Abstract
Circular RNAs reminiscent of viroids and the human hepatitis delta virus have been proposed as possible nonconventional pathogens responsible for Crohn’s disease and ulcerative colitis, two inflammatory bowel diseases. Consequently, RNA was extracted from various areas of intestinal tissues from individuals with either Crohn’s disease or ulcerative colitis as well as several appropriate control diseases, and analyzed by two-dimensional gel electrophoresis. No circular viroid-like RNAs (<1500 nucleotides) were detected, confirming a previous report that was limited to the investigation of small RNAs (<300 nucleotides). However, three small, unusually stable, linear RNAs were shown to be associated to both Crohn’s disease and ulcerative colitis tissues: a specific 28S ribosomal RNA cleavage product characterized previously; a 5.8S ribosomal RNA conformer; and a fragment homologous to transcripts from DNA CpG islands. The two last RNAs were detected prior to visible morphological tissue alterations, suggesting that they are produced early during the inflammation and that they have value as molecular diagnostic tools for the inflammatory bowel diseases. The potential cellular mechanisms producing these RNAs and their involvement in inflammatory bowel disease are discussed.
Keywords: ribosomal RNA, inflammatory bowel diseases, human intestine, inflammation, viroids
Introduction
Crohn’s disease (CD) and ulcerative colitis (UC) are two idiopathic inflammatory bowel diseases (IBD) of unknown aetiology (Podolsky 1991). Both diseases are believed to be due to a combination of factors, involving diet, genetic factors, immunologic responses, and environmental factors including either bacterial or viral infections; however, the progression of events remains unclear (Podolsky 1991; Thayer 1990). It has been proposed that an aggressive immunologic event initiated by an as yet unidentified antigen, or a conventionally presented antigen, in conjunction with a particular genetic background, may be involved in the lack of correct repression of the inflammation (Fewell and Snook 1990; Strober and Ehrhardt 1993). The suggestion that infectious agents are the cause for these diseases has been entertained for as long as they have been studied, but none of the classical infectious agents have been found thus far (Van Kruiningen et al. 1993; Hermon-Taylor 1993). The most serious infectious candidate isolated from CD tissues has been Mycobacterium paratuberculosis (Chiodini et al. 1984); however, it was demonstrated that such opportunistic microorganisms were not the causative agent and that they invaded the mucosa after ulceration, thereby contributing to the disease’s perpetuation (Yoshimura et al. 1987).
The appearance of the illness in clusters of small populations and its slow progress and resistance to antibiotic and antiviral medications have prompted the investigation of non-conventional RNA species as primary pathogenic agents of CD and UC. Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) provided evidence for the presence of small circle-like RNAs in tissues from individuals with either CD or UC (Pechan et al. 1987). More recently, Lundberg et al. (1995) performed similar experiments and did not observe any circular RNA between 75 and 300 nucleotides. In this study, we have investigated the presence of RNAs with slower electrophoretic mobility, within the range of 75 to over 1200 nucleotides, from the intestinal tissues of individuals with CD or UC or appropriate controls.
Materials and methods
Tissues and RNA extraction
After surgery, the tissues were washed with saline solution, and small sections from either ulcerated or nearby visually normal areas were isolated and rapidly snap-frozen in liquid nitrogen prior to storage at −70°C. Sections from the same areas were fixed for histopathological analysis. The number of tissues analyzed is reported in Table 1. RNA was extracted from tissues either using guanidinium thiocyanate (GITC; Trizol Reagent, Gibco-BRL) according to the manufacturer’s directions or via the acidified phenol procedure (Roe 1975). After acidified phenol extraction, diethylaminoethyl-Sepharose (Sigma Chemical Company) column chromatography was performed, and the RNA was ethanol precipitated, ethanol washed, dried, and stored at −70°C (Lundberg et al. 1995). As control, circular and linear transcripts of RNA corresponding to the peach latent mosaic viroid were synthesized and purified as described previously (Beaudry and Perreault 1995; Beaudry et al. 1995).
