitioning | ||||
| 4.90 | COG3116 | 11/11 | 17/17 | Cell division protein ftsL |
| 3.56 | COG0849 | 21/21 | 63/57 | Cell division protein ftsA |
| 3.48 | COG0552 | 29/29 | 90/90 | Cell division protein ftsY |
| 3.20 | COG3096 | 5/5 | 8/8 | Cell division protein mukB |
| 3.11 | COG3006 | 5/5 | 8/8 | Killing factor protein kicB |
| 3.02 | COG3095 | 5/5 | 8/8 | Killing protein supressor kicA |
| Carbohydrate transport and metabolism | ||||
| 3.36 | COG0205 | 25/23 | 70/58 | 6-Phosphofructokinase |
| 3.01 | COG0166 | 26/26 | 82/78 | Glucose-6-phosphate isomerase |
| General function prediction only | ||||
| 3.82 | COG4572 | 5/5 | 8/7 | Cation transport regulator ChaB |
| 3.82 | COG1084 | 8/8 | 16/16 | Predicted GTPase |
| 3.70 | COG1075 | 14/12 | 27/19 | Triacylglycerol lipase precursor |
| 3.33 | COG0795 | 27/14 | 80/40 | Membrane protein |
| 3.18 | COG2071 | 16/16 | 41/34 | Glutamine amidotransferase, class I |
| 3.12 | COG3081 | 7/7 | 12/12 | Nucleoid-associated protein |
| 3.03 | COG2607 | 9/9 | 17/17 | ATP-dependent protease |
| 3.02 | COG1823 | 8/8 | 17/17 | Na+/dicarboxylate symporter |
| Function unknown | ||||
| 5.60 | COG2001 | 23/23 | 44/44 | Hypothetical protein |
| 4.18 | COG3870 | 7/7 | 13/12 | Hypothetical protein |
| 4.00 | COG3862 | 5/5 | 9/8 | Hypothetical protein |
| 3.85 | COG1799 | 14/14 | 31/28 | Hypothetical protein |
| 3.73 | COG3025 | 10/10 | 17/16 | Hypothetical protein |
| 3.70 | COG1945 | 9/8 | 18/14 | Hypothetical protein |
| 3.65 | COG2302 | 9/9 | 21/21 | Hypothetical protein |
| 3.47 | COG0779 | 21/21 | 59/59 | Hypothetical protein |
| 3.41 | COG4095 | 6/5 | 12/11 | Hypothetical protein |
| 3.32 | COG2976 | 10/10 | 20/20 | Hypothetical protein |
| 3.24 | COG0762 | 17/17 | 47/45 | Hypothetical protein |
| 3.15 | COG4807 | 7/7 | 12/12 | Hypothetical protein |
| 3.08 | COG3665 | 6/5 | 7/6 | Hypothetical protein |
| 3.01 | COG4649 | 5/5 | 7/7 | Hypothetical protein |
The columns indicate the distance, in SDs, to the expected mean (see Materials and Methods), the COG number, the number of observed genes/organisms with curved signals, the total number of genes/organisms in the COG, and the function according to the classification of the COG database, respectively. Genes are subgrouped into general functional scopes.
Promoter prediction
Promoter sequences for the genes in significant COGs were predicted using the algorithm of Mulligan et al. (16). Weight matrices were derived from alignments of experimentally characterized promoters for σ70 (16) and σ54 (17). Images of the regions containing the best scoring promoters were obtained using the DIAMOD DNA curvature display software (18).
RESULTS
Analysis of COGs with conserved curvature signals
Sixty-eight of the 4391 COGs studied presented a statistically significant number of curvature signals (at least 3 SDs above the expected mean). These COGs were classified according to their global functional characterization (10) (Table 1). Experimental data supporting a role for DNA curvature in transcriptional regulation were searched for in the literature. Biologically relevant COGs with lower scores, yet still above 2 SDs, are available as supplementary information (http://www.ibt.unam.mx/biocomputo/curvature/table2.html) and considered in our discussion. Representative cases of the best scoring COGs are given below.