Table 1.
Detection of stable RNAs in intestinal tissues from individuals with IBD.
| RNA X | RNA Y | RNA Z | |
|---|---|---|---|
| Acidified phenol | |||
| Crohn’s disease | |||
| Lesion (10) | 2 | 7 | 9 |
| Normal (2) | 0 | 1 | 1 |
| Ulcerative colitis | |||
| Lesion (2) | 1 | 1 | 2 |
| Normal (1) | 0 | 1 | 1 |
| Diverticulitis (1) | 0 | 0 | 0 |
| Polyposis (1) | 0 | 0 | 0 |
| Adenocarcinoma (5) | 0 | 1 | 0 |
| Acid guanidinium thiocyanate | |||
| Crohn’s disease | |||
| Lesion (5) | 0 | 4 | 2 |
| Normal (1) | 0 | 1 | 0 |
| Ulcerative colitis | |||
| Lesion (1) | 0 | 1 | 1 |
| Diverticulitis (1) | 0 | 0 | 0 |
| Adenocarcinoma (4) | 0 | 1 | 0 |
Note: Numbers in parentheses indicate the total number of tissues analyzed.
Two-dimensional gel electrophoresis
RNA was analyzed by 5% 2D-PAGE, with the first dimension being run under native conditions and the second under denaturing conditions (50°C in the presence of 7 M urea), essentially as described by Schumacher et al. (1983). Under denaturing conditions circular RNAs display a slower electrophoretic mobility than do linear RNAs of equivalent size. For most gels, the dimensions were 20 cm × 18 cm × 1 mm thick. The ratio of acrylamide to bisacrylamide was 40:1 (w/w) in the first dimension and 20:1 in the second. RNA aliquots (200 mu;g) were resuspended in ~30 mu;L of either nanopure water or 10 mM Tris-HCl pH 8.0 – 1 mM EDTA (TE), and 10 mu;L loading buffer (0.25% xylene cyanol and bromophenol blue and 50% glycerol) was then added. Analytical gels were silver stained by two different procedures (Lundberg et al. 1995; Schumacher et al. 1986). The stained gels were air dried and photographed. RNA from preparative gels was visualized under ultraviolet light, following ethidium bromide staining (0.5 mu;g ethidium bromide/mL TE). Selected off-diagonal bands were excised from the gel, and the RNA were eluted by a “crush and soak” procedure and recovery was as described previously (Beaudry et al. 1995).
Electron microscopy
RNAs Y and Z (see below) were renatured, and then visualized by electron microscopy under denaturing conditions according to a microversion of the Kleinschmidt and Zhan technique (Kleinschmidt and Zhan 1959). Briefly, RNA was resuspended in 10 mu;L of 10 mM sodium phosphate pH 8.0 – 0.5 M deionized glyoxal solution and incubated at 37°C for 1 h. The resulting solutions were made up to 200 mu;L by addition of 100 mM Tris-HCl pH 8.5 – 20 mM EDTA –50% deionized formamide and cytochrome C type V (Sigma Chemical Company) to a final concentration of 1 mg/mL. The final solution was spread out on metallized plates, using a fresh solution of 10 mM Tris-HCl pH 8.5 – 1 mM EDTA – 20% formamide.
Enzymatic sequencing of RNA
Gel-purified RNAs were 5′ kinased in the presence of [γ-32P]ATP (3000 Ci/mmol (1 Ci = 37 GBq), Amersham Life Science) using T4 polynucleotide kinase (Pharmacia Biotech) both with and without prior dephosphorylation by calf intestinal alkaline phosphatase (Boehringer Mannheim) as described by the manufacturers. Reactions were stopped by the addition of 0.5 volumes of loading buffer (0.3% of bromophenol blue and xylene cyanol – 10 mM EDTA pH 7.5 –97.5% deionized formamide) and analyzed by 5% PAGE containing 100 mM Tris-borate, 1 mM EDTA pH 8.0, and 7 M urea. Selected gel spots were isolated and the RNA was eluted as described above. Partial sequences were determined by the rapid gel-sequencing method with various enzyme concentrations of ribonuclease T1 (G specific), Phy M (U and A specific), or Bacillus cereus (U and C specific) under partial hydrolysis conditions as described by the manufacturer (Pharmacia Biotech). Ladders were produced by partial RNA alkaline hydrolysis involving an incubation of 5′-labelled RNA in 50 mM sodium carbonate pH 9.2 – 1 mM EDTA solution for 3–5 min, followed by ethanol precipitation in the presence of 20 mu;g/mL oyster glycogen. RNA fragments were fractionated by either 5% or 8% PAGE gels.
Results
Detection of RNAs associated with CD and UC
To investigate the possible association of viroid-like RNAs with IBD, RNA was extracted from the ulcerated areas of intestinal tissues from individuals with active CD, UC, diverticulitis, familial polyposis, or adenocarcinoma and then fractionated by 2D-PAGE. Only resected tissues from individuals with unequivocal diagnosis were considered. Figure 1A shows as control the migration on 2D-PAGE of both circular and linear synthetic transcripts corresponding to the viroid responsible for the latent mosaic of peach trees. The minimal RNA quantity to allow detection of a spot was established as 0.5 ng (~10 fmol of 338-nucleotide RNA). Three RNA species from CD and UC individuals exhibit an off-diagonal migration compared with control tissues (Figs. 1B, 1C). The off-diagonal position of these RNA species was confirmed by longer gel migration of several samples. However, Fig. 1B was selected since the three RNA species were detected in a single gel (see below). Two of these RNAs were located in the region between the 5S (120 nucleotides) and the 5.8S (159 nucleotides) rRNA spots, while the third off-diagonal RNA was located near the region of the 7S RNA spot (~300 nucleotides). Recently, Lundberg et al. (1995) reported detection of these three RNAs in the intestinal tissues of individuals with IBD. We have adopted their nomenclature system and refer to these species as RNA X, Y, and Z, respectively (see Fig. 1B). Other streaks below RNA Z appeared off-diagonal (see Fig. 1B). However, these bands were only partially off-diagonal and were detected from a minimum number of specimens. Therefore, we did not undertake further characterization of these RNA species.
Fig. 1.
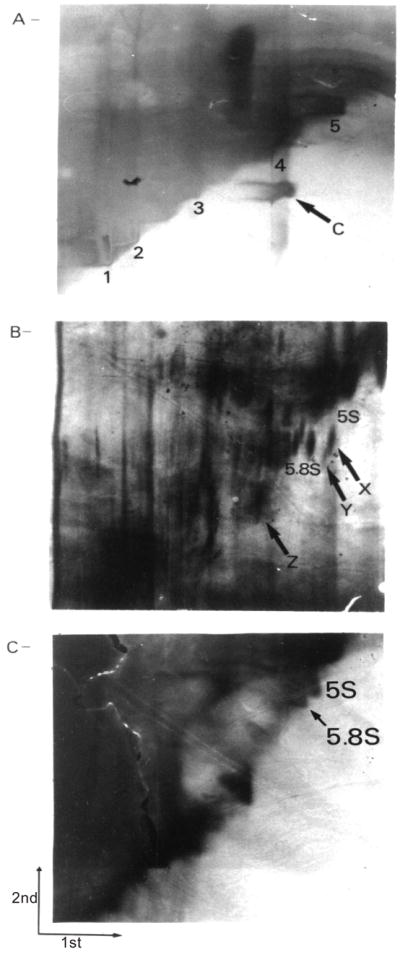
2D-PAGE fractionation of RNA extracted from tissues using the acidified phenol procedure. (A) Analysis of a mixture of both circular (338 nucleotides) and linear RNA peach latent mosaic viroids. C, circular transcripts; 1, 2, 3, 4, and 5, linear transcripts corresponding to 745, 621, 462, 338, and 283 nucleotides, respectively. In each case 1 mu;g of RNA was applied to the gel. (B) RNA extracted from the intestinal tissue of an individual affected by Crohn’s disease. On this panel, several species of RNA are identified. (C) RNA extracted from the intestinal tissue of an individual with a diverticulitis. This gel was overstained to ensure detection of any small amounts of off-diagonal migrating species. For Figs. 1B and 1C, 200 mu;g of RNA was applied to the gel. The directions of migration under the native (1st) and the denaturing (2nd) conditions are shown in Fig. 1C.