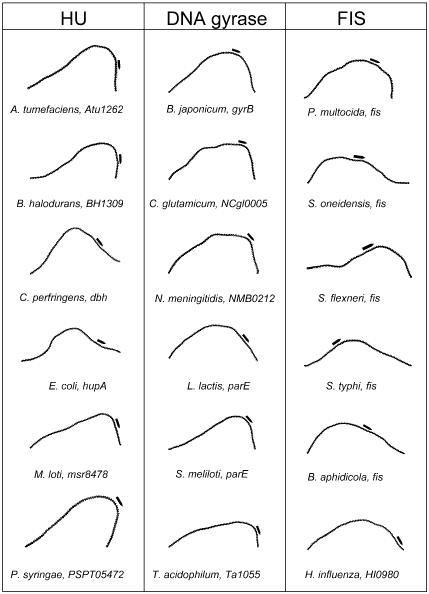
Proteins HU and IHF from COG0776. Sixty-four genes from 43 different organisms were found to have curvature signals in their regulatory regions. Both IHF and HU proteins are known to be key regulators of a broad spectrum of genes in several organisms. These proteins bind to curved DNA regions and further bend the DNA molecule (19). Autogenous transcriptional regulation for the hupA and hupB genes in E.coli has been demonstrated (20). Besides being autoregulated, the transcription of these genes has been found to be dependent on CRP (catabolite repression protein) and FIS (factor for inversion stimulation), both of which bind to curved DNA (21). In the case of the genes coding for the IHF dimer, himA and himD, autoregulation has also been demonstrated along with dependence on rpoS and ppGpp levels (22–24). Representative cases of curved upstream regions with predicted σ70 promoters are shown in Figure 1.
Figure 1.
DNA curvature plots of upstream regions for genes coding for DNA gyrase, HU and FIS orthologs. The sequence spanning from 400 nt upstream to the first codon was chosen for graphical representation, since the curvature is readily discernible for sequence fragments of this size. A black arrow indicates putative promoter regions. DNA curvature plots were obtained using the DIAMOD software (18), and the geometrical matrix was derived from nucleosome positioning preferences (14).
DNA gyrase subunits A and B from COG0188 and COG0187, respectively. Forty-five genes for subunit A were found in 37 organisms, and 45 genes for subunit B in 38 organisms. This protein is responsible for introducing negative supercoils in the DNA molecule, and its own transcription depends on the supercoiled state of the genome (25), and FIS (26). A bent DNA region between the –35 and –10 elements of the gyrA promoter has been described in Streptococcus pneumoniae, and it has been proposed that this region makes the promoter very sensitive to super-structural changes, allowing the presence of GyrA to regulate DNA supercoiling in the cell (27). Significant curvature signals have been previously predicted in several mycobacterial gyrase promoters (9). Representative cases from this COG are shown in Figure 1.
FIS protein-coding genes, from COG2901, with nine genes in nine organisms. Even though this COG’s score is below 3 SD units (see supplementary information at http://www.ibt.unam.mx/biocomputo/curvature/table2.html), the relevance of this global regulator cannot be overlooked. The FIS protein is a pleiotropic regulator involved in stringent control and directly regulating the expression of many ribosomal genes, polymerases and proteins related to cell division (26). FIS also plays a role in DNA replication and recombination (28). FIS is autoregulated in E.coli (29) and, through the repression of the gyrase genes, plays the role of a homeostatic topological regulator, counteracting the negative supercoiling of the genome (30). It is also known that this protein binds to bent DNA regions (31). In addition, FIS is regulated by IHF, which also binds to curved DNA (32).