The detection results following the extraction of RNAs with the acidified phenol procedure, which is most efficient for the extraction of molecules smaller than 5.8S rRNA (159 nucleotides) (Roe 1975), and with the acid guanidium procedure are presented in Table 1. RNA X was detectable in only a small percentage of samples and only after acidified phenol extraction. RNA Y was detected in a large proportion of both CD and UC ulcerated tissues regardless of the extraction procedure used. However, RNA Y was also detected in one inflammatory adenocarcinoma, suggesting that it is associated with the inflammation (see Discussion). RNA Z was detected in almost all RNA populations from CD and UC tissues submitted to the acidified phenol procedure, while it was only detected in half of both CD and UC tissues after total RNA extraction. With both extraction procedures, RNA Z was never detected in control samples. Using different electrophoretic conditions, RNA Z was detected in several additional samples of resected tissues from individuals with either CD or UC (J.-P. Perreault and S. Altman, unpublished data). Thus, the frequency of detection of the X, Y, and Z RNA species was subject to limitations of both the extraction procedure and the sensitivity of the silver stain. This conclusion is supported by the demonstration by Northern blot hybridization that RNA X was present in all tissues, albeit in different amounts (Lundberg et al. 1995). In the present study we also extracted RNA from visually normal tissues nearby the ulcerations in CD and UC individuals. In these normal tissues, RNA Y and RNA Z were detected from 1 CD and 1 UC individual, while RNA X was never detected. However, the amount of RNA Y and Z appeared lower in normal areas than in ulcerated tissues.
Despite investigations with longer migration times, we did not detect circle-like RNAs between 300 and >1200 nucleotides associated specifically with tissues from individuals suffering from IBD. In general, we observed that the electrophoresis of RNAs from both diverticulitis and familial polyposis exhibit a diagonal with more varied species of RNA than that from CD and UC tissues, even though their appearances are similar. In contrast, the diagonal of RNAs from individuals with adenocarcinoma had fewer small RNA species and the spots were less well defined, suggesting the presence of exonuclease activities.
Characterization of RNAs
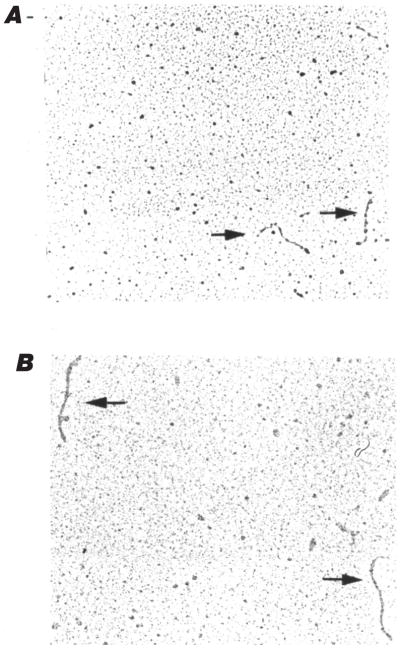
We have not pursued the investigation of RNA X because of its low frequency of detection. However, RNA X has been identified as a discrete cleavage product encompassing 126 nucleotides of the V2-9 region of 28S rRNA (Lundberg et al. 1995). This fragment forms a rod-like secondary structure that accounts for its slow electrophoretic mobility. In contrast with Lundberg et al. (1995), we examined RNAs Y and Z by electron microscopy under denaturing conditions. This procedure allows the direct visualization of macromolecules without any enzymatic treatment. RNAs Y and Z were observed to be single-stranded rod-like RNAs, with respective sizes of ~150 and ~250 nucleotides as estimated from comparison with RNA standards (Fig. 2). Thus, the off-diagonal mobility observed for these RNAs probably results from their capacity to fold into stable secondary and (or) tertiary structures. As a control, we have observed the circularity of covalently closed synthetic viroids (data not shown).