Transposases from COG3385, with 38 genes in six organisms. There is no evidence relating curved DNA regions with the transcriptional regulation of transposases, but the modulation of transposition events has been found to depend on global regulatory proteins such as H-NS, HU and IHF (33–35), all of them known to bind curved DNA. Interestingly, the role of DNA curvature involved in transposition has been confirmed for the insertion sequence IS231A in Bacillus thuringiensis, where one of the terminal repeats of the transposon Tn4430 was found to be an insertional hot spot, due to flanking curved DNA regions (36). Even though these data are not directly involved in transcriptional regulation, they confirm a general role for DNA curvature in transposition events. Within the context of this analysis, our data suggest that curved sites also play a role in transcriptional regulation of transposase genes. This idea is further supported by the presence of four other transposase COGs, with SD values above 2 (supplementary information).
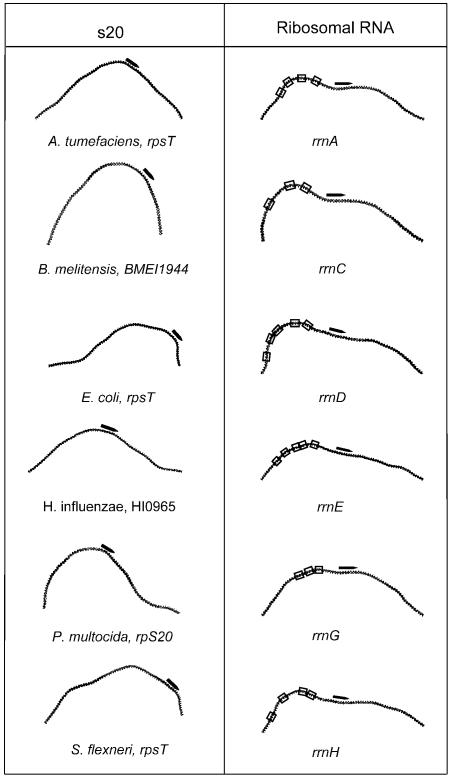
Translation-related COGs. Five COGs presented a number of genes with curvature signals above 3 SDs, and nine COGs above 2 SDs. 30S ribosomal proteins S20 and S16 from COG0268 and COG0228, with 23 genes in 23 organisms and 20 genes in 20 organisms, respectively, presented significant curvature signals. Although no direct evidence has been reported for the relevance of DNA curvature in the transcription of these, the role of FIS-regulated upstream activating sequences (UAS) has been documented for several ribosomal operons (37,38), and the DNA in the UAS is known to be bent in E.coli (39). Experimentally determined ribosomal promoters in E.coli present bent regions (40) (Fig. 2). Aspartyl-, histidyl- and isoleucyl-tRNA synthetases also presented curvature signals in 25, 26 and 23 genomes, respectively. The presence of conserved UAS, regulating bi-directional promoters of glutamyl-tRNA synthetase and the valU and alaW tRNA operon in E.coli, has already been demonstrated (41). COGs for the genes coding for translation initiation, elongation and peptide release factors were also present with scores above 2 SDs (supplementary information), along with other ribosomal genes.
Figure 2.
Curvature profiles for the upstream region of genes coding for ribosomal protein S20 orthologs in different organisms and experimentally characterized promoter regions for rRNAs in E.coli. Black arrows indicate putative and real promoters; FIS-binding sites identified by DNase I footprints are boxed.
Cell division-related genes. These were from COG3116 (ftsL), COG0552 (ftsY), COG3096 (mukB), COG3006 (kicB), COG0849 (ftsA), COG3095 (mukE) and, with lower scores, COG0772 (ftsW), COG3115 (zipA), COG4839 (ftsL-like) and COG0445 (gidA). In this case, we found 10 different COGs involved in genome replication and cell division. As in the previous groups, the curvature signals were present in an important number of genomes, 29 and 31 genomes in the cases of ftsY and ftsW, respectively (Table 1 and supplementary information). The time coordination requirements for such processes impose a highly regulated transcription schedule. Transcription is, in many cases, mediated by general DNA curvature-dependent regulators such as IHF, FIS and HU (28,42). Nevertheless, there is no direct experimental evidence relating DNA curvature to the transcription of these genes.