Fig. 2.
Electron microscopy of (A) RNA Y and (B) RNA Z under denaturing conditions. The arrows indicate RNA molecules.
The linearity of RNAs Y and Z was confirmed by 5′ end phosphorylation. RNA Y required a dephosphorylation step before it could be labelled (Fig. 3, lanes 1 and 2), confirming that it is linear and possesses a phosphate group at its 5′ end. RNA Z was directly 5′-kinased, while the inclusion of a preliminary dephosphorylation step led to a larger proportion of this species being labelled (Fig. 3, lanes 3 and 4). Thus, RNA Z is linear with either a 5′ hydroxyl group or a 5′ phosphate group. The 5′-kinased RNAs were eluted from the gel and sequenced. We established a partial sequence of RNA Y (5′GUGCGUCGAUGAAGAACG3′). A BLAST homology search (Altschul et al. 1990) in GenBank identified this species as human 5.8S rRNA (159 nucleotides; accession No. J01866). The sequenced region occurred from position 29 to 46 of the 5.8S rRNA. This result confirms the previous suggestions by Lunberg et al. (1995) that, based on a 3′ partial sequence homology, RNA Y is related to 5.8S rRNA. To explain the slower mobility of RNA Y in comparison with the 5.8S rRNA in the native conditions, we investigated whether this 5.8S rRNA is linked covalently to a protein as has been shown to occur with the tumor suppressor gene product p53 (Founoura et al. 1992). The introduction of a proteinase K treatment prior to electrophoresis did not alter the mobility of RNA Y, thereby indicating that it is not a covalently associated RNA–protein complex (data not shown). Taken together these results suggest that RNA Y is a 5.8S rRNA possessing a more compact conformation, which accounts for its faster migration under native conditions and its slightly slower migration under denaturing conditions.
Fig. 3.
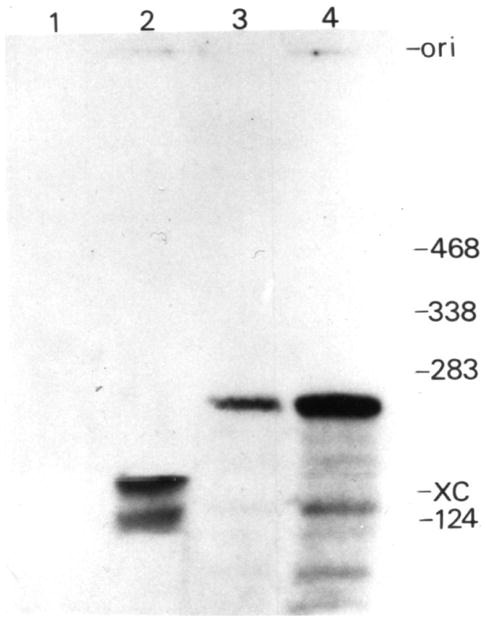
Enzymatic analysis of RNA Y and Z by 5′ end labelling. RNA were phosphorylated directly (lanes 1 and 3) or after a prior dephosphorylation (lanes 2 and 4); RNA Y (lanes 1 and 2) and RNA Z (lanes 3 and 4). Adjacent to the gel are the positions of RNA standards (length in nucleotides). ori, origin of migration; XC, position of xylene cyanol.
RNA Z appeared highly resistant to ribonuclease treatment. The concentration of ribonucleases required to observe hydrolysis was at least fivefold higher compared with RNA Y. A chemical hydrolysis performed at 96°C clearly showed that the isolated molecules were made entirely of RNA. Based on the overall picture of RNA Z susceptibility to RNase treatment, we estimate that ~75% of the nucleotides are either protected by double strands or tertiary foldings and this accounts for its slower electrophoretic mobility. Partial sequences of RNA Z were obtained starting 18 nucleotides (5′CGG(A/G)G(C/U)UGG(C/U)UGGGAUGGNAGAAUCU3′; N = A,G,C, or U) and ~100 nucleotides (5′GG(C/U)NGGNGG(C/U)AGG′3) from the 5′ end. A GenBank search for homologous sequences failed to identify the nature of this RNA. However, with their preference for GGY repeats our new sequences showed homology with CpG islands entries. These are defined as short stretches of DNA containing a high density of nonmethylated CpG dinucleotides, predominantly associated with coding regions (Cross et al. 1994). We designed several oligonucleotides based on the RNA Z partial sequences in order to perform reverse transcription – polymerase chain reaction using either Taq DNA polymerase or rTth polymerase. Under various conditions, no product related to RNA Z could be obtained, probably because its stable structure did not allow the annealing of primers (data not shown). In addition, the chemical sequencing protocol did not reveal more sequence from RNA Z.
Discussion
We have shown three RNAs that appear to be associated, albeit at different levels, with CD and UC diseases. These RNAs share one characteristic in that they all exhibit off-diagonal migration in two-dimensional electrophoresis even though they are linear in nature, as shown by electron microscopy and enzymatic labelling. These RNAs have the capacity to fold into relatively stable structures, which accounts for their unusual electrophoretic mobility. RNA X, an ~126-nucleotide molecule, is a discrete cleavage product of region V2-9 of the 28S rRNA (Lundberg et al. 1995). Our results suggest that RNA Y is a conformer of 5.8S rRNA. However, this does not dismiss the possibility that it may be a sequence variant expressed only in diseased cells, as suggested previously (Lundberg et al. 1995). Finally, our data suggest that RNA Z shows homologies with transcribed DNA CpG islands. The nature of this RNA species remains to be determined. When the acidified phenol procedure is used, RNA Z appears as the most interesting species for further diagnostic development, while RNA Y appears the most promising after total RNA extraction. Knowing the size of RNA Z, we analyzed several samples after total RNA extraction for the presence of this species by “return”-gel electrophoresis, a technique commonly used for viroid detection (data not shown). RNA Z was present in approximately half of the Crohn’s disease tissues, which confirms the results in Table 1. Furthermore, this approach confirms the limit of detection of RNA Z. Interestingly, RNAs Y and Z were not detected in nonintestinal tissues (data not shown), while their presence in normal areas of one CD and one UC individual suggests that detection of these RNAs may be an interesting feature for prognostic analyses. In contrast with a previous report (Lundberg et al. 1995), our results from the acidified phenol extraction protocol indicate that RNA Z has more potential than RNA Y as an IBD molecular marker. However, further evaluation, with a large tissue collection and a more sensitive method, is required to establish the use of RNAs Y and Z for IBD diagnosis.