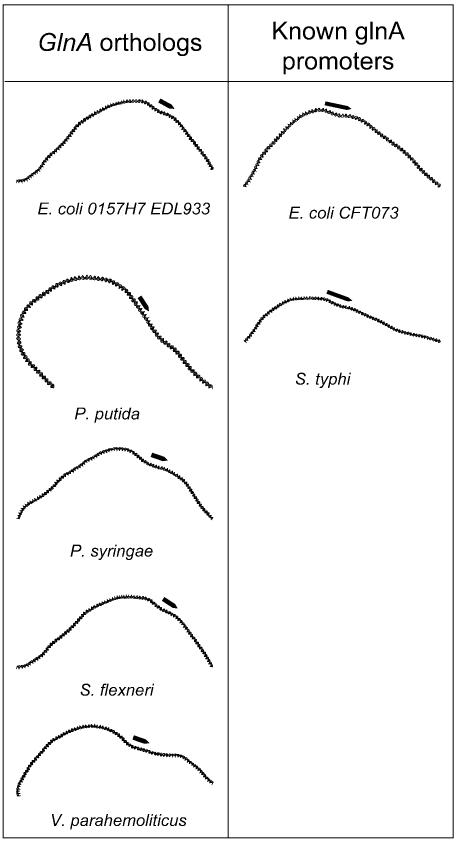
Glutamine synthetase from COG0174, with 46 genes from 31 organisms. This gene, with a σ54-dependent promoter, has been found to also depend on a bent region between the promoter and the enhancer site to initiate transcription in E.coli (4,11). Our finding of a curvature signal in the regulatory regions of these genes in 31 genomes indicates that this mechanism is widely conserved. In an attempt to further characterize the regulatory regions of these genes, we predicted σ54-dependent promoters for these MURs, using a weight matrix derived from the compilation of 85 experimentally determined sequences (17), and generated images of the curvature of these regions, including experimentally characterized glnA promoter sequences (Fig. 3).
Figure 3.
Curvature profiles of upstream regions for glutamine synthetase orthologs and experimentally characterized promoters for glnA genes in E.coli and S.typhi. Black arrows indicate putative promoters and RNA polymerase-protected regions.
Flagellum genes. These were from COG1344 (fliC) (38/20), COG1360 (motB) (22/19) and, with lower scores, COG4787 (flgF) (7/7), COG1291 (motA) (16/15), COG1377 (flhB) (15/14), COG1558 (flgC) (13/13) and COG1261 (flgA) (10/10). In S.enterica, the flagellar biosynthesis system is regulated by a master operon containing the specific flagellar σ factor, σ28, and the flagellin gene, both of which regulate the expression of the rest of the flagellar genes (43). The presence of regulatory sites for CRP and H-NS regulators was demonstrated for this operon in Salmonella typhimurium and E.coli (44,45), both known to bind curved DNA (1,46). In Vibrio cholerae and Campylobacter jejuni, the flagellar biosynthesis pathway is dependent on both σ54 and σ28 (47,48). Again, there is no direct experimental evidence relating curved DNA to the transcriptional regulation of these genes. It is important to consider that flagellar genes are commonly organized in operons, and, in this particular case, flgC and flgF share the same regulatory region in E.coli, N.europaea, R.solanacearum and S.typhi.
DISCUSSION
The structure of the DNA molecule, in this case measured as DNA curvature, has been found to play important roles in several biological processes, including DNA replication and packaging, chromosome segregation, recombination, transposition, virus integration and transcriptional regulation. Until recently, DNA curvature had been studied in the context of discrete DNA fragments and particular loci in single organisms, and no attempt had been made to analyze this feature in a broad genomic context. Here we present a first attempt to establish the conservation of DNA curvature as an element of transcriptional regulation in a comparative study across bacterial genomes.
The COG database was originally compiled based on protein sequence conservation among phylogenetically distinct organisms, implying that proteins in a given COG should share a common ancestor and therefore be functionally related. It is not far fetched to postulate that regulatory mechanisms could be conserved in some of these orthologous groups. With this in mind, we addressed the presence of conserved regulatory signals that we associate with DNA curvature. Among the 4391 COGs analyzed, 68 present a significant number of curvature signals spanning a broad range of biological functions, including, in many cases, phylogenetically distant organisms, with different genomic GC content, and no significant sequence conservation in their regulatory regions. The predictive capabilities of computer algorithms (13) and the accuracy of rotational and displacement matrices (14) was evident since our study reveals most of the genes regulated by curvature that have been previously described experimentally, such as hupA, hupB, himA, himD and glnA, among others. These results led us to extend the analysis to previously uncharacterized regions that present conserved signals in a genomic context.
Several COGs selected by our methods have the same global biological function, such as cell division. In this case, 10 different COGs were found to present curved DNA in the regulatory regions of their corresponding genes. This was an unexpected finding, since, to our knowledge, there is no experimental evidence relating DNA curvature and their transcriptional regulation. Global morphological changes are known to occur in the chromosome during the cell cycle, and the idea of an as yet uncharacterized curvature-dependent mechanism, or regulatory protein, playing a role in this process is feasible. In the same manner, our finding of several genes related to flagellum biosynthesis and motility with conserved curved motifs was also surprising. Until now, no experimental evidence exists relating the transcriptional regulation of flagellar genes to curved DNA, even though the dependence on curvature-recognizing regulatory proteins has been demonstrated in some cases (44,45). The genes of some poorly characterized transcriptional regulators such as the araC family COG4977 and the asnC family COG1522 and several uncharacterized COGs were also found to present a significant number of curvature signals. This information is interesting since we can now postulate the existence of conserved regulatory mechanisms a priori, and suggest that the experimental characterization of their expression should take the curved sites into account. In the case of transcriptional regulators in particular, our data might prove to be helpful for the future detection of genes under their control.
The group of global regulatory proteins HU, IHF and FIS merits special attention since all of them are non-specific DNA-binding proteins, involved not only in the control of gene expression, but also in other important biological processes such as recombination, DNA replication and organization of the bacterial chromosome. All these proteins are known to bind to curved DNA or induce a bend upon binding. Such conformational changes mediate or stabilize the binding of other regulators. These data support our observation of many orthologous groups, whose transcription is dependent on the global regulators mentioned. In addition, the genes of this group of proteins are known to be autoregulated, and therefore a conserved role of DNA curvature in their own regulatory regions is expected (20,22,29). This hypothesis was confirmed by the identification of numerous bacterial genomes sharing a curvature signal in the regulatory regions of the same proteins. Another point worth mentioning is the possibility that several complementary mechanisms could exist among different organisms regulating the same set of genes. The presence of a curved element could be one, but its absence could be partially or totally compensated by a flexible stretch and/or a site for a protein capable of forcing a bend in the DNA, resulting in a similar structural conformation. An example of this is the σ54-dependent gene glnH, in E.coli, whose transcription is activated by a bend induced by IHF (4). The role of curved or flexible loci in the DNA of upstream regions in genes regulated by σ54 promoters has been well established for many years (49). This property helps to bring the upstream specific regulator close to the promoter, thus enhancing the activation of σ54 promoters.
DNA curvature might not only serve as a signal for the recognition of regulatory proteins. It has been reported that in several highly expressed genes in E.coli, the curved sites could facilitate the formation of the open complex during transcription initiation. This is the case not only for proposed ribosomal proteins and aminoacyl-tRNA synthetases identified in our study, but also for experimentally characterized rRNA genes. This feature has been demonstrated also by artificial promoter constructs where a curved region is added upstream of the promoter, increasing the transcription rates by up to 10-fold (5). Here we should add a word of caution since promoter enhancement by phased A tracts, one type of evidence that is sometimes cited in support of a stimulatory role for DNA curvature, may actually represent sequence-specific recognition of ‘up’ elements by RNA polymerase, rather than DNA structure. In this report, we present evidence to show that curvature-mediated transcriptional activation is a common feature that is shared even in phylogenetically distant bacteria. Further studies of the regulatory mechanisms of these genes might demonstrate the prevalence of curvature-related transcriptional regulation.