The finding of both RNAs Y and Z in samples from normal areas near the ulcerations suggests that these RNAs are associated with pathological events that take place prior to the occurrence of morphological alterations. Considering that proinflammatory cytokine mRNA has been shown to be present in inactive as well as active CD tissues (Isaacs et al. 1992), we suggest that the RNAs X, Y, and Z could result from an immunological response that occurred in tissues of individuals with IBD. Moreover, this finding also suggests that these RNAs may be part of the process leading to either inflammation or cell necrosis. As mentioned earlier, RNA X is a discrete cleavage product of 28S rRNA, while RNA Y seems to be a 5.8S rRNA conformer. The detection of RNAs X and Y, which are components that interact together in a ribosome, leads us to suggest that the translational apparatus is specifically affected by the inflammatory process that occurs in tissues. Specific cleavage of the 28S rRNA has been shown to occur in myeloid leukemia cell apoptosis, and found to coincide with internucleosomal DNA fragmentation and cessation of cellular protein synthesis (Houge et al. 1995). This selective cleavage of the 28S rRNA has been associated with apoptosis-associated changes in ribosome conformation. We can envision specific RNA nuclease activities or conformational changes occurring within the ribosome of diseased cells and leading to a 5.8S rRNA conformer and a specific cleavage product of the 28S rRNA. Such specific RNA cleavage has been shown to occur in normal tissues, indicating that an active, but less abundant, nuclease is present (Lundberg et al. 1995; Houge et al. 1995). As observed in apoptosis, the inflammation leading to necrosis of cells in IBD appears as a more ordered process than originally believed. Further study of the localization of RNAs X, Y, and Z, and of their relationship to IBD pathogenesis will be instructive in this regard.
No circular RNA, resembling viroids or the human hepatitis delta virus genome, was detected in the present work. The detection of RNA species was limited by both the extraction and the staining procedures used. The maximum amount of sample successfully fractionable in a single gel was 200 mu;g, and the detection’s threshold was established to be 0.5 ng of such a RNA (e.g., ~10 fmol of synthetic viroid). Samples over 200 mu;g required a thicker gel, which decreased the sensitivity of silver staining. Therefore, the relative weight of a species must be higher than 1 to 4 × 106 to allow its detection under these conditions. Thus, to detect a viroid-like species either most of analyzed cells must be infected or the RNA must be actively replicated. Consequently, the lack of detection of any viroid-like RNA associated with CD and (or) UC does not negate the initial infectious agent hypothesis and more sensitive procedures will be required to test this hypothesis.
Acknowledgments
The authors thank Drs. J. Poisson, D. Ménard, and S. Langevin for their suggestions and Mrs. Beaudry and Mr. Magny for technical assistance. This research was supported by a grant from the Medical Research Council of Canada (MRC) to J.-P.P. G.R. was sponsored by graduate fellowships from the Fonds de la recherche en santé du Québec and the Natural Sciences and Engineering Research Council of Canada. J.-P.P. is an MRC scholar.
Abbreviations
- CD
Crohn’s disease
- UC
ulcerative colitis
- IBD
inflammatory bowel diseases
- 2D-PAGE
two-dimensional polyacrylamide gel electrophoresis
- GITC
guanidinium thiocyanate
- Tris
tris(hydroxymethyl)aminomethane
- TE
10 mM Tris-HCl pH 8.0 – 1 mM EDTA
Contributor Information
Guylaine Roy, Département de biochimie, Faculté de médecine, Université de Sherbrooke, Sherbrooke, QC J1H 5N4, Canada.
Stéphane Mercure, Département de biochimie, Faculté de médecine, Université de Sherbrooke, Sherbrooke, QC J1H 5N4, Canada.
Frédéric Beuvon, Département de pathologie, Faculté de médecine, Université de Sherbrooke, Sherbrooke, QC J1H 5N4, Canada.
Jean-Pierre Perreault, Département de biochimie, Faculté de médecine, Université de Sherbrooke, Sherbrooke, QC J1H 5N4, Canada.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beaudry D, Perreault JP. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 1995;23:3064–3066. doi: 10.1093/nar/23.15.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudry D, Bussière F, Lareau F, Lessard C, Perreault JP. The RNA of both polarities of the peach latent mosaic viroid self-cleaves in vitro solely by single hammerhead structures. Nucleic Acids Res. 1995;23:745–752. doi: 10.1093/nar/23.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodini RJ, Van Kruiningen HJ, Merkal RS, Thayer WR, Jessica AC. Characteristics of an unclassified mycobacterium species isolated from patients with Crohn’s disease. J Clin Microbiol. 1984;20:966–971. doi: 10.1128/jcm.20.5.966-971.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- Fewell DP, Snook FA. Immunology of ulcerative colitis and Crohn’s disease. In: Allan RN, Keighley MRB, Hawkins C, editors. Inflammatory bowel diseases. Churchill Livingstone; New York: 1990. pp. 127–146. [Google Scholar]
- Founoura BMA, Sorokina EA, David E, Carroll RB. p53 is covalently linked to 5.8S rRNA. Mol Cell Biol. 1992;11:5145–5151. doi: 10.1128/mcb.12.11.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermon-Taylor J. Causation of Crohn’s disease: the impact of clusters. Gastroenterology. 1993;104:643–646. doi: 10.1016/0016-5085(93)90438-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houge G, Robaye B, Eikhom TS, Golstein J, Mellgren G, Gjertsen BT, Lanotte M, Doskeland SO. Fine mapping of 28S rRNA sites specifically cleaved in cells undergoing apoptosis. Mol Cell Biol. 1995;15:2051–2062. doi: 10.1128/mcb.15.4.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs KL, Sartor RB, Haskill S. Cytokine messenger RNA profiles in inflammatory bowel disease mucosa detected by polymerase chain reaction amplification. Gastroenterology. 1992;103:1587–1595. doi: 10.1016/0016-5085(92)91182-4. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt AK, Zhan RK. Über deoxyribonukleinsäure-molekin in protein misschfilmen. Z Naturforsch B. 1959;14:770–779. [Google Scholar]
- Lundberg U, West AB, Altman S. Characterization of RNA with unusual electrophoretic mobility from tissues of patients with Crohn’s disease. FEBS Lett. 1995;371:345–350. doi: 10.1016/0014-5793(95)00946-7. [DOI] [PubMed] [Google Scholar]
- Pechan R, Kunert H, Gross HJ. Are small RNAs associated with Crohn’s disease? Z Naturforsch C. 1987;42:1006–1008. doi: 10.1515/znc-1987-7-848. [DOI] [PubMed] [Google Scholar]
- Podolsky DK. Inflammatory bowel disease. N Engl J Med. 1991;325:928–937. doi: 10.1056/NEJM199109263251306. [DOI] [PubMed] [Google Scholar]
- Roe BA. Studies of human tRNA. I The rapid, large scale isolation and partial fractionation of placenta and liver tRNA. Nucleic Acids Res. 1975;2:21–42. doi: 10.1093/nar/2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Randles JW, Riesner D. A two-dimensional electrophoretic technique for the detection of circular viroids and virusoids. Anal Biochem. 1983;135:288–295. doi: 10.1016/0003-2697(83)90685-1. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Meyer N, Riesner D, Weidelmann HL. Diagnostic procedures for detection of viroids and viruses with circular RNAs by “return”-gel electrophoresis. J Phytopathol. 1986;115:332–343. [Google Scholar]
- Strober W, Ehrhardt RO. Chronic intestinal inflammation: an unexpected outcome in cytokine or T cell receptor mutant mice. Cell. 1993;75:203–205. doi: 10.1016/0092-8674(93)80062-j. [DOI] [PubMed] [Google Scholar]
- Thayer WR. The aetiology of inflammation bowel disease: the role of infectious agents. In: Allan RN, Keighley MRB, Hawkins C, editors. Inflammatory bowel diseases. Churchill Livingstone; New York: 1990. pp. 147–163. [Google Scholar]
- Van Kruiningen HJ, Colombel JF, Cartun RW, Whitlock RH, Koopmans M, Kangro HO, Hoogkamp-Korstanje JAA, Lecomte-Houcke M, Devred M, Paris JC, Corot A. An in-depth study of Crohn’s disease in two French families. Gastroenterology. 1993;104:351–360. doi: 10.1016/0016-5085(93)90401-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura HH, Graham DY, Estes MK, Merkal RS. Investigation of association of mycobacteria with inflammatory bowel disease by nucleic acid hybridization. J Clin Microbiol. 1987;25:45–51. doi: 10.1128/jcm.25.1.45-51.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]