Our finding of several clusters of orthologous genes with a significant number of curvature signals in their upstream regions is coherent with the previous experimental characterization of a DNA curvature-related regulatory mechanism and provides additional evidence in several predicted cases. It is difficult or impossible to define a useful consensus for several DNA-binding proteins. In the case of HU and H-NS, we could not find a consensus and, although a weight matrix has been proposed for FIS binding sites, it is highly degenerate (50). For these examples, the conformation of the binding sites could be more important than the sequence per se, and one of the determinant properties for this conformation could be DNA curvature. All these facts point towards an important role for the structure of the DNA molecule in transcriptional regulation.
The collection of genes we found where DNA bending plays a role in transcriptional regulation seems to be, at first sight, a diverse group of unrelated genes. In some cases, such as σ54 promoters, as well as autoregulation of FIS, HU and related proteins, a rationale for the role of curvature is well established. The degree of supercoiling is known to be affected by oxygen availability and, in fact, supercoiling and its effects on the structure of the genome has been proposed as a general regulatory mechanism in response to the overall energy state of the cell (51). An integrated approach, combining sequence and structure, for the prediction of regulatory regions on the one hand, and a detailed analysis of microarray experiments, performed at different overall energy states, could contribute to the search for an integrated biological paradigm for all these different systems and genes, here proposed as being subjected to curvature-sensitive regulation.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Shirley Ainsworth for bibliographical assistance, and Ricardo Ciria, Abel Linares, Juan Manuel Hurtado and Alma Martinez for computer support. This work was supported by the UNAM-PAPIIT grant IN215402.
REFERENCES
- 1.Atlung T. and Ingmer,H. (1997) H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol., 24, 7–17. [DOI] [PubMed] [Google Scholar]
- 2.Espinosa-Urgel M. and Tormo,A. (1993) Sigma s-dependent promoters in Escherichia coli are located in DNA regions with intrinsic curvature. Nucleic Acids Res., 21, 3667–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pérez-Martin J., Rojo,F. and De Lorenzo,V. (1994) Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev., 58, 268–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmona M. and Magasanik,B. (1996) Activation of transcription at σ54-dependent promoters on linear templates requires intrinsic or induced bending of the DNA. J. Mol. Biol., 261, 348–356. [DOI] [PubMed] [Google Scholar]
- 5.Collis C.M., Molloy,P.L., Both,G.W. and Drew,H.R. (1989) Influence of the sequence-dependent flexure of DNA on transcription in E.coli. Nucleic Acids Res., 17, 9447–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jáuregui R., O’Reilly,F., Bolivar,F. and Merino,E. (1998) Relationship between codon usage and sequence dependent curvature of genomes. Microb. Comp. Genomics, 3, 243–253. [PubMed] [Google Scholar]
- 7.Gabrielian A.E., Landsman,D. and Bolshoy,A. (1999–2000) Curved DNA in promoter sequences. In Silico Biol., 1, 83–96. [PubMed] [Google Scholar]
- 8.Bolshoy A. and Nevo,E. (2000) Ecologic genomics of DNA: upstream bending in prokaryotic promoters. Genome Res., 10, 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalate R.N., Kulkarni,B.D. and Nagaraja,V. (2002) Analysis of DNA curvature in mycobacterial promoters using theoretical models. Biophys. Chem., 99, 77–97. [DOI] [PubMed] [Google Scholar]
- 10.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gralla J.D. (1996) Activation and repression of E.coli promoters. Curr. Opin. Genet. Dev., 6, 526–530. [DOI] [PubMed] [Google Scholar]
- 12.Moreno-Hagelsieb G. and Collado-Vides,J. (2002) A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics, 18 (Suppl. 1), 329–336. [DOI] [PubMed] [Google Scholar]
- 13.Goodsell D.S. and Dickerson,R.E. (1994) Bending and curvature calculations in B-DNA. Nucleic Acids Res., 22, 5497–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satchwell S.C., Drew,H.R. and Travers,A.A. (1986) Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol., 191, 659–675. [DOI] [PubMed] [Google Scholar]
- 15.Gabrielian A., Vlahovicek,K. and Pongor,S. (1997) Distribution of sequence-dependent curvature in genomic DNA sequences. FEBS Lett., 406, 69–74. [DOI] [PubMed] [Google Scholar]
- 16.Mulligan M.E., Hawley,D.K., Entriken,R. and McClure,W.R. (1984) Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res., 12, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrios H., Valderrama,B. and Morett,E. (1999) Compilation and analysis of σ(54)-dependent promoter sequences. Nucleic Acids Res., 27, 4305–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dlakic M. and Harrington,R.E. (1998) DIAMOD: display and modeling of DNA bending. Bioinformatics, 14, 326–331. [DOI] [PubMed] [Google Scholar]
- 19.Dickerson E.R., (1998) DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res., 26, 1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohno K., Wada,M., Kano,Y. and Imamoto,F. (1990) Promoters and autogenous control of the Escherichia coli hupA and hupB genes. J. Mol. Biol., 213, 27–36. [DOI] [PubMed] [Google Scholar]
- 21.Claret L. and Rouviere-Yaniv,J. (1996) Regulation of HU-α and HU-β by CRP and FIS in Escherichia coli. J. Mol. Biol., 263, 126–139. [DOI] [PubMed] [Google Scholar]
- 22.Mechulam Y., Blanquet,S. and Fayat,G. (1987) Dual level control of the Escherichia coli pheST–himA operon expression: tRNA-phe-dependent attenuation and transcriptional operator–repressor control by himA and the SOS network. J. Mol. Biol., 197, 453–470. [DOI] [PubMed] [Google Scholar]
- 23.Miller H.I., Kirk,M. and Echols,H. (1981) SOS induction and autoregulation of the himA gene for site-specific recombination in Escherichia coli. Proc. Natl Acad. Sci. USA, 78, 6754–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aviv M., Giladi,H., Schreiber,G., Oppenheim,A.B. and Glaser,G. (1994) Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol. Microbiol., 14, 1021–1031. [DOI] [PubMed] [Google Scholar]
- 25.Menzel R. and Gellert,M. (1987) Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc. Natl Acad. Sci. USA, 84, 4185–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Travers A., Schneider,R. and Muskhelishvili,G. (2001) DNA supercoiling and transcription in Escherichia coli: the FIS connection. Biochimie, 83, 213–217. [DOI] [PubMed] [Google Scholar]
- 27.Balas D., Fernandez-Moreira,E. and De La Campa,A.G. (1998) Molecular characterization of the gene encoding the DNA gyrase A subunit of Streptococcus pneumoniae. J. Bacteriol., 180, 2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polaczek P., Kwan,K., Liberies,D.A. and Campbell,J.L. (1997) Role of architectural elements in combinatorial regulation of initiation of DNA replication in Escherichia coli. Mol. Microbiol., 26, 261–275. [DOI] [PubMed] [Google Scholar]
- 29.Ninnemann O., Koch,C. and Kahmann,R. (1992) The E.coli fis promoter is subject to stringent control and autorregulation. EMBO J., 11, 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menzel R. and Gellert,M. (1983) Regulation of the genes for E.coli DNA gyrase: homeostatic control of DNA supercoiling. Cell, 34, 105–113. [DOI] [PubMed] [Google Scholar]
- 31.Schneider R., Travers,A., Kutateladze,T. and Muskhelishvili,G. (1999) A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol., 34, 953–964. [DOI] [PubMed] [Google Scholar]
- 32.Pratt T.S., Steiner,T., Feldman,L.S., Walker,K.A. and Osuna,R. (1997) Deletion analysis of the fis promoter region in Escherichia coli: antagonistic effects of integration host factor and Fis. J. Bacteriol., 179, 6367–6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiga Y., Sekine,Y., Kano,Y. and Ohtsubo,E. (2001) Involvement of H-NS in transpositional recombination mediated by IS1. J. Bacteriol., 183, 2476–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavoie B.D. and Chaconas,G. (1996) Transposition of phage Mu DNA. Curr. Top. Micribiol. Immunol., 204, 83–99. [DOI] [PubMed] [Google Scholar]
- 35.Chalmers R., Guhathakurta,A., Benjamin,H. and Kleckner,N. (1998) IHF modulation of tn10 transposition: sensory transduction of supercoiling status via a proposed protein–DNA molecular spring. Cell, 93, 897–908. [DOI] [PubMed] [Google Scholar]
- 36.Hallet B., Rezsohazy,R., Mahillon,J. and Delcour,J. (1994) IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol. Microbiol., 14, 131–139. [DOI] [PubMed] [Google Scholar]
- 37.Nilsson L., Vanet,A., Vijgenboom,E. and Bosch,L. (1990) The role of FIS in trans activation of stable RNA operons of E.coli. EMBO J., 9, 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plaskon R.R. and Wartell,R.M. (1987) Sequence distributions associated with DNA curvature are found upstream of strong E.coli promoters. Nucleic Acids Res., 15, 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verbeek H., Nilsson,L. and Bosch,L. (1991) FIS-induced bending of a region upstream of the promoter activates transcription of the E.coli thrU (tufB) operon. Biochimie, 73, 713–718. [DOI] [PubMed] [Google Scholar]
- 40.Hirvonen C.A., Ross,W., Wozniak,C.E., Marasco,E., Anthony,J.R., Aiyar,S.E., Newburn,V.H. and Gourse,R.L. (2001) Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J. Bacteriol., 183, 6305–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brun Y.V., Sanfacon,H., Breton,R. and Lapointe,J. (1990) Closely spaced and divergent promoters for an aminoacyl-tRNA synthetase gene and a tRNA operon in Escherichia coli. Transcriptional and post-transcriptional regulation of gltX, valU and alaW. J. Mol. Biol., 214, 845–864. [DOI] [PubMed] [Google Scholar]
- 42.Bahloul A., Boubrik,F. and Rouviere-Yaniv,J. (2001) Roles of Escherichia coli histone-like protein HU in DNA replication: HU-β suppresses the thermosensitivity of dnaA46ts. Biochimie, 83, 219–229. [DOI] [PubMed] [Google Scholar]
- 43.Givens J.R., McGovern,C. and Dombroski,A. (2001) Formation of intermediate initiation complex at pfliD and pflgM by σ28 RNA polymerase. J. Bacteriol., 183, 6244–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soutourina O., Kolb,A., Krin,E., Laurent-Winter,C., Rimsky,S., Danchin,A. and Bertin,P. (1999) Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol., 181, 7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kutzukake K. (1997) Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol. Gen. Genet., 254, 440–448. [DOI] [PubMed] [Google Scholar]
- 46.Hagerman P.J. (1990) Sequence-directed curvature of DNA. Annu. Rev. Biochem., 59, 755–781. [DOI] [PubMed] [Google Scholar]
- 47.Pouty M., Correa,N.E. and Klose,K. (2001) The novel σ54 and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol., 39, 1595–1609. [DOI] [PubMed] [Google Scholar]
- 48.Jagannathan A., Constantinidou,C. and Penn,C.W. (2001) Roles of rpoN, fliA and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol., 183, 2937–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kustu S., Santero,E., Keener,J., Popham,D. and Weiss,D. (1989) Expression of σ54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev., 53, 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finkel R.A. and Johnson,R.C. (1992) The Fis protein: it’s not just for DNA inversion anymore. Mol. Microbiol., 6, 3257–3265. [DOI] [PubMed] [Google Scholar]
- 51.Hatfield G.W. and Benham,C.J. (2002) DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet., 36, 175–203. [DOI] [PubMed] [Google Scholar]